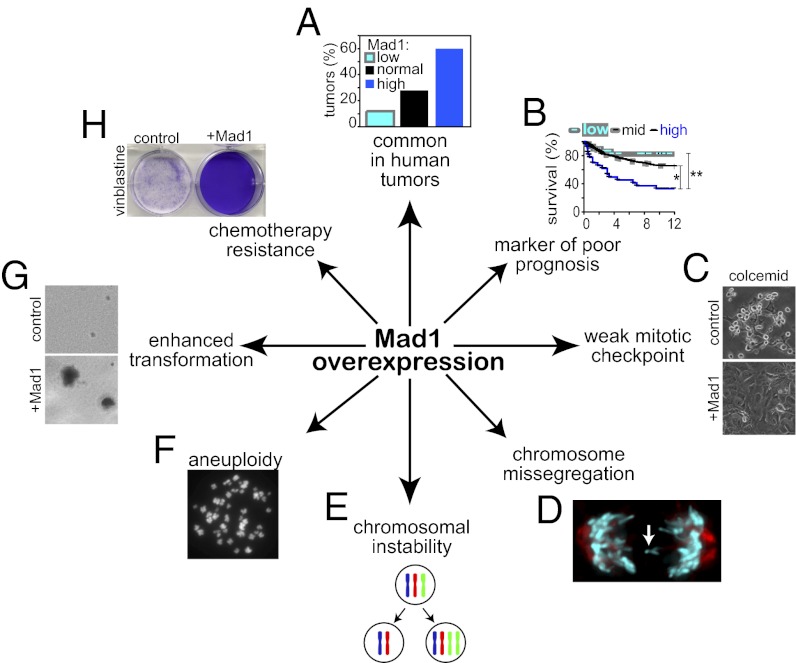
Abstract
The mitotic checkpoint is the major cell cycle checkpoint acting during mitosis to prevent aneuploidy and chromosomal instability, which are hallmarks of tumor cells. Reduced expression of the mitotic checkpoint component Mad1 causes aneuploidy and promotes tumors in mice [Iwanaga Y, et al. (2007) Cancer Res 67:160–166]. However, the prevalence and consequences of Mad1 overexpression are currently unclear. Here we show that Mad1 is frequently overexpressed in human cancers and that Mad1 up-regulation is a marker of poor prognosis. Overexpression of Mad1 causes aneuploidy and chromosomal instability through weakening mitotic checkpoint signaling caused by mislocalization of the Mad1 binding partner Mad2. Cells overexpressing Mad1 are resistant to microtubule poisons, including currently used chemotherapeutic agents. These results suggest that levels of Mad1 must be tightly regulated to prevent aneuploidy and transformation and that Mad1 up-regulation may promote tumors and cause resistance to current therapies.
Keywords: spindle assembly checkpoint, paclitaxel
Aneuploidy, an abnormal chromosome number that deviates from a multiple of the haploid, is a common characteristic of tumor cells, occurring in ∼85% of all human cancers (1). Some aneuploid cells also exhibit chromosomal instability (CIN), and have constantly evolving karyotypes. The mitotic checkpoint, also known as the spindle assembly checkpoint, is the major cell cycle checkpoint acting during mitosis to prevent aneuploidy and CIN (reviewed in refs. 2, 3). This checkpoint delays separation of the replicated chromosomes until each and every chromosome has made stable attachments to the microtubules of the mitotic spindle (4–6). This promotes accurate chromosome segregation and the production of genetically identical progeny. Reduction of individual components of the mitotic checkpoint, including Mad1, Mad2, Bub1, BubR1, Bub3, CENP-E, and Mps1, leads to aneuploidy and CIN in multiple systems (7–20).
The identification of mutations in two mitotic checkpoint genes in colorectal cancer cell lines suggested a possible mechanism for CIN in human tumors (21). However, extensive analysis has revealed that mitotic checkpoint genes are only rarely mutated in human cancers. Reduced expression is more common than mutation (22–27) (reviewed in ref. 1). Reduction of some mitotic checkpoint components, including Mad1, Mad2, Bub1, and CENP-E, increases spontaneous tumor incidence in mice (10, 11, 14, 28). Reduction of other mitotic checkpoint components, such as BubR1 and Bub3, does not increase spontaneous tumor incidence, although these animals can be more sensitive to tumors induced by carcinogens or concomitant loss of tumor suppressors (7, 8, 18, 28–30). In some cases, reduction of specific mitotic checkpoint components promotes tumors in certain contexts but suppresses them in others (18, 31). Interestingly, up-regulation of mitotic checkpoint components is as common, and in some cases more common, than down-regulation (1). Although effects of reduction of many mitotic checkpoint components have been studied, less is known about the consequences of overexpression of mitotic checkpoint components on mitosis, the mitotic checkpoint, aneuploidy, and transformation.
Mad1 is an ∼85-kD component of the mitotic checkpoint that remains tightly bound to Mad2 throughout the cell cycle (32). During interphase, Mad1 and Mad2 localize to the nucleus, nuclear envelope, and nuclear pores, where Mad1 has been implicated in nuclear import (33, 34). During mitosis, Mad1 localizes to kinetochores, where it serves as a docking site for Mad2. Each molecule of Mad1 recruits two molecules of Mad2 (35, 36). One molecule of Mad2 remains stably bound to Mad1, whereas the other rapidly cycles on and off kinetochores (37, 38). The second, rapidly cycling, molecule of Mad2 is converted from a large soluble pool of inactive Mad2 into an inhibitor of the Anaphase Promoting Complex (APC) bound to its specificity factor Cdc20 (APC-Cdc20), the target of the mitotic checkpoint (39, 40). In vitro, Mad1 fragments that lack the kinetochore targeting domain serve as competitive inhibitors of Mad2–Cdc20 complexes (41). In culture, a similar Mad1 fragment that lacks the kinetochore localization sequence abrogates mitotic checkpoint signaling without substantial mislocalization of Mad2 (42).
Mad1 mRNA is up-regulated in ∼90% of breast carcinomas and cell lines (43–45). In addition, the Mad1 genetic locus is amplified in ∼90% of small-cell lung cancer cell lines (46) and ∼55% of mantle-cell lymphoma cell lines (47). Elevated Mad1 mRNA expression is also associated with tumor progression in prostate and colorectal cancer (48, 49). However, it is unclear whether Mad1 protein is overexpressed in tumors and whether this is a cause or a consequence of transformation. In this study, we find that Mad1 protein is frequently overexpressed in human tumors and that up-regulation of Mad1 weakens mitotic checkpoint signaling and enhances transformation.
Results
Mad1 Is Overexpressed in Human Cancer.
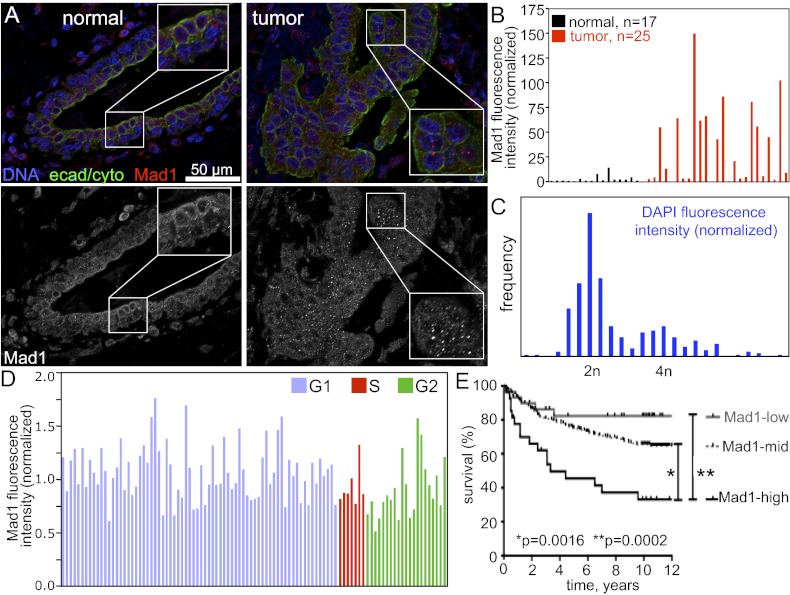
To determine whether Mad1 is overexpressed in human tumors, we compared the expression level of Mad1 protein in normal vs. cancerous tissue by using a breast cancer tumor microarray containing 16 normal breast specimens and 25 breast tumor samples. In normal tissue, Mad1 localized to the nuclear envelope and nucleus (Fig. 1A, Left). Quantitative immunofluorescence microscopy revealed that Mad1 was frequently overexpressed in tumor samples (Fig. 1A, Right). Fifteen of 25 tumor samples (60%) expressed Mad1 at levels greater than fivefold more than those in normal samples, whereas three of 25 tumors (12%) had Mad1 levels <0.5 fold that of normal samples (Fig. 1B). The remaining seven tumors (28%) had levels similar to normal samples (0.5 < n < 5). Overexpression of Mad1 was substantial, with 11 of 25 cancers (44%) having Mad1 expression levels >35 fold greater than those of normal controls. The highest-expressing tumors had a Mad1 expression level >100 fold greater than the median level of expression in noncancerous controls (Fig. 1B). High-expressing cells often exhibited Mad1 puncta (Fig. 1A, Right, Inset), similar to those reported previously in cells transiently overexpressing Mad1 to be annulate lamellae, storage compartments for nuclear pore components (34).
Fig. 1.
