Abstract
RAS is frequently mutated in human cancers and has opposing effects on autophagy and tumorigenesis. Identifying determinants of the cellular responses to RAS is therefore vital in cancer research. Here, we show that autophagic activity dictates the cellular response to oncogenic RAS. N-terminal Apoptosis-stimulating of p53 protein 2 (ASPP2) mediates RAS-induced senescence and inhibits autophagy. Oncogenic RAS-expressing ASPP2(Δ3/Δ3) mouse embryonic fibroblasts that escape senescence express a high level of ATG5/ATG12. Consistent with the notion that autophagy levels control the cellular response to oncogenic RAS, overexpressing ATG5, but not autophagy-deficient ATG5 mutant K130R, bypasses RAS-induced senescence, whereas ATG5 or ATG3 deficiency predisposes to it. Mechanistically, ASPP2 inhibits RAS-induced autophagy by competing with ATG16 to bind ATG5/ATG12 and preventing ATG16/ATG5/ATG12 formation. Hence, ASPP2 modulates oncogenic RAS-induced autophagic activity to dictate the cellular response to RAS: to proliferate or senesce.
Active mutations of RAS, one of the first oncogenes identified, occur in about 20% of human tumors (1). Oncogenic RAS can transform cells and promote tumorigenesis, although it can also induce senescence and suppress tumor growth (2). Senescent cells are arrested and incapable of responding to mitogens, although they are viable and metabolically active. Senescence is characterized by dramatic cellular remodelling, which is energetically demanding. Autophagy, a genetically regulated process responsible for the turnover of cellular proteins and damaged or superfluous organelles, is a stress response involved in energy homeostasis (3). It was shown that autophagy is a critical mediator of oncogenic RAS-induced senescence, suggesting a negative role of autophagy in tumorigenesis (4). In contrast, a number of recent studies showed that active RAS requires autophagy to maintain its oncogenic function in tumorigenesis, arguing for a positive role of autophagy in tumorigenesis (5–8). The underlying reason for the conflicting observations remains unclear.
RAS activation inhibits autophagy by activating the PI3K/AKT/mammalian target of rapamycin (mTOR) pathway (9), and rapamycin, an mTOR inhibitor, is a potent inducer of autophagy. RAS also inhibits autophagy by reducing Beclin-1 expression in intestinal epithelial cells, an important mediator of autophagy (10). However, RAS was reported to induce autophagy by increasing the expression of key components of the autophagy machinery, such as ATG5 (11) and Beclin-1 (12). These reports suggest that RAS signaling performs a finely regulated balancing act to control autophagy. Identification of switching molecules that determine the cellular responses to RAS is thus needed urgently.
The tumor suppressor p53 is one of the most well-established pathways by which RAS mediates cellular senescence. A recent study also showed that p53 is able to regulate autophagic activity by inducing the expression of LC3 (13). However, it remains unknown whether the ability of p53 to induce autophagy is required to mediate oncogenic RAS-induced senescence. Interestingly, an NMR study showed that the N-terminal ASPP2, a known activator of p53, shares high structure similarity with LC3 and ATG12 (14). Additionally, we showed recently that ASPP2 deficiency can bypass oncogenic RAS-induced senescence independent of p53 (15). ASPP2 was first identified as a p53 binding protein, with subsequent studies showing that it enhances p53-induced apoptosis in vitro (16) and p53-mediated tumor suppression in vivo (17). Nonetheless, recent studies showed that ASPP2 is able to suppress cell proliferation through p53-independent pathways. By binding to Par3, ASPP2 is able to maintain the integrity of cell polarity and suppress excessive growth of neural progenitors (18). The ability of ASPP2 to suppress cell proliferation independent of p53 is conserved because dASPP inhibits cell proliferation in Drosophila independent of p53 (19). Knowing that the N terminus of ASPP2 shares structure similarity to ATG12 and LC3 and that this region of ASPP2 does not bind p53, we hypothesized that ASPP2 may mediate RAS-induced senescence by regulating autophagic activity independent of p53.
Results
N-Terminal ASPP2 Mediates RAS-Induced Senescence and Inhibits Autophagy.
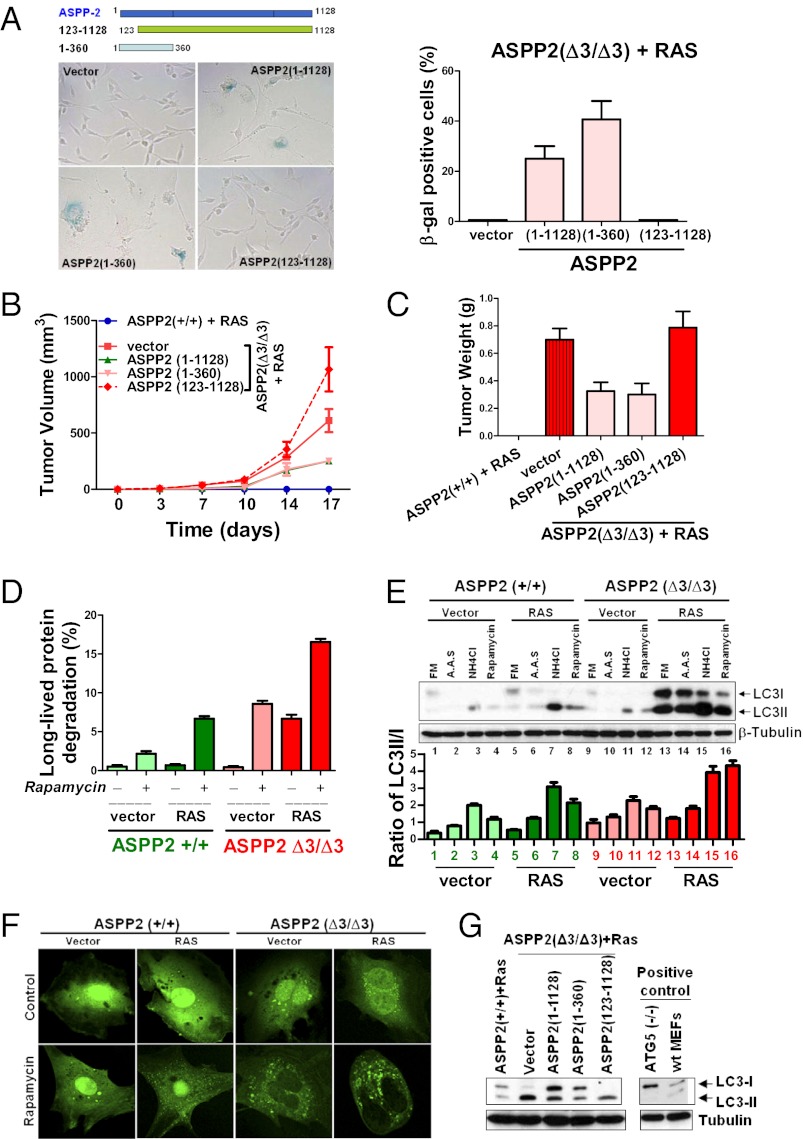
