Abstract
A central goal of chromatin biology is to reveal how posttranslational histone marks modulate gene expression; however, relatively little is known about the spatial or temporal dynamics of these marks. We previously showed that a dynamic model of histone mark nucleation, propagation, and turnover fits the mean enrichment profiles from 99% of noncentromeric histone H3 lysine 9 trimethylation (H3K9me3) domains in mouse embryonic stem cells without the need for boundary or insulator elements. Here we report the full details of this “inherently bounded” model of histone modification dynamics and describe several dynamic features of the model using H3K9me3 as a paradigm. By analyzing the kinetic and structural constraints that drive formation of inherently bounded domains, we find that such domains are optimized when the rates of marking and turnover are comparable. Additionally, we find that to establish such domains, propagation of the histone marks must occur primarily through local contacts.
Keywords: epigenetics, heterochromatin, methylation
Chromatin is a dynamic environment subject to spatial and temporal regulation through a number of factors. A central focus of this regulation is the posttranslational modification of histone proteins, whose modification states are associated with a variety of transcription states at a given genetic locus. Unfortunately, there are few quantitative models regarding the dynamics of histone marking and therefore little basis for describing the structure, stability, or fluctuations of histone modification domains.
One particular posttranslational histone modification, histone H3 lysine 9 trimethylation (H3K9me3), has served as a paradigm to study how histone marks are propagated in live cells to induce local silencing. In mammalian cells, H3K9me3 is associated with heterochromatic and transcriptionally silent regions (1–4). H3K9me3-based repression requires Heterochromatin Protein 1 (HP1), which mediates cis-spreading of the H3K9me3 mark by binding existing H3K9me3 sites (5–7), oligomerizing to bind neighboring nucleosomes (8, 9), and recruiting H3K9-specific histone methyltransferases (HMTs) (10–13) to induce outward spreading of the mark.
Unless opposed, this positive feedback would expand H3K9me3-containing domains without bounds. Several groups have described “insulator elements” in flies, yeast, and other organisms, which prevent further expansion of H3K9me3 marking (14). The presence of these insulator elements gives rise to distinct chromatin boundaries, which suggests chromatin may be packaged into modular, independent structural domains near these elements (15). Additionally, in fission yeast, the mating-type locus is silenced through expansion of H3K9me3 over an ∼20-kbp domain with relatively sharp borders (16, 17). The distinct structure of this histone modification domain, with a large plateau and definite borders, indicates that the expansion of H3K9me3 marks is blocked or subject to increased turnover at the border of the domain, consistent with a boundary element.
In mammals, however, such expansive H3K9me3 plateaus in coding regions are rare. In mouse embryonic stem cells, the vast majority of noncentromeric H3K9me3 domains span less than 10 kbp, with most less than 5 kbp (18, 19). These domains also typically show peaked patterns of enrichment, rather than broad, flat plateaus (20). We recently showed that small-molecule–mediated recruitment of HP1α to the Oct4 (Pou5f1) locus gives rise to a symmetric and peaked H3K9me3 domain in both mouse ES cells and fibroblasts (20). This domain grew continuously over time to between 2 and 10 kbp while maintaining its peaked shape, allowing us to obtain kinetic measurements on the dynamics of H3K9me3 propagation in vivo. Additionally, the enrichment profile and dynamics of this domain were consistent with those of H3K9me3 domains throughout the mouse genome. In our previous work, we showed that this synthetic H3K9me3 domain and >99% of all euchromatic H3K9me3 domains were well described by an “inherently bounded” model of histone modification dynamics (20), where propagation of the H3K9me3 mark was opposed by random turnover.
In this model, histone modification domains like H3K9me3 do not require insulator or boundary elements because the spreading of the mark is inherently constrained by mark and/or histone lifetime. Because the average enrichment profiles of >99% of mammalian H3K9me3 domains are well described by this model, we suggested that these domains are in fact inherently bounded and that our model provides quantitative predictions into the kinetics and dynamics of H3K9me3 marking throughout the genome. Here, we focus on the kinetic and structural constraints that lead to the characteristic pattern of peaked mark enrichment within the domains. We also report the full details of our dynamic model, using H3K9me3 as a paradigm.
Results
Reaction Scheme and Assumptions.