Overexpression of Mad1 is common in tumors and is a marker of poor prognosis. (A) Images of normal breast tissue (Left) and breast tumor tissue (Right) stained for DNA (blue), a mixture of e-cadherin and cytokeratin to identify epithelial cells (green) and Mad1 (red). Black and white images of Mad1 are shown in the bottom row. Inset: Enlargement of boxed region shows Mad1 puncta previously identified as annulate lamellae (34). (Scale bar, 50 μm.) (B) Quantification of Mad1 fluorescence intensity pictured in A. Mad1 fluorescence intensity in 16 normal breast samples, a normal spleen sample, and 25 breast tumor samples was quantified in a region of interest containing the nucleus and perinuclear region to ensure inclusion of nuclear envelope staining. Values shown are normalized to the median value in normal tissues (Materials and Methods). (C) Histogram of DAPI fluorescence intensity in DLD1 control cells. (D) Mad1 quantification in control DLD1 cells shows that Mad1 levels do not vary more than twofold from the average value during the cell cycle. Cell cycle stages were identified based on DAPI fluorescence intensity shown in C. (E) Kaplan–Meier survival curves of patients with breast cancer expressing high (black), intermediate (mid; gray dashed), or low (gray) levels of Mad1. Significance was calculated by using a Wilcoxon log-rank test (SI Appendix, Table S1).
Expression of some mitotic checkpoint components is elevated during mitosis, making it difficult to determine whether increased expression in tumors is simply a result of a higher rate of proliferation. However, we do not believe this is the reason for increased expression of Mad1 in tumors for several reasons. First, Mad1 is expressed throughout the cell cycle, making it unlikely that protein levels vary 35 to 100 fold during a typical division. Second, the Mad1 promoter has been reported to be predominantly active during G1, suggesting that Mad1 protein levels could be expected to be reduced in cells with a high mitotic index (50). However, as assessed by immunoblotting, Mad1 protein levels do not exhibit cell cycle regulated expression in yeast (51) or human (34). Consistent with this, Mad1 fluorescence intensity does not vary more than twofold from its average value in cells determined to be in G1, S, or G2 by DAPI fluorescence intensity (Fig. 1 C and D). Finally, none of the tumors in the microarray had a mitotic index greater than 1.8%. We conclude that primary cancers frequently overexpress Mad1 protein and that this is not simply caused by an increased rate of proliferation.
Mad1 Overexpression Is a Marker of Poor Prognosis.
To determine the relevance of Mad1 overexpression in human tumors, we screened a microarray dataset containing 242 breast tumors from the Stockholm and Uppsala cohorts described by Ivshina et al. (52) [Gene Expression Omnibus (GEO) accession no. GSE4922]. Mad1 expression level in the tumors was classified as high, low, or intermediate if Mad1 expression was twofold higher, lower, or within twofold of the median expression level in the tumors, respectively. Patients displayed a Mad1 expression dependent decrease in 12-y survival. Patients with high Mad1 expression had only a 33% survival rate at 12 y, whereas patients with intermediate levels of Mad1 had a 65% survival rate, and patients with low expression of Mad1 displayed greater than 80% survival (Fig. 1E). Mad1 status showed a positive correlation with lymph node involvement, tumor size, and grade (SI Appendix, Table S1). Interestingly, there is a significant correlation between Mad1 expression levels and p53 status in this cohort such that Mad1-high tumors were more likely to be p53 mutant than Mad1-low tumors. There was no correlation between Mad1 expression levels and estrogen receptor status, a key determinant of patient outcome. Thus, Mad1 overexpression is a marker of poor prognosis in breast cancer that is independent of hormone status.
Up-Regulation of Mad1 Causes Aneuploidy.
Well-established and characterized breast cancer cell lines (e.g., MCF-7, MDA-MB-231, T47D, SK-BR-3) are extensively aneuploid, complicating studies of chromosome segregation. Interestingly, Mad1 expression has been reported to be up-regulated in colon adenocarcinomas compared with colon adenomas (GEO accession no. GSE8067) (49) and well-characterized chromosomally stable colon cancer cell lines do exist. To determine if Mad1 overexpression is also a marker for poor prognosis in colon cancer, we screened a microarray dataset containing 83 stage II and III colorectal cancers from the cohort described by Sveen et al. (53) (GEO accession no. GSE24549). Patients with high Mad1 expression had only a 37% survival rate at 10 y, whereas patients expressing low levels of Mad1 had a 62% survival rate (SI Appendix, Fig. S1A). For these reasons, we determined the effects of Mad1 overexpression in a chromosomally stable colorectal cancer cell line (DLD1).
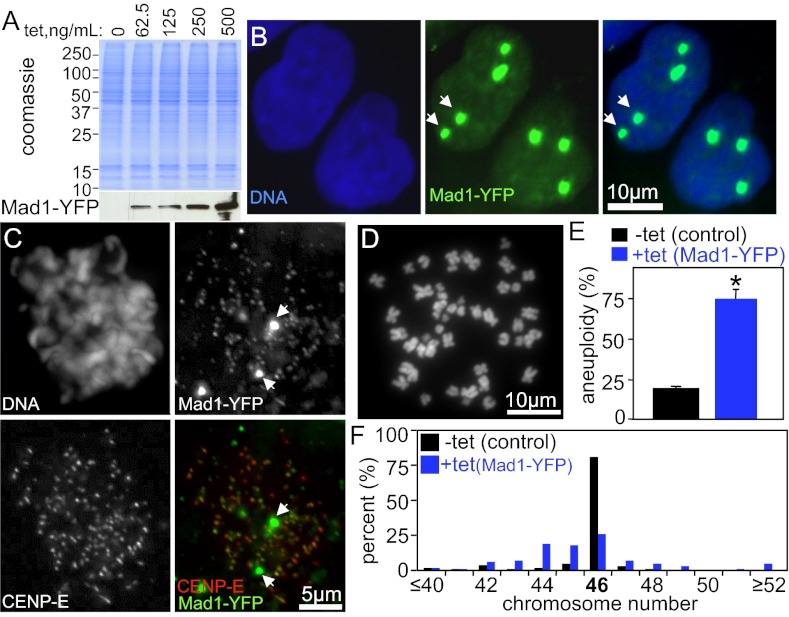
DLD1 cells stably expressing tet-inducible Mad1 or Mad1-YFP were generated. Addition of tetracycline induced expression of Mad1-YFP in a dose-dependent manner (Fig. 2A). A single administration of tetracycline was sufficient to induce Mad1-YFP protein for at least 4 d (SI Appendix, Fig. S1B). The level of overexpression was measured by immunoblotting serially diluted extracts (SI Appendix, Fig. S1 C and D) and by quantitative immunofluorescence (SI Appendix, Fig. S1 E and F), which gave similar results. A dose of tetracycline that induced Mad1-YFP (SI Appendix, Fig. S1C) or untagged Mad1 (SI Appendix, Fig. S1D) to a level equivalent to the median fold change of Mad1 overexpression in human tumors (Fig. 1 A and B) was chosen for further analysis.
Fig. 2.
Up-regulation of Mad1 causes aneuploidy. (A) Tet-inducible expression of Mad1-YFP. Coomassie staining was used as a loading control. (B) Mad1-YFP (green) localizes to nuclei in interphase, like endogenous Mad1. Arrows indicate that Mad1-YFP also localizes to additional sites previously identified as annulate lamellae (34). (C) Mad1-YFP (green) localizes to kinetochores (marked with CENP-E, red) during mitosis, as well as to additional sites (arrows). (D) Chromosome spread of DLD1 cell. (E) DLD1 cells expressing Mad1-YFP for 48 h have higher levels of aneuploidy than control cells (n = 100 cells from two independent experiments; *P < 0.05, t test). (F) Chromosome numbers in control and Mad1-YFP–expressing cells show near-diploid aneuploidy with minimal tetraploidy (n = 100 cells from two independent experiments; SI Appendix, Fig. S2 A–C, shows results with untagged Mad1).
Like endogenous Mad1 (SI Appendix, Fig. S1E), Mad1-YFP localized to the nucleus and nuclear envelope in interphase cells (Fig. 2B) and to kinetochores during mitosis (Fig. 2C). Kinetochore localization was confirmed by colocalization with CENP-E (Fig. 2C). Expression of untagged Mad1 resulted in a similar localization pattern during interphase (SI Appendix, Fig. S1E) and mitosis (SI Appendix, Fig. S1 G–I). Although expression of Mad1 was elevated, kinetochore recruitment was not substantially increased (SI Appendix, Fig. S1 G and H). In addition to exhibiting the expected localization pattern, overexpressed Mad1 and Mad1-YFP apparently saturated the available binding sites and localized to additional sites as well (Fig. 2 B and C, arrows, and SI Appendix, Fig. S1E), as seen in the tumor microarray (Fig. 1A, Right). These sites were previously identified as annulate lamellae, storage compartments for NPC proteins, on the basis of their colocalization with nuclear pore components (34). Consistent with this, overexpressed untagged Mad1 partially colocalized at presumptive annulate lamellae and at the nuclear envelope with the mAb414 antibody, which recognizes the FG repeats found in multiple NPC proteins (SI Appendix, Fig. S1E).
To determine the effect of Mad1 overexpression on ploidy, chromosome number was determined in chromosome spreads (also known as metaphase spreads; Fig. 2D) from cells with or without induction of Mad1-YFP. Control DLD1 cells were predominantly diploid, whereas cells overexpressing Mad1-YFP exhibited significant aneuploidy (Fig. 2E). Examination of the number of chromosomes per cell identified frequent gain and loss of small numbers of chromosomes with little evidence of tetraploidy (Fig. 2F). Similar results were obtained in cells overexpressing untagged Mad1 (SI Appendix, Fig. S2 A–C).