Oncogenic RAS induces senescence in primary fibroblasts. We showed recently that ASPP2 deficiency bypasses oncogenic RAS-induced senescence independent of p53 (15). To identify the region of ASPP2 that mediates RAS-induced senescence, two ASPP2 mutants were generated: ASPP2(1–360) and ASPP2(123–1,128). ASPP2(1–360) does not bind p53 but contains a ubiquitin-like fold similar to ATG12 or LC3 and binds Par3 (18, 20). ASPP2(123–1,128) binds p53 and Par3 but lacks the ubiquitin-like fold and represents a naturally occurring ASPP2 splice variant (21) (Fig. 1A, Upper). Retroviruses expressing full-length or truncated ASPP2 were infected into HRAS V12-expressing ASPP2(Δ3/Δ3) mouse embryonic fibroblasts (MEFs). As expected, reintroducing ASPP2(1–1,128) induced senescence in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs. Senescence associated-β-galactosidase (SA-β-gal) activity was detected in around 30% or 41% of cells infected with retroviruses expressing ASPP2(1–1,128) or ASPP2(1–360), respectively (Fig. 1A), whereas SA-β-gal activity was not detected in ASPP2(123–1,128)-infected cells. Reintroducing ASPP2(1–1,128) or ASPP2(1–360) also significantly repressed the tumorigenicity of HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs in vivo, reflected by the size and weight of the tumors (Fig. 1 B and C), whereas ASPP2(123–1,128) had no effect. The lack of complete repression of tumor growth and size was consistent with the percentage of SA-β-gal–positive cells observed, and it was most likely caused by the low infection efficiency. Nevertheless, the results demonstrate that N-terminal ASPP2(1–123) is required to mediate senescence and to suppress the tumorigenicity of oncogenic HRAS independent of p53.
Fig. 1.
N-terminal ASPP2 mediates oncogenic RAS-induced senescence and inhibits autophagy. (A) (Upper) Schematic representation of truncated ASPP2 mutants. HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs, infected with retroviruses expressing full-length or mutant ASPP2 as indicated, were subjected to SA-β-gal staining to identify senescent cells. Images were taken with a ×20, air objective lens. (Right) Percentages of positive cells are shown in the graph. Error bars indicate SD. (B and C) HRAS V12-expressing ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs infected with retroviruses expressing either full-length or truncated ASPP2 mutants were injected s.c. into the flanks of nude mice. (B) Tumor volume was monitored twice weekly over 17 d. Each point represents the mean volume ± SD of four tumors. (C) After 17 d, tumors were removed and weighed. Results are shown as the mean ± SD of tumor weights. (D) ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs with or without HRAS V12 expression were treated with 50 μg/mL rapamycin for 24 h to induce autophagy. Long-lived protein degradation was scored as the percentage of trichloroacetic acid-soluble counts out of total radioactivity incorporated in a standard protein degradation assay. Error bars represent SD of three independent experiments. (E) Induction of LC3II in ASPP2(Δ3/Δ3) MEFs. Western blot shows LC3 expression in lysates from ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs, with or without HRAS V12 expression, treated with the indicated compounds. β-Tubulin was used as a loading control. A.A.S., amino acid starvation; FM, full medium containing 1% FBS. (Lower) Ratio of LC3II against LC3I was calculated by densitometry. Error bars indicate SD. (F) Representative confocal images of EGFP-LC3–infected MEFs, treated with or without 50 μg/mL rapamycin for 24 h. Images were taken with a ×63, oil-immersion objective lens. (G) Western blot of LC3 expression in lysates from HRAS V12-expressing ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs infected with retroviruses expressing full-length ASPP2 or truncated mutants as indicated. Positive controls of LC3 were obtained from ATG5(−/−) and WT (wt) MEFs.
Because N-terminal ASPP2(1–123) contains the ubiquitin-fold sharing motif with high structural similarity to ATG12 and LC3 (14), we tested whether ASPP2 may influence autophagic activity. Autophagy is the principal method by which long-lived proteins are degraded. Interestingly, oncogenic RAS significantly enhanced basal protein degradation by about eightfold in ASPP2(Δ3/Δ3) but not in ASPP2(+/+) MEFs. Moreover, rapamycin induced long-lived protein degradation under all conditions, but the most profound increase of about 17-fold was observed in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs (Fig. 1D). Biochemically, the autophagosome-associated lipidated form of LC3II can be distinguished from unmodified LC3 (LC3I) by immunoblotting. Oncogenic RAS induced lipidated LC3II expression in ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs in response to amino acid starvation or rapamycin treatment (Fig. 1E, compare lanes 2 and 4 vs. lanes 6 and 8 and lanes 14 and 16). However, the maximum increase was observed in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs. LC3II expression was further elevated in the presence of NH4Cl, which prevents autophagic degradation, indicating that autophagic flux is increased. There was little difference in the amount of basal LC3II in ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs (Fig. 1E, compare lanes 1 and 9), suggesting that this effect is predominantly RAS-dependent. Autophagic activity was further analyzed by introducing GFP-tagged LC3. On treatment with rapamycin, around 50% of HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs showed punctate GFP-LC3–positive autophagic vesicles, compared with 15% in control ASPP2(Δ3/Δ3) MEFs. In HRAS V12-expressing ASPP2(+/+) MEFs, only 20% of cells produced a similar pattern of GFP-LC3 on rapamycin treatment (Fig. 1F and Fig. S1A).