We model the dynamics of an individual genetic locus as a discrete one-dimensional lattice. We consider that each lattice position corresponds to an individual nucleosome, analogous to the “beads on a string” picture of chromatin. The lattice  spans from
spans from 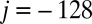 to
to  , which corresponds to 256 + 1 nucleosomes or 51.4 kbp of DNA if nucleosomal spacing is 200 bp. To make our model as general as possible, we consider that each position j along the lattice may occupy one of two states: an unmodified
, which corresponds to 256 + 1 nucleosomes or 51.4 kbp of DNA if nucleosomal spacing is 200 bp. To make our model as general as possible, we consider that each position j along the lattice may occupy one of two states: an unmodified 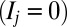 state or a modified
state or a modified 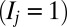 state. Briefly, three features define the standard model:
state. Briefly, three features define the standard model:
i) Nucleation: The origin (position 0) corresponds to a target site that is uniquely able to nucleate H3K9me3 modification. This nucleation occurs at rate
 .
.ii) Propagation: Our general kinetic scheme integrates the series of processes that propagate H3K9me3 into a single net propagation rate
 . In our kinetic model,
. In our kinetic model,  describes the net rate of H3K9me3 addition at nucleosomes immediately adjacent to H3K9me3-marked sites. In this way, spreading of H3K9me3 along the chromosome occurs through linear propagation of the mark to neighboring nucleosomes, consistent with proposed spreading via HP1 oligomerization (5, 6, 8, 21, 22). Mechanistically, this propagation likely requires multiple steps (e.g., recruitment of HP1, oligomerization, recruitment of HMT, enzymatic methylation, etc.); however, for simplicity, we treat propagation as subject to a single rate-limiting step with rate
describes the net rate of H3K9me3 addition at nucleosomes immediately adjacent to H3K9me3-marked sites. In this way, spreading of H3K9me3 along the chromosome occurs through linear propagation of the mark to neighboring nucleosomes, consistent with proposed spreading via HP1 oligomerization (5, 6, 8, 21, 22). Mechanistically, this propagation likely requires multiple steps (e.g., recruitment of HP1, oligomerization, recruitment of HMT, enzymatic methylation, etc.); however, for simplicity, we treat propagation as subject to a single rate-limiting step with rate 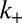 .
.iii) Turnover: Unlike propagation of the mark, turnover is equally likely everywhere, and thus
 describes the stochastic turnover of H3K9me3 at any marked nucleosome. Mechanisms of mark turnover include enzymatic catalysis (e.g., demethylation), histone turnover, or any other process that results in removal of the mark. For simplicity, we treat turnover as subject to a single rate-limiting step with rate
describes the stochastic turnover of H3K9me3 at any marked nucleosome. Mechanisms of mark turnover include enzymatic catalysis (e.g., demethylation), histone turnover, or any other process that results in removal of the mark. For simplicity, we treat turnover as subject to a single rate-limiting step with rate  .
.
We developed a simple probabilistic description of histone marking based on the kinetics of nucleation, propagation, and turnover. Monte Carlo simulations based on this model allowed us to determine the kinetic and structural constraints that promote formation of inherently bounded domains.
Implementation.
The stochastic trajectory of a given genetic locus is carried out by iteration of the following processes at each time step (Fig. 1):
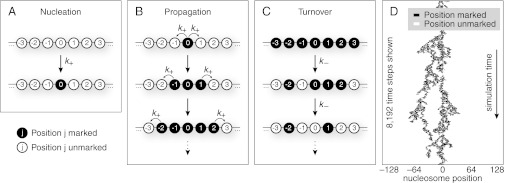
Fig. 1.
An inherently bounded histone modification domain arises from three processes. (A) A target site is marked at rate  , which nucleates the histone modification domain. (B) Neighboring unmarked nucleosomes are marked at rate
, which nucleates the histone modification domain. (B) Neighboring unmarked nucleosomes are marked at rate  to propagate the domain. (C) Random turnover occurs at rate
to propagate the domain. (C) Random turnover occurs at rate  and is equally likely everywhere. Turnover reverses the marks placed during nucleation and propagation. When all three processes occur simultaneously, an inherently bounded steady-state histone modification domain arises. (D) Simulation by iterating these three processes results in a stochastic, bounded steady-state modification domain. Shown here are 8,192 time steps from a simulation where the relative propagation rate
and is equally likely everywhere. Turnover reverses the marks placed during nucleation and propagation. When all three processes occur simultaneously, an inherently bounded steady-state histone modification domain arises. (D) Simulation by iterating these three processes results in a stochastic, bounded steady-state modification domain. Shown here are 8,192 time steps from a simulation where the relative propagation rate 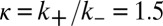 . In an individual simulation (analogous to an individual locus in a cell), the marking can transiently bifurcate to form multiple smaller domains. Accumulation of marks is limited by turnover.
. In an individual simulation (analogous to an individual locus in a cell), the marking can transiently bifurcate to form multiple smaller domains. Accumulation of marks is limited by turnover.
i) Nucleation: If the target site is unmarked
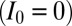 , it is converted to a marked state (
, it is converted to a marked state ( with probability
with probability 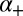 (where
(where  ).