Cells Overexpressing Mad1 Exhibit Misaligned and Lagging Chromosomes.
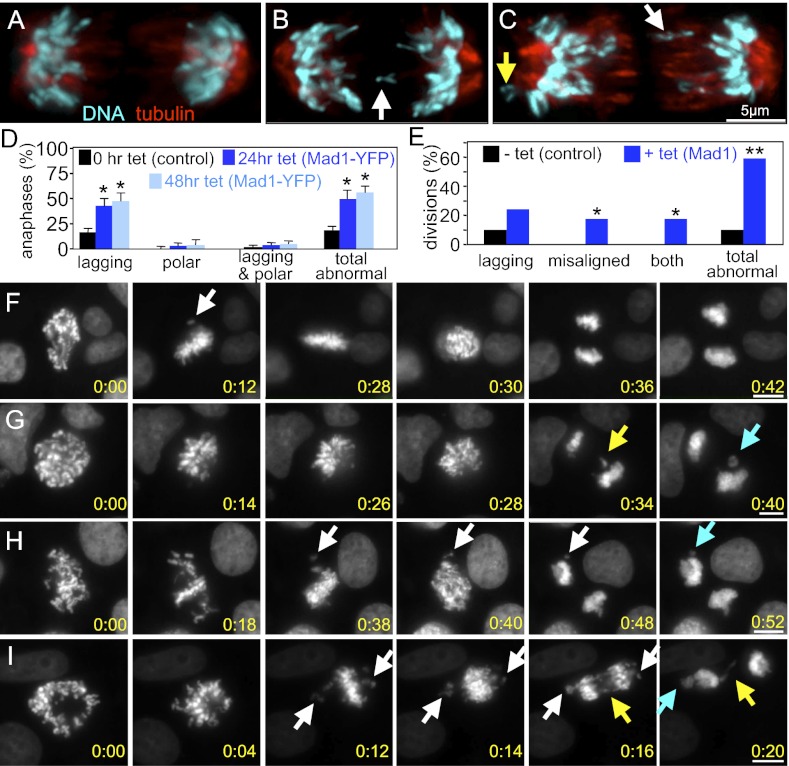
Consistent with the observed aneuploidy, examination of fixed cells overexpressing Mad1-YFP revealed a high frequency of lagging chromosomes in anaphase and telophase (Fig. 3 A–D; note white arrows in Fig. 3 B and C). Polar chromosomes, which remained near one of the spindle poles in anaphase and appeared to be misaligned upon chromosome disjunction, were only rarely detected (Fig. 3 C and D, yellow arrow). Similar results were obtained when examining cells overexpressing untagged Mad1 (SI Appendix, Fig. S2 D and E).
Fig. 3.
Elevated expression of Mad1 causes CIN from lagging and misaligned chromosomes. Control (A) and Mad1-YFP–expressing (B and C) DLD1 cells in anaphase. White arrows indicate lagging chromosomes. Yellow arrow indicates polar chromosome. (D) Fixed cells expressing Mad1-YFP have high levels of lagging chromosomes in anaphase (n > 100 cells from each of four independent experiments; *P < 0.05, t test). SI Appendix, Fig. S2 D and E, shows results with untagged Mad1. (E) Quantification of segregation errors observed during time-lapse microscopy of DLD1 cells stably expressing histone H2B-RFP, as shown in F–I (*P < 0.05, t test). (F–I) Still images from time-lapse acquisition of H2B-RFP in control (F) and Mad1-overexpressing (G–I) cells (Movies S1, S2, S3, and S4). Panel 1, First image after nuclear envelope breakdown. Panel 3, Image immediately before anaphase onset, shown in panel 4. Control cells waited for the last chromosome to align (F, panel 2, arrow) before segregating chromosomes in anaphase. (G–I) Cells induced to overexpress Mad1 for 24 h show lagging chromosomes (yellow arrows) and/or misaligned chromosomes at anaphase onset (white arrows). Some of the lagging and misaligned chromosomes form micronuclei (turquoise arrows). (Scale bars, 10 μm.) The time from nuclear envelope breakdown is shown in hours:minutes in the bottom right corner of each image.
To observe chromosome segregation defects more closely, cells stably expressing histone H2B-RFP were observed by live cell microscopy. Control cells exhibited predominantly normal divisions in which they waited for the last chromosome to align before segregating their chromosomes equally to produce genetically identical daughters (Fig. 3 E and F and Movie S1). Cells overexpressing Mad1 exhibited a variety of phenotypes of differing severity. Approximately one quarter (24.1%) of cells overexpressing Mad1 had lagging chromosomes in anaphase and telophase with no other observable defects (Fig. 3 E and G and Movie S2). In addition to cells with lagging chromosomes, time-lapse analysis revealed a population of Mad1-overexpressing cells (17.1%) that entered anaphase in the presence of misaligned chromosomes (Fig. 3 E and H and Movie S3). Another 17.2% entered anaphase with misaligned chromosomes and exhibited lagging chromosomes as well (Fig. 3 E and I and Movie S4), bringing the percentage of cells with lagging chromosomes in anaphase and telophase to 41.4%, similar to that observed in our fixed-cell analysis. All told, 58.6% of Mad1-overexpressing cells missegregated chromosomes during mitosis, consistent with a CIN phenotype (Fig. 3E).
Overexpression of Mad1 Disrupts Mitotic Timing.
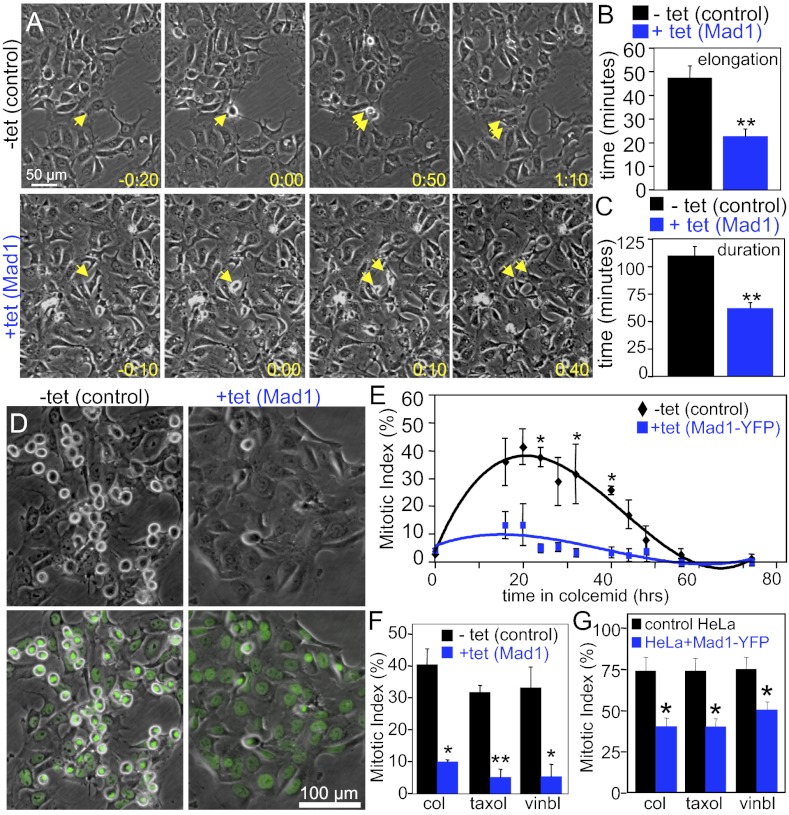
Ectopic expression of Mad2 has been shown to delay the metaphase-to-anaphase transition (54). To determine whether overexpression of Mad1 had a similar or different effect, the timing of mitosis was determined by phase-contrast microscopy. Whereas control cells elongated in an average of 47.7 min and completed mitosis (scored as the time from rounding to flattening) in 109.3 min, cells overexpressing Mad1 elongated in 22.7 min and completed mitosis in 61.8 min (Fig. 4 A–C and Movies S5 and S6). Thus, Mad1-overexpressing cells traversed mitosis in approximately half the time of control cells.
Fig. 4.
Excess Mad1 causes a weakened mitotic checkpoint. (A) Stills from 10× phase-contrast movies show control cells (Upper) and cells overexpressing Mad1 (Lower) in interphase (Left), as they round up in mitosis (Center Left), elongate (Center Right), and readhere at the end of mitosis (Right). Numbers show time in hours:minutes that corresponds with the division of the cells denoted by arrows. 0:00 is the time at which the cells round up in mitosis (Movies S5 and S6). (Scale bar, 50 μm.) (B) The time from cell rounding to cell elongation (which roughly corresponds to anaphase onset) is shortened in cells expressing increased levels of Mad1. (C) The duration of mitosis, as measured from cell rounding to cell flattening, is decreased in cells with excess Mad1 (B and C, n > 70; **P < 0.001, t test). (D) Phase-contrast (Upper) or phase-contrast and Hoechst (Lower) images of control and Mad1-overexpressing cells treated with colcemid for 20 h. Note that the frequency of rounded, mitotic cells is significantly lower in cells overexpressing Mad1. (E) Mad1-YFP–expressing cells do not accumulate in mitosis like control cells in response to colcemid (n > 500 cells from each of two independent experiments; *P < 0.05, t test). (F) Reduced mitotic index in Mad1-overexpressing cells treated with the microtubule poisons colcemid (col), paclitaxel (Taxol), or vinblastine (vinbl) for 20 h (n > 250 cells from each of three independent experiments; *P < 0.05 and **P < 0.001, t test). (G) Mad1-YFP expression also causes deficits in mitotic checkpoint signaling in HeLa cells treated with colcemid (col), paclitaxel (Taxol), or vinblastine (vinbl) for 24 h (n > 500 cells from each of four independent experiments; *P < 0.05).