The ability of ASPP2 and its mutants to inhibit autophagy was also tested in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs. Expression of ASPP2(1–1,128) or ASPP2(1–360) significantly inhibited rapamycin-induced autophagy, as demonstrated by a reduction in the LC3II/I ratio (Fig. 1G and Fig. S1B), whereas ASPP2(123–1,128) failed to do so under the same conditions. These results suggest that N-terminal ASPP2 may mediate RAS-induced senescence via its ability to inhibit autophagy.
Elevated ATG5/ATG12 Expression Accompanies the Bypass of RAS-Induced Senescence.
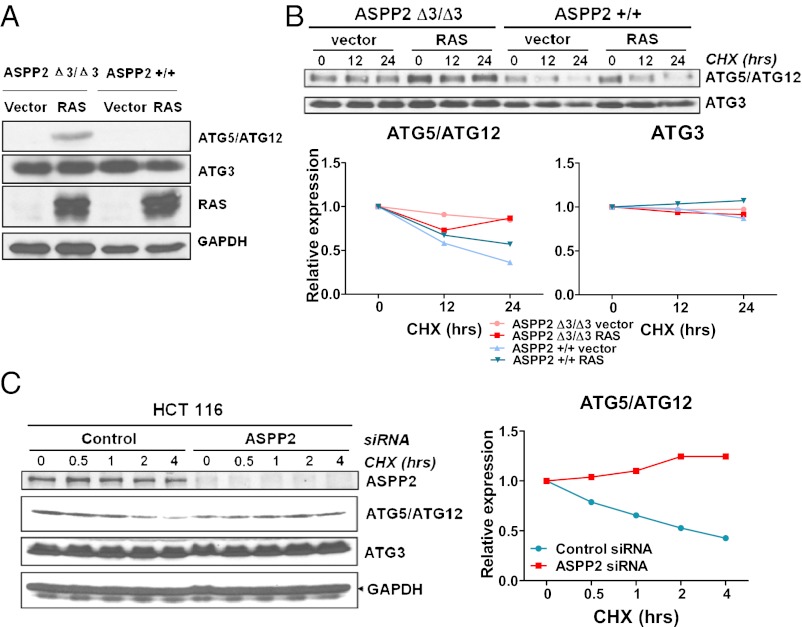
The involvement of autophagy in the bypass of RAS-induced senescence was further examined in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs. Expression levels of key autophagy genes were examined in early and late passages of HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs. ATG5/ATG12 and ATG3 expression remained unchanged 1 wk after HRAS V12 infections in both ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs (Fig. S2A). A small increase in expression of ATG5/ATG12 was observed 2 wk after HRAS V12 infection in ASPP2(Δ3/Δ3) MEFs (Fig. S2B). At 3 wk after infection, the expression of ATG5/ATG12 was significantly enhanced in these cells (Fig. 2A), with no observed increase in ATG5 mRNA (Fig. S2C). In contrast to ATG5/ATG12, the expression levels of ATG3 (Fig. 2A and Fig. S2 A and B) and Beclin-1 (Fig. S2D) were not affected in MEFs.
Fig. 2.
Increased expression and stability of ATG5/ATG12 in ASPP2(Δ3/Δ3) MEFs that have escaped oncogenic RAS-induced senescence. (A) Expression of ATG5/ATG12 is induced by HRAS V12 in late passage of ASPP2(Δ3/Δ3) MEFs. Western blot of ASPP2(+/+) or ASPP2(Δ3/Δ3) MEF lysates, with or without HRAS V12 expression, shows the expression level of ATG5/ATG12 or ATG3 and RAS. GAPDH was used as a loading control. (B) ATG5/ATG12, but not ATG3, is more stable in ASPP2(Δ3/Δ3) MEFs than in ASPP2(+/+) MEFs. ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs were infected with or without RAS and then treated with 10 μg/mL cycloheximide (CHX) for 12 h or 24 h as indicated. Western blots were used to detect ATG5/ATG12 and ATG3 levels. Graphs show the relative expression of ATG5/ATG12 or ATG3 with the indicated treatments. (C) ATG5/ATG12, but not ATG3, is more stable in ASPP2 siRNA-transfected HCT116 cells than in control cells. HCT116 cells were transfected with control siRNA or ASPP2 siRNA for 4 d, followed by treatment with 10 μg/mL CHX for the indicated time. Western blots were used to detect ATG5/ATG12 and ATG3. The graph shows the relative expression of ATG5/ATG12 with the indicated treatments.
To understand how ASPP2 deficiency induces ATG5/ATG12 expression, the half-life of ATG5/ATG12 was determined in oncogenic RAS-expressing ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs. The half-life of ATG5/ATG12 was longer in ASPP2(Δ3/Δ3) MEFs than in ASPP2(+/+) MEFs (Fig. 2B). Similar results were also observed in human HCT116 cells, which contain endogenous oncogenic RAS, on RNAi-mediated depletion of ASPP2 (Fig. 2C). ASPP2 may therefore inhibit autophagy by specifically affecting the stability of ATG5/ATG12 on RAS activation. The appearance of elevated ATG5/ATG12 in late-passage HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs also suggests that elevated autophagy activity may provide survival signals for the bypass of RAS-induced senescence.