).ii) Propagation: For each marked nucleosome j on the lattice, if the
 position is unmarked
position is unmarked  , it is converted to a marked state
, it is converted to a marked state 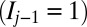 with probability
with probability  . Similarly, if the
. Similarly, if the  position is unmarked
position is unmarked  , it is converted to a marked state
, it is converted to a marked state 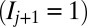 with probability
with probability  . Mark propagation thus proceeds outward in both directions from each marked site.
. Mark propagation thus proceeds outward in both directions from each marked site.iii) Turnover: For each marked nucleosome j on the lattice, conversion to an unmarked state
 occurs with probability
occurs with probability  (where
(where  ).
).iv) Time evolution: Simulation time t is incremented by
 .
.
The above procedure treats propagation and turnover as homogeneous Poisson processes with exponentially distributed lifetimes. We also allow all simulations to evolve using periodic boundary conditions; therefore, there is no boundary implicit in the simulations.
Constraints on the Establishment of an Inherently Bounded Domain.
When these processes are allowed to evolve, a stochastic domain of histone marks is established at the target site (Fig. 1D). To characterize which regions of the parameter space support the establishment of an inherently bounded H3K9me3 domain, we held  constant and varied
constant and varied  between
between  and
and 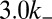 . We chose values of,
. We chose values of,  ,
,  , and
, and  such that
such that 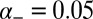 , and
, and  varied between 0.005 and 0.15. We found it convenient to define the ratio
varied between 0.005 and 0.15. We found it convenient to define the ratio
 |
to convey the relative rate of mark propagation and turnover. By allowing the model to evolve constrained by different values of κ, we discovered that an inherently bounded domain was established only when  (Fig. 2). At values of
(Fig. 2). At values of  , H3K9me3 marks spread without bounds (Fig. 2 D and H and Fig. S1). Here the simulations undergo a second-order phase transition (Fig. 3A) to a new state where marks occupy the entire locus.
, H3K9me3 marks spread without bounds (Fig. 2 D and H and Fig. S1). Here the simulations undergo a second-order phase transition (Fig. 3A) to a new state where marks occupy the entire locus.
Fig. 2.
The relative propagation rate κ determines the spatial extent of inherently bounded domains. (A–D) Snapshots from simulations obtained using different values of κ. Each row represents the state of an individual trajectory at a single time step. Black pixels indicate that the position is marked, and white pixels indicate the position is unmarked. When 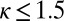 , a localized steady-state H3K9me3 domain arises that is inherently limited by mark and/or histone turnover and does not spread. At values of
, a localized steady-state H3K9me3 domain arises that is inherently limited by mark and/or histone turnover and does not spread. At values of 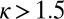 , marking spreads to occupy the entire simulated space. (E–H) For each value of κ, the mean mark density is plotted at each position (±SD in gray). For A–H, the value of κ is given above each plot. See also Fig. S1.
, marking spreads to occupy the entire simulated space. (E–H) For each value of κ, the mean mark density is plotted at each position (±SD in gray). For A–H, the value of κ is given above each plot. See also Fig. S1.
Fig. 3.
Structural features vary with the relative propagation rate. (A) As the relative propagation rate κ increases, the mean number of marks increases. Above  , the number of marks undergoes a second-order phase transition and marks occupy the entire locus. (B) The mean mark density at the target site monotonically increases with the relative propagation rate κ. (C) The specificity of marking S (defined in the main text) is sharply reduced when the relative propagation rate
, the number of marks undergoes a second-order phase transition and marks occupy the entire locus. (B) The mean mark density at the target site monotonically increases with the relative propagation rate κ. (C) The specificity of marking S (defined in the main text) is sharply reduced when the relative propagation rate  . (D) The efficiency score, which is a product of target-site mark density and specificity (see main text), shows a peak between
. (D) The efficiency score, which is a product of target-site mark density and specificity (see main text), shows a peak between  .
.
Intensity and Specificity of Marking.
Although the H3K9me3 domains established according to our model are, on the average, symmetric and centered around the target site, the relative propagation rate κ plays an important role in determining the overall mark density and spatial specificity of the domain. At low values of κ, the average domain is not very densely marked but is extremely specific, because the only marking takes place at or very near to the target site. As κ increases, the target site is more and more likely to be marked (Fig. 3B).