Increased Expression of Mad1 Weakens the Mitotic Checkpoint.
The high percentage of cells entering anaphase with misaligned chromosomes, coupled with the reduced mitotic timing, suggested that the mitotic checkpoint is weakened by increased expression of Mad1. As an initial method of examination, the status of the mitotic checkpoint was determined by measuring the mitotic index after challenge with microtubule poisons. Mitotic index was measured in live cells treated with the DNA-binding dye Hoechst 33258 by phase-contrast and fluorescence microscopy (Fig. 4D). As expected, control cells accumulated in mitosis in response to loss of microtubules caused by treatment with colcemid, with the maximal mitotic index occurring 20 h after drug addition (Fig. 4E). However, cells expressing Mad1-YFP exhibited a maximal mitotic index approximately fourfold lower (Fig. 4E). A similar failure to accumulate in mitosis in response to colcemid was observed in cells overexpressing untagged Mad1 (Fig. 4F). Importantly, Mad1-overexpressing cells also failed to accumulate in mitosis in response to the clinically useful microtubule poisons paclitaxel (Taxol) and vinblastine. These drugs represent microtubule poisons with different mechanisms, as paclitaxel stabilizes microtubules, whereas vinblastine results in net loss of spindle microtubules, similar to colcemid (Fig. 4F). To determine whether this effect was unique to DLD1 cells, we tested the effect of Mad1-YFP expression on the mitotic checkpoint in HeLa cells. Like DLD1 cells, HeLa cells stably expressing Mad1-YFP showed a significantly decreased mitotic index in response to colcemid, paclitaxel, and vinblastine (Fig. 4G).
Defects in mitotic checkpoint signaling are sufficient to cause chromosome missegregation (9, 14, 19). However, multiple mitotic checkpoint components, including Bub1, BubR1, and CENP-E, also function in chromosome congression (13, 55–58). To determine whether overexpression of Mad1 inhibited congression, DLD1 cells were treated with the proteasome inhibitor MG132 to prevent sister chromatid disjunction and accumulate cells in metaphase. Cells overexpressing untagged Mad1 aligned their chromosomes to form metaphase plates at a rate indistinguishable from control cells (SI Appendix, Fig. S2F), suggesting that overexpression of Mad1 does not cause defects in congression. To further examine chromosome alignment in cells up-regulating Mad1, we treated cells expressing histone H2B-RFP and GFP-tubulin with monastrol to induce monopolar spindles. The monastrol was then washed out and replaced with MG132. Spindle assembly and chromosome congression after monastrol washout were observed by time-lapse microscopy. Compared with controls, cells expressing elevated levels of Mad1 were not delayed in aligning chromosomes under these conditions (SI Appendix, Fig. S2G).
Congression defects are often associated with a decrease in kinetochore-microtubule attachments, which can be detected by a reduction in interkinetochore distance (13, 17, 56, 59, 60). As a further test of whether Mad1 overexpression negatively influenced kinetochore-microtubule attachments, we measured the interkinetochore distance in control cells and those that overexpress untagged Mad1. No difference in interkinetochore distance was observed in Mad1-overexpressing cells (1.11 ± 0.16 μm; n = 139) compared with control cells (1.13 ± 0.18 μm; n = 134; P = 0.3245; SI Appendix, Fig. S2 H and I). This is consistent with recent data showing no role for Mad1 in kinetochore-microtubule turnover, at least when Mad1 is depleted by siRNA (61). Thus, the chromosome missegregation phenotype in Mad1-overexpressing cells appears to occur as a result of a deficit in mitotic checkpoint signaling coupled with a decrease in the duration of mitosis, as opposed to defects in kinetochore–microtubule interactions.
Overexpressed Mad1 Mislocalizes Mad2 from Kinetochores.
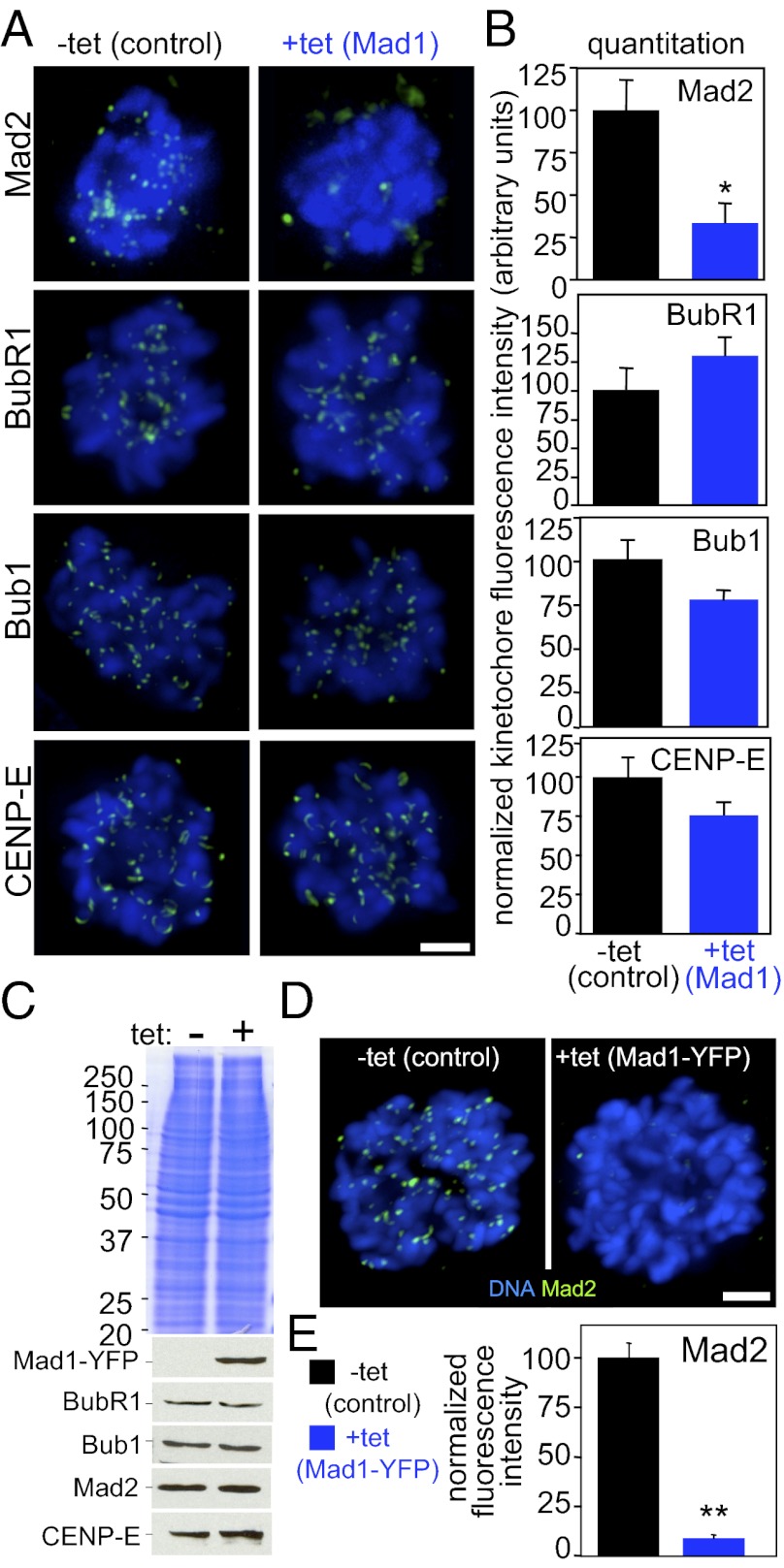
To determine the mechanism underlying the deficit in mitotic checkpoint signaling, kinetochore recruitment of several mitotic checkpoint components was determined. Overexpression of Mad1 did not affect kinetochore localization of Bub1, BubR1, or CENP-E (Fig. 5 A and B). Although a portion of the Mad1 binding partner Mad2 remained at kinetochores (SI Appendix, Fig. S1I), quantitative immunofluorescence revealed that the majority of Mad2 was mislocalized from kinetochores (Fig. 5 A and B) and colocalized with overexpressed Mad1 at nonchromosomal sites (SI Appendix, Fig. S3A). Mad2 kinetochore recruitment was reduced to approximately one third the level in control cells (Fig. 5B). Similar results were observed in an independent clone of cells overexpressing Mad1 (SI Appendix, Fig. S3B).
Fig. 5.
Mad2 is mislocalized from kinetochores in cells expressing excess Mad1. (A) Quantitative immunofluorescence of Mad2, BubR1, Bub1, CENP-E (green), and DNA (blue) in cells treated with colcemid for 1 h. (Scale bar, 2.5 μm.) (B) Quantification of kinetochore fluorescence intensity (n = 37–119 cells from at least three independent experiments; *P < 0.05, t test). SI Appendix, Fig. S3B, shows quantification of an independent clone of Mad1-overexpressing cells. (C) Mad1-YFP expression does not affect expression levels of other mitotic checkpoint components including BubR1, Bub1, Mad2, and CENP-E. Coomassie stain was used as a loading control. (D) Mad2 localizes to kinetochores during mitosis in control cells, but is largely mislocalized in cells expressing Mad1-YFP. (Scale bar, 5 μm.) (E) Quantification of Mad2 kinetochore fluorescence in Mad1-YFP–expressing cells (n > 32 cells from three independent experiments; **P < 0.001, t test).