Levels of Autophagy Dictate the Cellular Response to Oncogenic RAS.
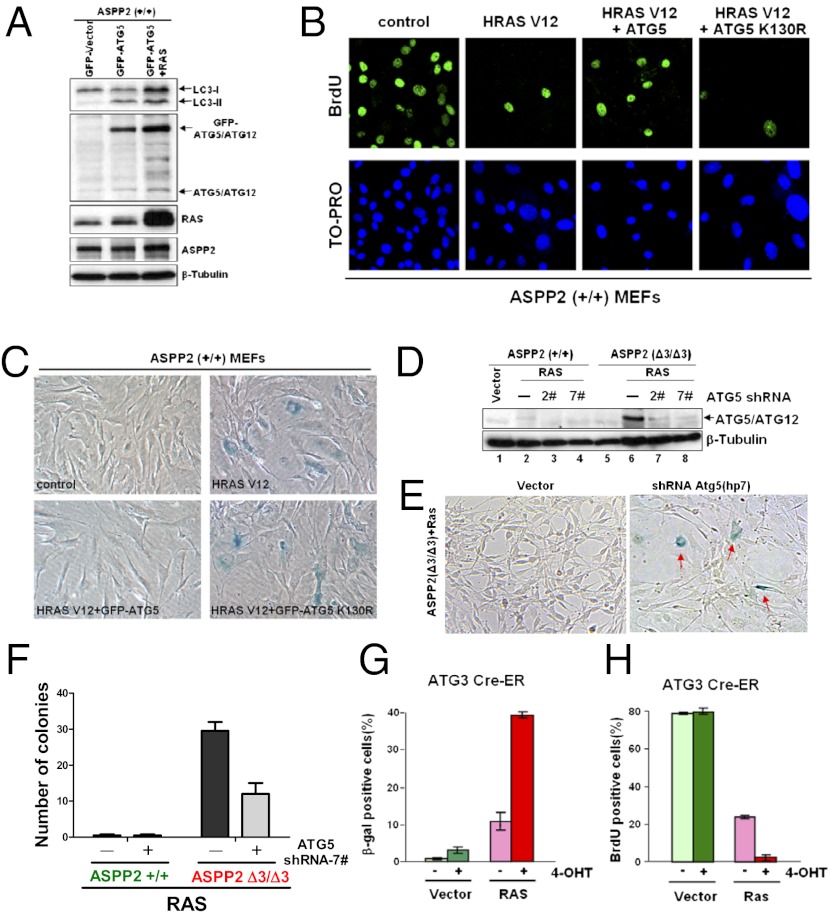
To test whether high levels of autophagic activity may be sufficient to bypass oncogenic RAS-induced senescence, ATG5 was introduced into ASPP2(+/+) MEFs to enhance autophagy in the presence or absence of HRAS V12. Overexpression of ATG5 induced autophagy, as demonstrated by detecting elevated amounts of LC3II (Fig. 3A), which was increased further on coexpression of HRAS V12. Remarkably, HRAS V12 failed to induce senescence in ATG5-expressing MEFs. ATG5- and HRAS V12-expressing ASPP2(+/+) MEFs grew rapidly on top of each other, presenting a typical transformed cell phenotype (Fig. S3A). Overexpression of ATG5 K130R, an ATG5 mutant that can neither bind ATG12 nor induce autophagy, failed to bypass RAS-induced senescence measured as either SA-β-gal activity or BrdU labeling (Fig. 3 B and C and graphs in Fig. S3 B–D); this bypass therefore requires ATG5 autophagic activity. In contrast, there was a minimal difference in the number of SA-β-gal–positive senescent cells or BrdU-labeled positive cells between ATG5 K130R-expressing MEFs and control HRAS V12-expressing ASPP2(+/+) MEFs (Fig. 3 B and C and graphs in Fig. S3 C and D). These data suggest that autophagic activation contributes to the bypass of oncogenic RAS-induced senescence.
Fig. 3.
High levels of autophagy bypass oncogenic RAS-induced senescence, whereas a reduced level of autophagy sensitizes to it. (A) Western blot shows the expression of LC3I/LC3II, endogenous ATG5/ATG12, GFP-ATG5/ATG12, RAS, and ASPP2 in ASPP2(+/+) MEFs with the indicated treatments. β-Tubulin expression levels demonstrate equal loading. (B and C) Overexpression of ATG5, not the ATG5 K130R mutant, is sufficient to overcome oncogenic RAS-induced senescence. ASPP2(+/+) MEFs with the indicated infections were stained for BrdU incorporation (B) or SA-β-gal activity (C) 1 wk after infection. Images for BrdU incorporation were taken with a ×63, oil-immersion objective lens while images for SA-β-gal activity were taken with a ×20, air objective lens. (D) Western blot analysis of lysates from indicated MEFs demonstrates that the amount of ATG5/ATG12 protein is reduced by two independent shRNA constructs (2# and 7#) against ATG5. (E) Knockdown of ATG5 restores senescence in oncogenic HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs, as indicated by the presence of SA-β-gal–stained senescent cells (red arrows). Images were taken with a ×20, air objective lens. (F) Graph shows the number of colonies formed by HRAS V12-expressing ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs infected with control shRNA or shRNA against ATG5 (7#). (G and H) Deletion of ATG3 accelerates RAS-induced senescence. ATG3 Cre-ER MEFs were first treated without or with 200 nM 4-OHT for 4 d, followed by infection with or without HRAS V12 for 1 wk. Graphs show the percentage of SA-β-gal–positive (G) or BrdU-positive (H) cells with indicated infections.
To confirm that the levels of autophagy are crucial in dictating the cellular response to RAS activation, ATG5 expression was depleted using shRNAs against ATG5 (Fig. 3D). This led to an increased number of enlarged flat cells with positive SA-β-gal staining in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs, in contrast to a minimal effect on ASPP2(+/+) MEFs (Fig. 3E and Fig. S3E). In addition, depletion of ATG5 significantly reduced the number of colonies formed by HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs from 30 ± 3 to 12 ± 4 per well (Fig. 3F). To confirm further that it is autophagic activity and not ATG5 itself that dictates RAS-induced senescence, ATG3 Cre-estrogen receptor (ER) MEFs were used (22). ATG3 expression is deleted on the addition of 4-hydroxytamoxifen (4-OHT) in ATG3 Cre-ER MEFs (Fig. S3F). ATG3 deletion enhanced oncogenic RAS-induced senescence, as demonstrated by a fourfold increase in the percentage of SA-β-gal–positive cells after 4-OHT addition and a comparable decrease in the percentage of BrdU-positive cells, compared with MEFs expressing ATG3 (Fig. 3 G and H; cell images in Fig. S3 G and H). These data demonstrate that autophagic activity dictates the cellular response to oncogenic RAS, with elevated autophagy leading to a bypass of RAS-induced senescence and a reduction in, or a lack of, autophagy sensitizing to it.
We then asked how high levels of autophagic activity help bypass oncogenic RAS-induced senescence. Yang et al. (8) reported recently that autophagy inhibition in pancreatic cancer results in increased reactive oxygen species (ROS) production and DNA damage. Hence, we tested whether high levels of autophagy may overcome RAS-induced senescence by reducing ROS production and the level of DNA damage. This was tested by measuring ROS levels in ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs using MitoSOX (Invitrogen) staining. Oncogenic RAS induced ROS production in both ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs. However, ASPP2 status did not affect the ability of RAS to induce ROS production (Fig. S3I). DNA damage is the other inducer of senescence; this occurs predominantly through its ability to induce the p53/p19Arf/p21waf1 pathway. We found in our cell system that overexpression of ATG5 did not prevent oncogenic RAS from inducing p53/p19Arf/p21waf1 expression (Fig. S3J). However, this induction failed to induce senescence in these cells. These data suggest that increased autophagic activity uses other mechanisms independent of p53 to bypass RAS-induced senescence.
ASPP2 Inhibits Autophagy by Preventing ATG16/ATG5/ATG12 Complex Formation.
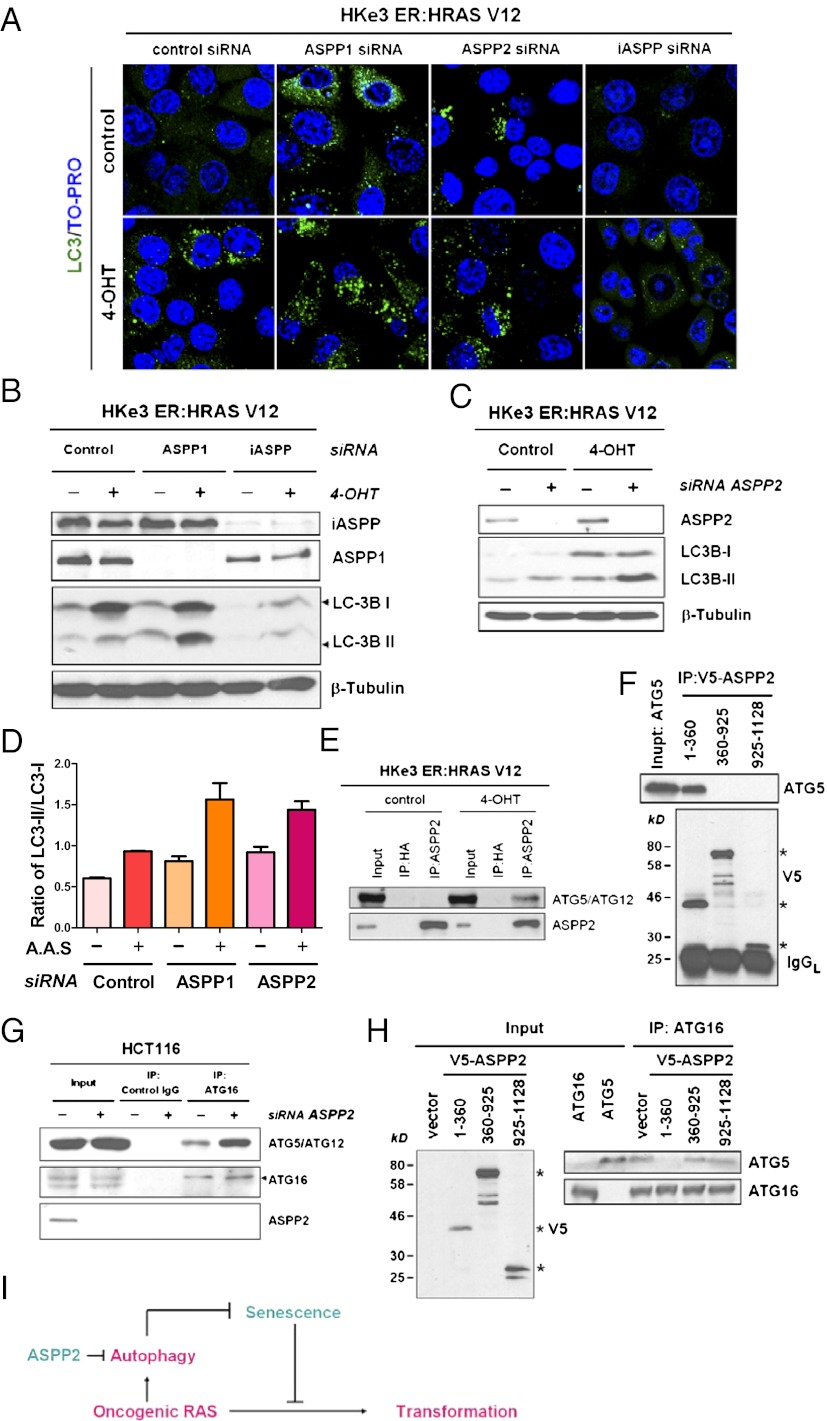
To investigate how ASPP2 inhibits RAS autophagy and to demonstrate that this property of ASPP2 is not specific to MEFs, we used a pair of isogenic human colon cancer cell lines: HCT116 and its isogenic counterpart HKe3, which was created by genetic disruption of the activated KRAS allele in HCT116 (23). HKe3 cells were transduced with a regulatable RAS construct made up of mutant HRAS fused to the ER ligand-binding domain that is conditionally responsive to 4-OHT: HKe3 ER:HRAS V12 cells (24, 25). Further, knowing that the N-terminal ASPP2 region is required and sufficient to inhibit RAS-induced autophagy, we hypothesized that the newly identified property of ASPP2 should also exist in ASPP1 but not in inhibitor of apoptosis-stimulating protein of p53 (iASPP). This is based on the fact that ASPP1 shares high sequence similarity to ASPP2 in its N terminus, whereas iASPP does not. This hypothesis was tested using HKe3 ER:HRAS V12 cells. Knockdown of ASPP1 or ASPP2 alone using RNAi resulted in a detectable increase in the number of cells expressing punctate LC3, an indicator of autophagy (Fig. 