Removal of Mad2 from kinetochores was not caused by loss of Mad2 protein, as Mad2, BubR1, Bub1, and CENP-E are all expressed at equivalent levels in Mad1-YFP–expressing cells as compared with controls (Fig. 5C). Mad1-YFP–expressing cells also showed reduction of Mad2 at kinetochores (Fig. 5 D and E) and relocalization of Mad2 with overexpressed Mad1-YFP (SI Appendix, Fig. S3C).
Normally, Mad2 is expressed in molar excess over Mad1 (35, 38, 62). Consistent with this, in control DLD1 cells, all detectable Mad1 is immunoprecipitated with Mad2 (SI Appendix, Fig. S3D, lanes 7 and 8) whereas only a fraction of detectable Mad2 immunoprecipitates with Mad1 (SI Appendix, Fig. S3D, lanes 3 and 4). However, in Mad1-overexpressing cells, nearly all Mad2 is immunoprecipitated with Mad1 (SI Appendix, Fig. S3D, lanes 5 and 6) whereas a portion of Mad1 remains soluble after immunoprecipitation of Mad2 (SI Appendix, Fig. S3D, lanes 9 and 10). Thus, the pool of free Mad2 that is not bound to Mad1 is diminished in Mad1-overexpressing cells.
Overexpression of Mad1 Enhances Transformation.
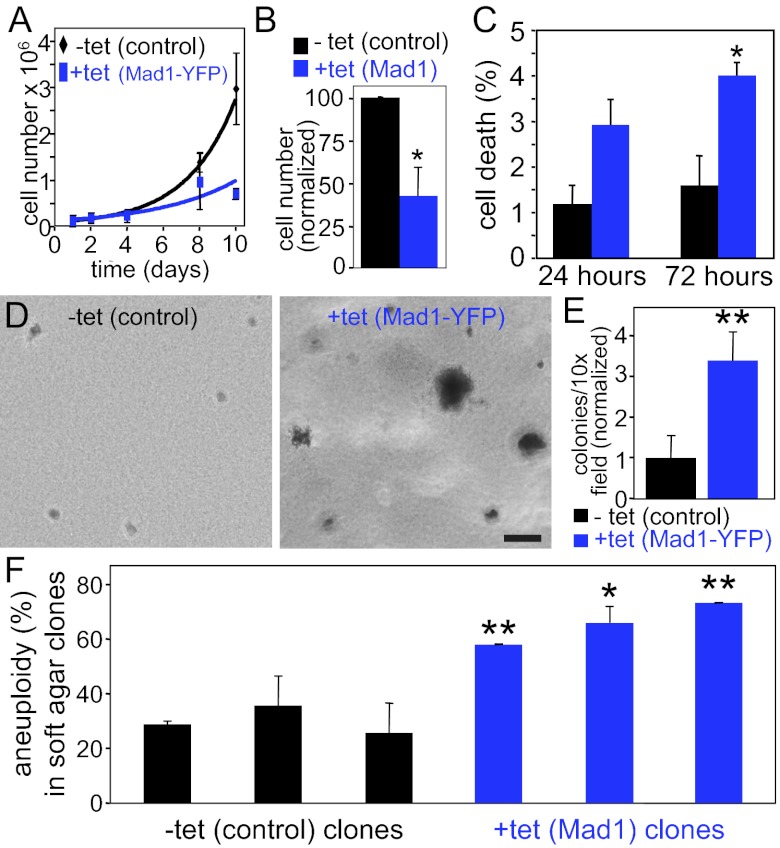
Up-regulation of Mad1 or Mad1-YFP reduced population growth rates compared with control cells (Fig. 6 A and B). This proliferative disadvantage was coupled with an increase in cell death (Fig. 6C), consistent with recent reports that clonal survival is inhibited in chromosomally stable cancer cell lines forced to exhibit CIN (63, 64). Interestingly, the surviving cells are significantly more likely to exhibit anchorage independent growth in soft agar, a marker of transformation. Although control cells exhibited limited growth in soft agar (Fig. 6D, Left, and Fig. 6E), ectopic expression of Mad1-YFP caused a 3.4-fold increase in the number of anchorage-independent colonies (Fig. 6D, Right, and Fig. 6E). Similar results were observed in cells overexpressing untagged Mad1 (SI Appendix, Fig. S3 E and F). This is reminiscent of a previous report that overexpression of Mad2 inhibits growth of cells in culture, but drives tumorigenesis in mice (65).
Fig. 6.
Up-regulation of Mad1 enhances transformation. (A) Expression of Mad1-YFP inhibits cell growth (n = 2). (B) Overexpression of untagged Mad1 for 10 d causes a proliferative defect (n = 3; *P < 0.05, t test). (C) Cells overexpressing Mad1 have a higher incidence of cell death. Cell death was scored in live cells stained with Hoechst based on cellular and DNA morphology (n ≥ 230 cells from each of three independent experiments; *P < 0.05, t test). (D and E) Cells expressing Mad1-YFP are more likely to exhibit anchorage independent growth in soft agar, a characteristic of transformation. (D) Images of cells and colonies in soft agar. (Scale bar, 100 μm.) (E) Quantification of colony growth after 10 to 12 d in soft agar (n = 3; **P < 0.001, t test). (F) Mad1-overexpressing anchorage-independent colonies exhibit increased aneuploidy. Three independent clones of control DLD1 cells and cells overexpressing untagged Mad1 were cored from soft agar and scored for aneuploidy using chromosome spreads (n > 50 cells from each clone; *P < 0.05 and **P < 0.001, t test).
If aneuploidy has a role in driving anchorage independent growth, colonies from Mad1-overexpressing cells should be more aneuploid than colonies from parental cells. To test this, we scored soft agar colonies for aneuploidy by using chromosome spreads. Three colonies each of control and Mad1-overexpressing cells were examined. In all cases, Mad1-overexpressing cells were significantly more aneuploid than controls (Fig. 6F), consistent with the hypothesis that aneuploid cells contribute to anchorage independent growth.
One potential concern is that, to date, the limited number of CIN cancer cell lines tested for mitotic checkpoint activity based on their ability to prevent anaphase onset in the presence of misaligned chromosomes (HT29, Caco2, MCF-7, SW480, and SW837) have all had an intact mitotic checkpoint (63, 66). Instead, these cells exhibit CIN as a result of lagging chromosomes likely caused by merotelic attachments. However, this does not demonstrate that mitotic checkpoint defects do not occur in any cancer cells or cell lines. Indeed, MDA-MB-231 breast cancer cells exhibit a weakened mitotic checkpoint. Eight of 49 (16%) MDA-MB-231 cells with bipolar spindles that expressed histone H2B-RFP and GFP-tubulin entered anaphase with misaligned chromosomes when examined by time-lapse microscopy (SI Appendix, Fig. S4 and Movie S7).
Cells Expressing High Levels of Mad1 Are Resistant to Microtubule Poisons.
The role of the mitotic checkpoint in the response to drugs that cause mitotic arrest remains controversial (67). Multiple groups have reported that a functional mitotic checkpoint is required for sensitivity to microtubule poisons and that a weakened mitotic checkpoint confers resistance (68–71). However, numerous other groups have found that cells with a weakened mitotic checkpoint are more sensitive to microtubule poisons (72–74) or that there is no difference (66, 75, 76). One recent hypothesis is that mitotic checkpoint impairment causes resistance to microtubule poisons in the short term (48–72 h), but does not confer resistance in longer-term studies (4–11 d) (75).
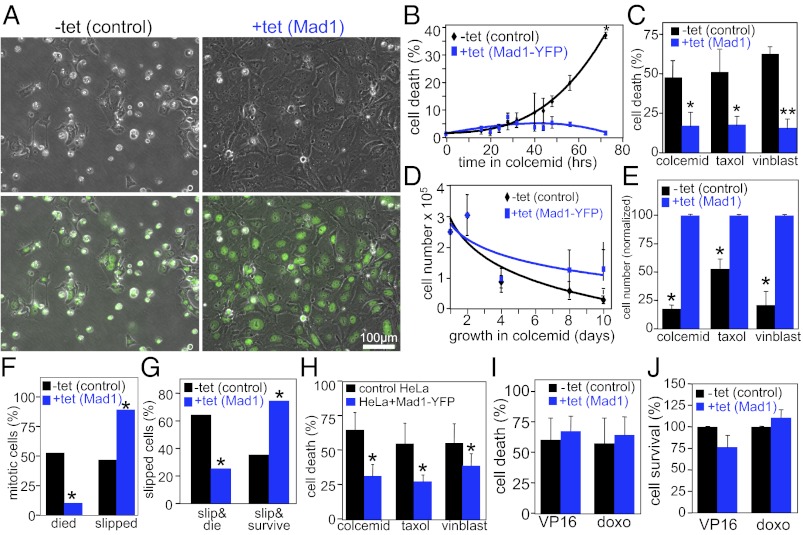
We tested the sensitivity of cells expressing increased levels of Mad1 to microtubule poisons at both short and long time points. Interestingly, up-regulation of Mad1 caused resistance to microtubule poisons after 72 h and 10 d of treatment. Death was determined based on DNA and cellular morphology by using phase-contrast and fluorescence microscopy of live cells treated with Hoechst 33258 (Fig. 7A). At 72 h after treatment with the microtubule poison colcemid, 36.5 ± 0.8% of control cells exhibited cell death, as opposed to 1.8 ± 0.1% of cells expressing Mad1-YFP (Fig. 7B). Overexpression of untagged Mad1 yielded similar results, with 45% to 65% death in control cells treated with colcemid, vinblastine and paclitaxel, but only 15% to 18% death in cells expressing increased levels of Mad1 (Fig. 7C).