4A and Fig. S4A). Moreover, RAS activation, together with ASPP1 or ASPP2 depletion, had a synergistic effect on autophagy, reflected by a significant increase in the number of cells expressing punctate LC3 (Fig. 4A and Fig. S4A). Consistent with the observed morphological changes, we also observed an increase in the ratio of LC3II/I in ASPP1- or ASPP2-deficient HKe3 cells on RAS activation (Fig. 4 B and C). iASPP knockdown did not induce autophagy, because LC3II production was not induced, but rather resulted in a decrease in the expression levels of both LC3I and LC3II (Fig. 4B). It remains unknown why iASPP depletion results in a decrease in LC3 expression. Nonetheless, these results demonstrate that only ASPP1/ASPP2 but not iASPP inhibits RAS-induced autophagy. The ability of ASPP1/ASPP2 to inhibit endogenous oncogenic RAS-induced autophagy was further tested in the HCT116 cell line expressing endogenous mutant RAS. In HCT116 cells, ASPP1/2 knockdown promoted basal and amino acid starvation-induced autophagic activity, indicated by an increased ratio of LC3II/I (Fig. 4D and Fig. S4B). These results are in keeping with the increased stabilization of ATG5/12 attributable to ASPP2 knockdown shown in Fig. 2C and demonstrate that the ability of ASPP2 to inhibit RAS-induced autophagy is a general phenomenon. ASPP1/ASPP2 may inhibit RAS-induced autophagy via its N terminus, because iASPP does not have this domain. The data are also in agreement with the observation in MEFs showing that ASPP2 (1–123) is required for inhibition of RAS-induced autophagy.
Fig. 4.
ASPP2 inhibits autophagy by binding to ATG5/ATG12, and thus prevents the formation of a complex of ATG5/ATG12/ATG16. (A) Immunofluorescent staining of LC3 (green) in HKe3 ER:HRAS V12 cells with the indicated treatments. TO-PRO-3 (Invitrogen) was used to stain nuclei (blue). Images were taken with a ×63, oil-immersion objective lens. HKe-3 ER:HRAS V12 cells were transfected with control siRNA or siRNA against ASPP1, iASPP (B), or ASPP2 (C) for 3 d, followed by treatment without (control) or with 100 nM 4-OHT for 1 d. (D) Depletion of ASPP1 or ASPP2 promotes amino acid starvation (A.A.S.)-induced autophagy. HCT116 cells were transfected with the indicated siRNA for 4 d, followed by amino acid starvation for 5 h. The graph shows the ratio of LC3II/I. Error bars indicate SD. (E) ASPP2 binds ATG5/ATG12 in HKe3 ER:HRAS V12 cells on RAS activation. Total cell lysates from HKe3 ER:HRAS V12 cells treated without (control) or with 4-OHT (1 d) were immunoprecipitated with an anti-ASPP2 antibody or control IgG. HA, hemagglutinin; IP, immunoprecipitation. (F) Coimmunoprecipitation experiment using in vitro translated ATG5 and V5-tagged ASPP2 fragments. V5 antibody was used to immunoprecipitate ASPP2. ATG5 and ASPP2 levels were then analyzed by SDS/PAGE/immunoblotting using antibodies against ATG5 or V5. Stars indicate the correct size of ASPP2 fragments. The IgG light chain (IgGL) is labeled. (G) ASPP2 depletion enhances the binding between ATG16 and ATG5/ATG12 in HCT116 cells. HCT116 cells were transfected with control siRNA or siRNA against ASPP2 for 4 d. Total cell lysates from HCT116 cells with the indicated treatments were immunoprecipitated with an anti-ATG16 antibody or control IgG. (H) N-terminal ASPP2 competes with ATG16 to bind ATG5 in vitro. In vitro translated ATG16, ATG5, and V5-tagged ASPP2 fragments were mixed. ATG16 antibody was used to immunoprecipitate ATG16. ATG16 and ATG5 levels were then analyzed by SDS/PAGE/immunoblotting using antibodies against ATG16 or ATG5. Stars indicate the correct size of the ASPP2 fragments. (I) Diagram summarizes the role of ASPP2 in regulating autophagic activity, thereby dictating the cellular response to oncogenic RAS (details are provided in Discussion).
The high structural similarity between N-terminal ASPP2 and ATG12 (14) suggests that ASPP2 may bind ATG5/ATG12 to affect autophagy directly. This possibility was tested in HKe3 ER:HRAS V12 cells. We observed that in these cells, on RAS activation, ASPP2 translocated from cell/cell junctions to the cytoplasm (Fig. S4C) and it also coimmunoprecipitated with ATG5/ATG12 (Fig. 4E). In addition, in vitro translated ASPP2(1–360) was able to coimmunoprecipitate with in vitro translated ATG5 (Fig. 4F), suggesting that N-terminal ASPP2 may bind ATG5 directly. The formation of a complex between ATG16 and ATG5/ATG12 plays an essential role in autophagy (26). Knowing that the ASPP2/ATG5/ATG12 complex was best detected in HKe3 cells on RAS activation, the impact of ASPP2 on this complex formation was also tested in HCT116 cells. ASPP2 knockdown enhanced the formation of the complex between ATG16 and ATG5/ATG12 in HCT116 cells (Fig. 4G). Similar results were observed in HKe3 ER:HRAS V12 cells (Fig. S4D). To investigate whether the binding of ASPP2 and ATG16 to the ATG5/12 complex was mutually exclusive, an in vitro translated ASPP2 fragment was incubated with in vitro translated ATG16 and ATG5 and complexes were precipitated with anti-ATG16. No ATG5 was detected in the presence of the ASPP2(1–360) fragment, but ATG5 was coprecipitated when the other two fragments were present (Fig. 4H). Together, these results suggest that ASPP2 may compete with ATG16 to form the complex with ATG5/ATG12.
Discussion
Human tumors frequently express activated RAS because of mutations. RAS inhibitors were thus developed. However, because of the complexity of the RAS pathway, therapies are not successful with most RAS inhibitors (1). The identification of molecules that determine the cellular response to RAS is therefore timely for the development of better strategies to treat mutant RAS-expressing tumors. Here, we show that ASPP2 is one such regulator that determines the cellular response to RAS signaling by controlling autophagic activity and cellular senescence.
ASPP1 and ASPP2 are common activators of p53 (16). They share high sequence similarity in their N terminus, which has similar structural folds to ATG12 (14). ATG12 covalently modifies ATG5. The ATG5/ATG12 conjugate further forms a complex with ATG16, which plays an essential role in autophagy. The present study suggests that it is this unique structural feature of ASPP2 that allows it to compete with ATG16 to bind to ATG5/ATG12 in cells on RAS activation. As a result, ASPP2 inhibits RAS-induced autophagy by preventing the formation of the ATG16/ATG5/ATG12 trimeric complex. In epithelial cells, ASPP2 binds and colocalizes with Par3 at the tight/adherens junctions in vitro and in vivo (18, 20). Here, we show that on RAS activation, ASPP2 translocates from cell/cell junctions to the cytoplasm. How RAS induces ASPP2 translocation is not yet known, but RAS-induced cytoplasmic ASPP2 is able to bind to ATG5/ATG12 and prevent ATG16 from complexing with ATG5/ATG12. The functional importance of the proposed ASPP2-ATG5/ATG12 interaction in regulating autophagy is supported by the findings that (i) ASPP2 expression status affected the stability of ATG5/ATG12 and (ii) a significant increase in the expression levels of ATG5/ATG12 conjugates, but not ATG3, was only seen in late-passage HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs.
The observed increase in ATG5/ATG12 expression in RAS-expressing ASPP2(Δ3/Δ3) MEFs that escaped RAS-induced senescence argues strongly that autophagy provides a survival signal that allows cells to bypass RAS-induced cellular senescence. This agrees with recent findings in mouse and human tumor models showing that transformation by oncogenic RAS causes an addiction to survival signals provided by autophagy (7, 8) (Fig. 4I). One of the important questions is how elevated autophagy provides signals to maintain tumor growth. High levels of nuclear cyclin D1/CDK4 kinase activity are essential for cells to phosphorylate retinoblastoma protein (Rb) and to bypass cell cycle arrest and cellular senescence. We observed recently that ASPP2 uses a p53/p19Arf/p21waf1 independent pathway to mediate RAS-induced senescence by inhibiting RAS-induced nuclear accumulation of small ubiquitin-like modifier (SUMO)-modified cyclin D1 (15). Because ASPP2 deficiency enhances RAS-induced autophagy and elevated autophagy is sufficient to bypass RAS-induced senescence, it is possible that elevated autophagic activity may use the same pathway as that of ASPP2 deficiency to bypass RAS-induced senescence. Future studies are needed to investigate whether autophagic activity may dictate this cellular response by directly modulating the cell cycle machinery.
Although the results presented here seem to contradict those of Young et al. (4), whose data suggested that autophagy, and its consequent protein turnover, mediates the acquisition of the senescence phenotype (4), the discrepancy could be explained by our findings that the levels of autophagic activity are critical in dictating the cell’s response to RAS-induced senescence. We observed that when RAS induced small increases in autophagic activity, this was accompanied by senescence in ASPP2 WT MEFs. In ASPP2(Δ3/Δ3) MEFs, however, RAS-induced autophagic activity was over twofold higher than that observed in RAS-expressing ASPP2 WT MEFs, illustrating how ASPP2’s status may explain why HRAS V12 induces autophagy in some cells and fails to do so in others (9, 27). The observed large increase in autophagic activity in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs was the result of a significant increase in ATG5/ATG12 expression, and this increase was most profound in HRAS V12-expressing ASPP2(Δ3/Δ3) MEFs that escaped senescence. This initial finding led us to hypothesize that different levels of autophagic activity may dictate the cellular response to RAS. This conclusion is supported by the following two findings. First, we showed that elevated autophagy is sufficient to bypass RAS-induced senescence, as demonstrated by the finding that overexpressing WT ATG5, but not the autophagy-defective ATG5 mutant ATG5 K130R, can bypass RAS-induced senescence in ASPP2 WT MEFs. Second, we showed that a significant reduction or lack of autophagy can predispose cells to RAS-induced senescence. This is demonstrated by the finding that ATG5 shRNAs or deletion of ATG3 sensitizes MEFs to RAS-induced senescence. These data support the notion that different levels of autophagic activity dictate the cellular response to RAS-induced senescence: High levels of autophagy bypass senescence, low levels accompany it, and a significant reduction or lack of autophagy sensitizes cells to it. This agrees with several recent reports (5–8) showing that activated RAS requires autophagy to maintain tumorigenesis in vivo.