Fig. 7.
Resistance to microtubule poisons from up-regulation of Mad1. (A) Phase-contrast images alone (Upper) or overlaid with Hoechst images (Lower) of control and Mad1-YFP–expressing cells treated with colcemid for 72 h. (B) Cell death, as scored in A, in control and Mad1-YFP–expressing cells exposed to colcemid for up to 72 h (n > 500 cells from each of two independent experiments; *P < 0.05, t test). (C) Cell death, as scored in A, of control and Mad1-overexpressing cells treated with colcemid, paclitaxel (Taxol), or vinblastine (vinblast) for 72 h (n > 250 cells from at least three independent experiments; *P < 0.05 and **P < 0.001, t test). (D) Cell numbers, as scored by hemacytometer, of control and Mad1-YFP–expressing cells exposed to colcemid (n = 2). (E) Cell numbers of control and Mad1-overexpressing cells exposed to microtubule poisons colcemid, paclitaxel (Taxol), or vinblastine (vinblast) for 10 d (n ≥ 3; *P < 0.05, t test). Cell number is normalized to the number of Mad1-overexpressing cells after drug treatment. (F) Cells expressing elevated levels of Mad1 exhibit less death from mitosis and more slippage than controls when treated with paclitaxel. (G) Control DLD1 cells that slip out of mitosis in paclitaxel are more likely to die than survive, whereas Mad1-overexpressing cells are more likely to survive than to die. (F and G, n = 66 cells from two independent experiments; *P < 0.001, χ2 test). (H) Expression of Mad1-YFP in HeLa cells results in resistance to 48 h treatment with colcemid, paclitaxel (Taxol), and vinblastine (vinblast; n > 500 cells from each of four independent experiments; *P < 0.001, χ2 test). (I and J) Mad1 overexpression does not affect sensitivity to DNA damaging drugs. (I) Cell death was determined as in A after 72 h of treatment with the topoisomerase II inhibitors etoposide (VP16; 15 μM) or doxorubicin [doxo; 500 ng/mL; n > 350 cells from each of four independent experiments; P = 0.6830 (etoposide) and P = 0.5233 (doxorubicin)]. (J) Cell survival was determined after 10 d of treatment with etoposide (VP16; 5 μM) or doxorubicin (doxo; 100 ng/mL) by hemacytometer (n = 3).
To determine whether up-regulation of Mad1 merely delayed cell death, cell number was determined over a 10-d exposure to colcemid. Cells expressing Mad1-YFP were significantly more viable than control cells over the time course (Fig. 7D). Similarly, cells expressing elevated levels of Mad1 were significantly more viable after 10 d of treatment with colcemid, paclitaxel and vinblastine (Fig. 7E). Mad1-overexpressing cells remained viable 1 wk after drug washout (SI Appendix, Fig. S5A), and proliferated into colonies when plated at low density (SI Appendix, Fig. S5B).
To better understand the short-term effects of Mad1 up-regulation on long-term mitotic arrest, cells were observed by time-lapse microscopy for ≥65 h after addition of microtubule poison. Cells expressing elevated levels of Mad1 were significantly less likely than controls to die from mitosis and were significantly more likely to slip out of mitosis (Fig. 7F). Of cells that slipped out of mitosis to form tetraploid G1 cells, most cells overexpressing Mad1 (75%) survived the duration of the movie, at least 30 h after slippage, whereas most control cells that slipped (64%) died during the period of observation (Fig. 7G).
To rule out the possibility that this effect of Mad1 up-regulation is unique to DLD1 cells, HeLa cells stably expressing Mad1-YFP were tested for their response to colcemid, paclitaxel and vinblastine. As in DLD1 cells, expression of Mad1-YFP in HeLa cells led to reduced cell death after treatment with microtubule poisons (Fig. 7H).
Based on the effects of Mad1 overexpression on mitosis, it is likely that Mad1 up-regulation causes resistance to microtubule poisons by reducing mitotic checkpoint activity. The effects on mitosis would not be predicted to affect the response to DNA damaging agents. To test this, rates of cell death and cell survival were measured after treating DLD1 cells with the topoisomerase II poisons doxorubicin (Adriamycin) and VP16 (etoposide). Cell death and survival rates in Mad1-overexpressing cells were indistinguishable from rates in control cells (Fig. 7 I and J). Thus, up-regulation of Mad1 decreases sensitivity to microtubule poisons currently used to treat human cancers without affecting the response to DNA damaging agents.
Discussion
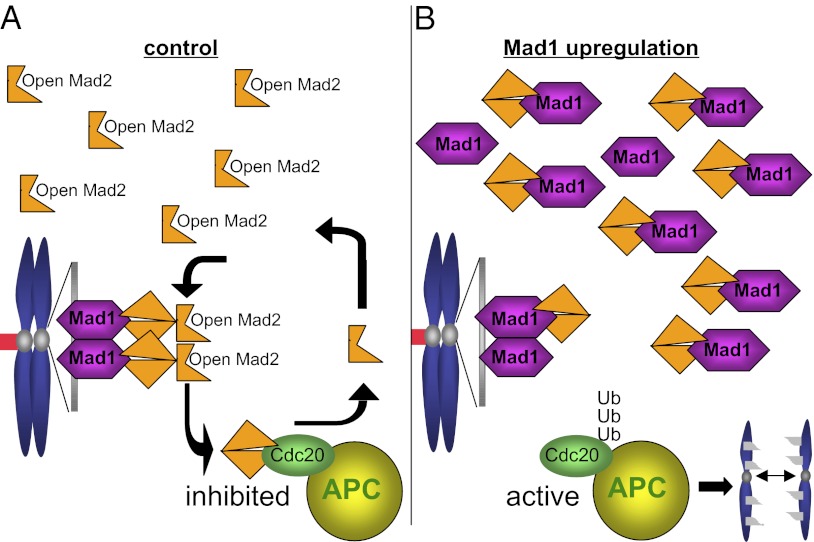
Our results are consistent with a model in which up-regulation of Mad1 weakens mitotic checkpoint signaling by titrating the soluble pool of Mad2 (Fig. 8). Normally, endogenous Mad2 is expressed in excess of endogenous Mad1 (35, 38, 62), permitting a soluble pool of Mad2. This soluble pool is converted into active inhibitors of APC-Cdc20 after transiently binding Mad1–Mad2 heterodimers at unattached kinetochores (Fig. 8A) (36). DLD1 cells that overexpress Mad1 to a level equivalent to the median overexpression in tumors contain an excess of Mad1 over Mad2. When expressed at this level, Mad1 saturates available kinetochore binding sites and accumulates in the cytoplasm (Figs. 2C and 8B and SI Appendix, Fig. S3 A and C). Mad2 then binds to cytoplasmic Mad1 instead of cycling on and off Mad1–Mad2 heterodimers at kinetochores. This weakens mitotic checkpoint signaling, resulting in premature activation of APC-Cdc20, aneuploidy, and CIN (Fig. 8B). This model is consistent with a study that found that the meiotic checkpoint in Xenopus extracts requires both Mad1-bound and Mad1-free Mad2 (62).
Fig. 8.
Up-regulation of Mad1 weakens mitotic checkpoint signaling by titrating Mad2. (A) In control cells, Mad2 exists in molar excess over Mad1 (32, 38, 62), permitting a large soluble pool of Mad2, which exists in an open conformation. Open Mad2 from the soluble pool rapidly cycles on and off Mad1–Mad2 heterodimers at unattached kinetochores, which contain Mad2 in a closed conformation. Mad2 that cycles off of unattached kinetochores then inhibits the APC in the context of its specificity factor Cdc20, and delays mitotic progression until all chromosomes are properly attached. (B) When Mad1 is expressed at levels equal to or greater than Mad2, Mad1-binding sites on kinetochores are saturated, and Mad1 localizes to additional, non-kinetochore sites. Mad2 binds to kinetochore- and non–kinetochore-bound Mad1 in the closed conformation, severely depleting the soluble pool of open Mad2. Without the soluble pool of Mad2 to be converted into APC-Cdc20 inhibitors at unattached kinetochores, APC-Cdc20 becomes active, resulting in premature anaphase onset and chromosome missegregation.
Multiple mitotic checkpoint proteins, including Bub1, BubR1, and CENP-E, have been shown to independently participate in chromosome congression (13, 55–58). However, we find that Mad1-overexpressing cells do not have defects in chromosome alignment or in kinetochore–microtubule attachments. This is consistent with overexpression of Mad1 weakening mitotic checkpoint signaling by titrating Mad2, as Mad2-depleted cells do not exhibit defects in congression, and missegregate chromosomes as a result of mitotic checkpoint defects coupled with a reduction in the duration of mitosis (13, 70, 77).
It has recently been shown that CIN cancer cell lines do not necessarily have weakened mitotic checkpoints (63, 66, 78). However, we show here that weakening of the mitotic checkpoint as a result of up-regulation of Mad1 does cause CIN. We also show a CIN cancer cell line with a mitotic checkpoint deficit. Thus, the status of the mitotic checkpoint in a given cell line cannot be predicted merely based on CIN, and must be determined empirically.
Mice expressing reduced levels of Mad1 develop spontaneous and carcinogen-induced tumors with increased frequency (10). The tumor microarray datasets with patient outcome we analyzed contained only tumor samples. Thus, we were unable to compare Mad1 levels in these tumors with Mad1 levels in control tissue. It is unclear whether patients with low Mad1 and increased survival express lower levels of Mad1 than healthy controls. However, mounting evidence suggests that overexpression of mitotic checkpoint components may be more detrimental, and more physiologically relevant, than underexpression. Multiple mitotic checkpoint components, including Mad1, Mad2, and Bub1, are frequently up-regulated in human cancers (43, 79, 80). Indeed, more than half of the breast cancers in our study expressed Mad1 protein at levels more than fivefold those in control breast samples (Fig. 1 A and B). Mouse models overexpressing the mitotic checkpoint components Mad2 (65) and Bub1 (80) have now been reported. Overexpressing Mad2 is significantly more tumorigenic than reducing its expression, and produces tumors with shorter latencies and higher penetrance (14, 65). Similarly, mice overexpressing Bub1 develop tumors with higher frequency and shorter latencies than mice with reduced levels of Bub1 (11, 80). Here we show that up-regulation of Mad1 is prevalent in human tumors with poor outcome and enhances transformation, aneuploidy, and CIN in culture. Together, this suggests that up-regulation of mitotic checkpoint components, rather than down-regulation, may be a more clinically relevant mechanism of promoting transformation and tumorigenesis.
Microtubule poisons are currently used to treat a host of cancers, including those of the lung, breast, ovaries, and testes. However, a substantial number of patients do not benefit from their use. Currently, there are no biomarkers available to predict patient response to these drugs. A clinically useful biomarker to predict response could significantly improve patient outcomes while simultaneously decreasing serious side effects and treatment costs. As excess Mad1 confers resistance to the clinically relevant microtubule poisons paclitaxel and vinblastine, up-regulation of Mad1 may have therapeutic relevance as a predictive biomarker.
Materials and Methods
Cell Culture.
DLD1 colorectal cancer cells were grown in DMEM supplemented with 10% (vol/vol) FBS, 2 mM L-glutamine, and 50 μg/mL penicillin/streptomycin at 37 °C and 5% CO2. FBS was selected to be low in tetracycline by lot testing. Human Mad1 and Mad1-YFP in the pcDNA/FRT/TO vector were integrated into TRex DLD1 cells, which contain a single FRT site and express the Tet repressor, by cotransfection with the Flp recombinase. Transfected cells were selected with 400 μg/mL hygromycin. Single clones were isolated with cloning cylinders. RFP-tagged histone H2B (H2B-RFP) was stably integrated into TRex DLD1 cells by transfection. Stable integrants were selected with 2 μg/mL puromycin. Single clones were isolated with cloning cylinders. Induction with 0.25 μg/mL tetracycline was sufficient to induce Mad1 and Mad1-YFP at levels equivalent to the median level of expression in primary human cancers. Unless otherwise specified, colcemid was used at 100 ng/mL, paclitaxel at 5 μM, vinblastine at 4 μg/mL, and tet at 0.25 μg/mL.
Chromosome Spreads.
Cells in 6-cm dishes were accumulated in mitosis by treatment with 100 ng/mL colcemid for 3 to 5 h. Cells in the media, as well as adherent cells, were collected by centrifugation and resuspended in 75 mM KCl for 9 min at room temperature. Three drops of freshly made fixative agent, consisting of methanol and acetic acid at a 3:1 ratio, were added to cells, which were then collected by centrifugation. Cells were resuspended in a residual volume by flicking. A total of 4 mL of fresh fixative agent was added dropwise while mixing cells after every 0.5 mL. Cells were washed again with fresh fixative agent twice more before being resuspended in ∼400 μL fixative agent. A total of 70 μL of cells were dropped onto precleaned microscope slides from a height of 2 ft. Slides were dried slowly in a fume hood for 10 s and then dried quickly on a hot plate at 74 °C for 30 s. Slides were stained with 1 μg/mL DAPI, washed with PBS solution, mounted using Vectashield, and sealed with nail polish. Chromosome spreads were imaged by using a 100×, 1.4-NA objective.
Immunofluorescence and Immunoblotting.
For immunofluorescence, cells were washed with MicroTubule Stabilizing Buffer [MTSB; 100 mM K-Pipes, pH 6.9, 30% (wt/vol) glycerol, 1 mM EGTA, 1 mM MgSO4] prewarmed to 37 °C, preextracted for 5 min with warm 0.5% Triton X-100 in MTSB at 37 °C, and fixed with warm 4% formaldehyde in MTSB. Cells were blocked in Triton Block [2.5% (vol/vol) FBS, 200 mM glycine, and 0.1% Triton X-100 in PBS solution] overnight at 4 °C. Antibodies were diluted in Triton Block. DNA was detected with DAPI, and cells were mounted with Vectashield. Staining was performed with antibodies to Mad1 (1:100 or newly generated, as detailed later; gift of A. Musacchio, IFOM-IEO, Milan, Italy), Mad2 (1:200) (81), CENP-E (Hpx; 1:200) (82), BubR1 (5F9; 1:200) (83), and Bub1 (1:200). YFP was visualized directly.
Images were acquired on a Nikon Ti-E inverted microscope using a CoolSNAPHQ2 camera driven by Nikon Elements software. Chromosome spread images are single z-planes acquired by using a 100× oil immersion objective (NA 1.4). All other images of tissue culture cells are from 0.2 μM z-stacks collected by using a 100× oil immersion objective (NA 1.4) and subsequently deconvolved by using the AQI 3D Deconvolution module in Elements. Unless otherwise specified, 2D maximum projections are shown. Maximum projections were assembled in Elements. Overlays were generated in Photoshop. Quantitative imaging was performed using identical exposure times on samples prepared identically and imaged in a single sitting. With the exception of the Mad1 and Mad2 quantification shown in Fig. 5A and SI Appendix, Figs. S1H and S3B, quantification was performed on 3D z-stacks by using the volume measurement tool in Elements. For analysis of Mad1 and Mad2, quantification was performed on maximum projections in areas identified as kinetochores by localization of BubR1 (for Mad2) or Bub1 (for Mad1). The fluorescence intensity of Mad1 and Mad2 at kinetochores was calculated by subtracting the average of the background signal in the four quadrants surrounding the DNA from the mean intensity of Mad1 or Mad2 at Bub1 or BubR1-positive kinetochores.
For immunoblotting, equal numbers of cells were resuspended in ELB lysis buffer (250 mM NaCl, 0.1% Nonidet P-40, 50 mM Hepes, pH 7, 5 mM EDTA) and 5× sample buffer. Proteins were separated by SDS/PAGE, transferred to nitrocellulose, and probed with antibodies at the same concentrations used for immunofluorescence.
Immunohistochemistry.
Five-micrometer sections of a formalin-fixed, paraffin-embedded tissue microarray (gift of A. Friedl, University of Wisconsin, Madison, WI) were subjected to antigen retrieval in citrate buffer, serum-blocked, and stained with rabbit anti-Mad1 antibody (described in the following paragraph), a mixture of e-cadherin and cytokeratin antibodies to identify epithelial cells (Dako), and DAPI overnight at 4 °C. Alexa Fluor-conjugated secondary antibodies were used. Images were acquired on a Nikon Ti-E inverted microscope by using a CoolSNAPHQ2 camera driven by Nikon Elements software. Images were acquired using identical exposure times in a single imaging session. z-stacks (0.2 μM) were collected by using a 40× dry objective (0.75 NA) and deconvolved by using the AQI 3D Deconvolution module in Elements. Maximum projections were quantified in Elements. DAPI signal was used to set a threshold defining a binary mask including the nucleus and perinuclear region to ensure inclusion of nuclear envelope staining. This binary mask was converted to a region of interest in which all Mad1 signal was quantified. Quantification was based on the modal pixel intensity in all DAPI-positive regions following a background correction. Background was considered to be the modal pixel value of the dimmest 0.1% of pixels. Values were exported to Microsoft Excel for further analysis. In Excel, the median value of Mad1 signal intensity in normal tissue samples (16 breast and one spleen) was determined and used to normalize each individual sample. Signal of individual samples (tumors and controls) is displayed as fold change relative to the median value of the normal samples.
Anti-Mad1 antiserum was generated by immunizing rabbits (Covance) with a GST fusion to amino acids 329 to 621 of human Mad1 expressed from a pGEX-KG vector (gift of T. Yen, Fox Chase Cancer Center, Philadelphia, PA). Polyclonal antibodies were affinity-purified from serum by binding to a column of Mad1 amino acids 329 to 621 following removal of the GST tag with thrombin.
Live Cell Microscopy.
For determination of mitotic index and cell death, Hoechst 33572 was added to media. After a 20-min incubation to permit uptake into cells, cells were imaged using a 10×, 0.13-NA and/or a 20×, 0.1-NA phase-contrast objective. Hoechst was imaged by using a DAPI filter.