The identified growth inhibitory property of ASPP2’s N terminus presents us with a potentially unique therapeutic target. Deregulated autophagic activities are known to be critical in neuronal degenerative diseases, liver disease, and heart disease (28). The role of autophagy in cancer therapy is also of great importance because autophagy is often induced in chemotherapy- or radiation-treated tumor cells (29). Studies have shown that autophagy has opposing effects on cell survival and death and that these effects are likely to be cell type-dependent (30). Finally, down-regulation of ASPP2 is frequently observed in human tumors and is linked to poor prognosis (16, 31). The knowledge that ASPP2 inhibits autophagy, enhances cellular senescence, and inhibits tumor growth, in addition to its role in mediating p53-induced apoptosis, supports the development of autophagy inhibitors (e.g., chloroquine and its derivatives) (8) as therapeutic agents with which to treat cancers with reduced ASPP2 expression and mutated RAS.
Materials and Methods
MEFs were prepared from day 13.5 embryos from XTV-ASPP2(+/Δ3) intercrosses. Genotyping was performed by PCR using specific primers (17). All animal procedures were approved by local ethical review and licensed by the UK Home Office. For autophagy activity assays, measurement of the degradation of long-lived proteins, GFP-LC3 immunofluorescence, and LC3II/I Western blotting were performed. Analysis of senescence in cell culture was performed using a senescence β-gal staining kit (Cell Signaling) or BrdU incorporation experiments (15). To quantify SA-β-gal– or BrdU-positive readings, at least 200 cells were counted in random fields in each of the duplicated wells.
More details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Julian J. Lum for the pKD-control-GFP and pKD-Atg5-GFP constructs, Prof. Kevin Ryan for pBabe-GFP-ATG5, Dr. Noboru Mizushima for the gift of the Atg5(−/−) MEFs, Senji Shirasawa for providing HKe3 cells and Dr. Julian Downward for providing HKe3 ER:HRAS V12 cells. We thank Dr. Claire Beveridge and Evelyn Harvey for critical reading of the manuscript. We thank the Ludwig Institute for Cancer Research Ltd., European Union, and Association for International Cancer Research for supporting this work. C.G.G. and A.M.T. were supported by the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120193109/-/DCSupplemental.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson S, Ryan KM. Autophagy: An adaptable modifier of tumourigenesis. Curr Opin Genet Dev. 2010;20(1):57–64. doi: 10.1016/j.gde.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lock R, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22(2):165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MJ, et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem. 2011;286:12924–12932. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- 10.Yoo BH, et al. Oncogenic ras-induced down-regulation of autophagy mediator Beclin-1 is required for malignant transformation of intestinal epithelial cells. J Biol Chem. 2010;285:5438–5449. doi: 10.1074/jbc.M109.046789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byun JY, et al. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis. 2009;30:1880–1888. doi: 10.1093/carcin/bgp235. [DOI] [PubMed] [Google Scholar]
- 12.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Scherz-Shouval R, et al. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc Natl Acad Sci USA. 2010;107:18511–18516. doi: 10.1073/pnas.1006124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tidow H, Andreeva A, Rutherford TJ, Fersht AR. Solution structure of ASPP2 N-terminal domain (N-ASPP2) reveals a ubiquitin-like fold. J Mol Biol. 2007;371:948–958. doi: 10.1016/j.jmb.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Wang XD, et al. SUMO-modified nuclear cyclin D1 bypasses Ras-induced senescence. Cell Death Differ. 2011;18:304–314. doi: 10.1038/cdd.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuels-Lev Y, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 17.Vives V, et al. ASPP2 is a haploinsufficient tumor suppressor that cooperates with p53 to suppress tumor growth. Genes Dev. 2006;20:1262–1267. doi: 10.1101/gad.374006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sottocornola R, et al. ASPP2 binds Par-3 and controls the polarity and proliferation of neural progenitors during CNS development. Dev Cell. 2010;19:126–137. doi: 10.1016/j.devcel.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Langton PF, Colombani J, Aerne BL, Tapon N. Drosophila ASPP regulates C-terminal Src kinase activity. Dev Cell. 2007;13:773–782. doi: 10.1016/j.devcel.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Cong W, et al. ASPP2 regulates epithelial cell polarity through the PAR complex. Curr Biol. 2010;20:1408–1414. doi: 10.1016/j.cub.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 21.Naumovski L, Cleary ML. The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M. Mol Cell Biol. 1996;16:3884–3892. doi: 10.1128/mcb.16.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia W, He Y-W. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 23.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260(5104):85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 24.Dajee M, Tarutani M, Deng H, Cai T, Khavari PA. Epidermal Ras blockade demonstrates spatially localized Ras promotion of proliferation and inhibition of differentiation. Oncogene. 2002;21:1527–1538. doi: 10.1038/sj.onc.1205287. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Critical role for transcriptional repressor Snail2 in transformation by oncogenic RAS in colorectal carcinoma cells. Oncogene. 2010;29:4658–4670. doi: 10.1038/onc.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kundu M, Thompson CB. Autophagy: Basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 27.Psahoulia FH, et al. Quercetin mediates preferential degradation of oncogenic Ras and causes autophagy in Ha-RAS-transformed human colon cells. Carcinogenesis. 2007;28:1021–1031. doi: 10.1093/carcin/bgl232. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo Y, Kondo S. Autophagy and cancer therapy. Autophagy. 2006;2(2):85–90. doi: 10.4161/auto.2.2.2463. [DOI] [PubMed] [Google Scholar]
- 30.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lossos IS, Natkunam Y, Levy R, Lopez CD. Apoptosis stimulating protein of p53 (ASPP2) expression differs in diffuse large B-cell and follicular center lymphoma: Correlation with clinical outcome. Leuk Lymphoma. 2002;43:2309–2317. doi: 10.1080/1042819021000040017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.