To measure mitotic timing, cells were grown in CO2-independent media (Invitrogen) supplemented with 10% FBS, 2 mM L-glutamine, and 50 μg/mL penicillin/streptomycin and overlaid with sterile mineral oil. Images were acquired at 10-min intervals on a Nikon Ti-E inverted microscope with a heated chamber and an automated stage at 37 °C. Images were acquired using a 10×, 0.13-NA objective and perfect focus.
Chromosome segregation was observed in cells stably expressing histone H2B-RFP with or without GFP-tubulin grown in 35-mm dishes with glass bottoms in CO2-independent media (Invitrogen) supplemented with 10% FBS, 2 mM L-glutamine, and 50 μg/mL penicillin/streptomycin and overlaid with sterile mineral oil. Five z-planes and a single DIC image were acquired every 2 min by using a 60×, 1.4-NA oil-immersion objective. Maximum projections of in-focus planes were assembled in Elements, exported as jpg files, and converted to mov files in QuickTime with a play rate of 6 frames/s.
Soft Agar Assays.
Soft agar assays were performed by plating 1 mL of a 1:1 mix of 2× media and 1.2% agar per well of a 12-well plate. A total of 2.5 × 104 cells in a 1:1 mix of 2× media and 0.6% agar were layered on top of the bottom agar after it had solidified.
Microarray Data Processing.
Breast.
Bioinformatic analyses on the microarray data were performed using MultiExperiment Viewer 4.5.1. Tumor gene expression data were obtained from the National Center for Biotechnology Information GEO database. Analysis of dataset GSE4922 was performed to determine the aggressiveness of tumors. Expression data were log2-transformed and median-centered by samples (columns) and genes (rows), and the tumors were then ranked by Mad1 expression per tumor expressed as fold vs. median Mad1 expression across all tumors. Tumors expressing greater than or equal to twofold Mad1 expression greater than the median Mad1 expression across all tumors were classed as Mad1-high. Mad1-intermediate tumors express less than twofold higher or lower Mad1 expression compared with the median. Tumors expressing less or equal to twofold Mad1 expression were labeled Mad1-low. All samples from this dataset that included survival information were included in our analysis, for a total of 242 outcome-associated tumors. Kaplan–Meier plots were generated by using Prism, and curves were compared by using the log-rank test.
Colon.
Tumor gene expression data were obtained for GSE24549 from the National Center for Biotechnology Information GEO database. Expression data were log2-transformed and stratified into quartiles based on Mad1 expression. The survival of patients with the highest and lowest levels of Mad1 expression were compared. Kaplan–Meier plots were generated and compared by using MSTAT.
Supplementary Material
Acknowledgments
The authors thank Dr. A. Friedl, Dr. A. Musacchio, and Dr. T. Yen for sharing tumor microarray slides, Mad1 antibody, and a Mad1 plasmid, respectively. This work was supported in part by National Institutes of Health Grants 1R01CA140458 (to B.A.W.), GM088151 (to A.A.) and T32 GM008688 (E.M.C.B. and L.M.Z.) and American Cancer Society Grant IRG-58-011-48 (to B.A.W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 13148 (volume 109, number 33).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201911109/-/DCSupplemental.
References
- 1.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 3.Rieder CL. Mitosis in vertebrates: The G2/M and M/A transitions and their associated checkpoints. Chromosome Res. 2011;19:291–306. doi: 10.1007/s10577-010-9178-z. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 5.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 9.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 10.Iwanaga Y, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 11.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 15.Perera D, et al. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Poss KD, Nechiporuk A, Stringer KF, Lee C, Keating MT. Germ cell aneuploidy in zebrafish with mutations in the mitotic checkpoint gene mps1. Genes Dev. 2004;18:1527–1532. doi: 10.1101/gad.1182604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Putkey FR, et al. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 18.Rao CV, et al. Colonic tumorigenesis in BubR1+/-ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci USA. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren CD, et al. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol Biol Cell. 2002;13:3029–3041. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver BA, et al. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cahill DP, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 22.Jeong SJ, et al. Transcriptional abnormality of the hsMAD2 mitotic checkpoint gene is a potential link to hepatocellular carcinogenesis. Cancer Res. 2004;64:8666–8673. doi: 10.1158/0008-5472.CAN-03-3455. [DOI] [PubMed] [Google Scholar]
- 23.Ohshima K, et al. Mutation analysis of mitotic checkpoint genes (hBUB1 and hBUBR1) and microsatellite instability in adult T-cell leukemia/lymphoma. Cancer Lett. 2000;158:141–150. doi: 10.1016/s0304-3835(00)00512-7. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang B, Knauf JA, Ain K, Nacev B, Fagin JA. Mechanisms of aneuploidy in thyroid cancer cell lines and tissues: Evidence for mitotic checkpoint dysfunction without mutations in BUB1 and BUBR1. Clin Endocrinol (Oxf) 2002;56:341–350. doi: 10.1046/j.1365-2265.2002.01475.x. [DOI] [PubMed] [Google Scholar]
- 25.Percy MJ, et al. Expression and mutational analyses of the human MAD2L1 gene in breast cancer cells. Genes Chromosomes Cancer. 2000;29:356–362. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1044>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 26.Sato M, et al. Infrequent mutation of the hBUB1 and hBUBR1 genes in human lung cancer. Jpn J Cancer Res. 2000;91:504–509. doi: 10.1111/j.1349-7006.2000.tb00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigeishi H, et al. Expression of Bub1 gene correlates with tumor proliferating activity in human gastric carcinomas. Pathobiology. 2001;69:24–29. doi: 10.1159/000048754. [DOI] [PubMed] [Google Scholar]
- 28.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalitsis P, et al. Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer. 2005;44:29–36. doi: 10.1002/gcc.20215. [DOI] [PubMed] [Google Scholar]
- 30.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 31.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- 35.Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 37.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 38.Shah JV, et al. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 39.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sironi L, et al. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canman JC, et al. Anaphase onset does not require the microtubule-dependent depletion of kinetochore and centromere-binding proteins. J Cell Sci. 2002;115:3787–3795. doi: 10.1242/jcs.00057. [DOI] [PubMed] [Google Scholar]
- 43.Yuan B, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 44.Cheng AS, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turashvili G, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coe BP, et al. Gain of a region on 7p22.3, containing MAD1L1, is the most frequent event in small-cell lung cancer cell lines. Genes Chromosomes Cancer. 2006;45:11–19. doi: 10.1002/gcc.20260. [DOI] [PubMed] [Google Scholar]
- 47.de Leeuw RJ, et al. Comprehensive whole genome array CGH profiling of mantle cell lymphoma model genomes. Hum Mol Genet. 2004;13:1827–1837. doi: 10.1093/hmg/ddh195. [DOI] [PubMed] [Google Scholar]
- 48.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Carvalho B, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 50.Iwanaga Y, Jeang KT. Expression of mitotic spindle checkpoint protein hsMAD1 correlates with cellular proliferation and is activated by a gain-of-function p53 mutant. Cancer Res. 2002;62:2618–2624. [PubMed] [Google Scholar]
- 51.Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivshina AV, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 53.Sveen A, et al. Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Med. 2011;3:32. doi: 10.1186/gm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 56.Meraldi P, Sorger PK. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 58.Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2005;7:93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 59.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McEwen BF, et al. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabeche L, Compton DA. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr Biol. 2012;22:638–644. doi: 10.1016/j.cub.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung E, Chen RH. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol Biol Cell. 2002;13:1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 67.Yamada HY, Gorbsky GJ. Spindle checkpoint function and cellular sensitivity to antimitotic drugs. Mol Cancer Ther. 2006;5:2963–2969. doi: 10.1158/1535-7163.MCT-06-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kasai T, Iwanaga Y, Iha H, Jeang KT. Prevalent loss of mitotic spindle checkpoint in adult T-cell leukemia confers resistance to microtubule inhibitors. J Biol Chem. 2002;277:5187–5193. doi: 10.1074/jbc.M110295200. [DOI] [PubMed] [Google Scholar]
- 69.Sudo T, Nitta M, Saya H, Ueno NT. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 2004;64:2502–2508. doi: 10.1158/0008-5472.can-03-2013. [DOI] [PubMed] [Google Scholar]
- 70.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swanton C, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 72.Lee EA, et al. Inactivation of the mitotic checkpoint as a determinant of the efficacy of microtubule-targeted drugs in killing human cancer cells. Mol Cancer Ther. 2004;3:661–669. [PubMed] [Google Scholar]
- 73.Sihn CR, Suh EJ, Lee KH, Kim TY, Kim SH. p55CDC/hCDC20 mutant induces mitotic catastrophe by inhibiting the MAD2-dependent spindle checkpoint activity in tumor cells. Cancer Lett. 2003;201:203–210. doi: 10.1016/s0304-3835(03)00465-8. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: Model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kienitz A, Vogel C, Morales I, Müller R, Bastians H. Partial downregulation of MAD1 causes spindle checkpoint inactivation and aneuploidy, but does not confer resistance towards taxol. Oncogene. 2005;24:4301–4310. doi: 10.1038/sj.onc.1208589. [DOI] [PubMed] [Google Scholar]
- 77.Orr B, Bousbaa H, Sunkel CE. Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells. Mol Biol Cell. 2007;18:850–863. doi: 10.1091/mbc.E06-07-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 80.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kops GJ, et al. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown KD, Wood KW, Cleveland DW. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J Cell Sci. 1996;109:961–969. doi: 10.1242/jcs.109.5.961. [DOI] [PubMed] [Google Scholar]
- 83.Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci. 2001;114:4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]