Conversely, the overall specificity of the marking decreases when κ increases, because the domain spreads outward from the target site. To quantify the effect of κ on the overall specificity of the domain, we introduce a specificity score S,
 |
where 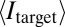 is the mean mark density at the target site, and
is the mean mark density at the target site, and 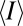 is the mean mark density over the entire lattice. Because the target site always has the maximum mean density throughout the lattice, this score returns a value of 0 when marking is perfectly uniform across the locus (no specificity) and returns a value approaching 1 when marking occurs exclusively at the target site (perfect specificity). When
is the mean mark density over the entire lattice. Because the target site always has the maximum mean density throughout the lattice, this score returns a value of 0 when marking is perfectly uniform across the locus (no specificity) and returns a value approaching 1 when marking occurs exclusively at the target site (perfect specificity). When  , the domain shows a high degree of specificity (Fig. 3C). The simulations undergo a phase transition for values of
, the domain shows a high degree of specificity (Fig. 3C). The simulations undergo a phase transition for values of  , into a new state that shows very poor specificity. In this state, H3K9me3 marks spread without bounds.
, into a new state that shows very poor specificity. In this state, H3K9me3 marks spread without bounds.
We next considered an “efficiency” measure E, which weighs the contribution of target site-specific histone mark density and specificity by multiplying these two factors together:
Conveniently, this measure also corresponds to the contrast between the mean mark density at the target site and the mean mark density over the entire locus. We found that marking efficiency is maximized when  (Fig. 3D). Thus, histone mark density and specificity are jointly optimized when the rates of mark propagation and turnover are coupled such that κ falls between 1.0 and 1.5.
(Fig. 3D). Thus, histone mark density and specificity are jointly optimized when the rates of mark propagation and turnover are coupled such that κ falls between 1.0 and 1.5.
Dynamics and Stability of Inherently Bounded Domains.
An important aspect of our model is that each simulation undergoes fluctuations in the number and distribution of marks throughout its trajectory. To characterize these fluctuations and to examine the stability of inherently bounded domains, we investigated several dynamic properties of the domains under varying values of κ. To focus on the dynamics of bounded domains, we restrict the remainder of our analysis to values of  . To address the role of κ on the dynamics of the domains, we examined how κ affected fluctuations in the total number of marks
. To address the role of κ on the dynamics of the domains, we examined how κ affected fluctuations in the total number of marks 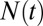 :
:
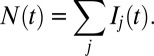 |
The total number of marks 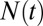 is not static, but constantly fluctuates about its mean value
is not static, but constantly fluctuates about its mean value  (Fig. 4A). We characterized the scale of domain size fluctuations by calculating the root mean-squared (rms) fluctuation of
(Fig. 4A). We characterized the scale of domain size fluctuations by calculating the root mean-squared (rms) fluctuation of  under different values of κ:
under different values of κ:
Fig. 4.
Dynamic features vary with the relative propagation rate. (A) The total number of marks 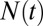 fluctuates throughout each individual simulation about the average value
fluctuates throughout each individual simulation about the average value  . (B) The absolute root mean-squared (rms) fluctuations in
. (B) The absolute root mean-squared (rms) fluctuations in  increase monotonically as κ increases. (C) When rms fluctuations are normalized by the average number of marks
increase monotonically as κ increases. (C) When rms fluctuations are normalized by the average number of marks  , the relative rms fluctuations decrease, indicating that higher values of κ lead to greater stability on a relative basis. (D) Autocorrelation of
, the relative rms fluctuations decrease, indicating that higher values of κ lead to greater stability on a relative basis. (D) Autocorrelation of  shows that domains with higher values of κ maintain correlation over longer times, providing stability and memory of the domain over a longer time.
shows that domains with higher values of κ maintain correlation over longer times, providing stability and memory of the domain over a longer time.
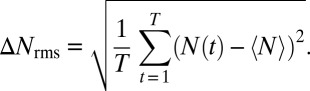 |
The absolute scale of these fluctuations increases as κ rises (Fig. 4B). However, when normalized by the mean number of marks 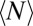 , these relative rms fluctuations instead decrease with increasing values of κ (Fig. 4C). Therefore, although an increased relative propagation rate κ induces larger fluctuations in absolute numbers of marks, these fluctuations are disproportionately smaller when normalized by the overall size of the domain.
, these relative rms fluctuations instead decrease with increasing values of κ (Fig. 4C). Therefore, although an increased relative propagation rate κ induces larger fluctuations in absolute numbers of marks, these fluctuations are disproportionately smaller when normalized by the overall size of the domain.
To examine the stability and memory of the domain, we calculated the autocorrelation function of  in trajectories with different values of κ. The autocorrelation function
in trajectories with different values of κ. The autocorrelation function  was calculated using the definition (23)
was calculated using the definition (23)
 |
where t is in units of time steps and T is the total number of time steps used for calculating autocorrelation. We found autocorrelation rose monotonically with increasing values of κ (Fig. 4D); thus, raising the relative propagation rate causes the domains to maintain correlation with their current states over a longer time, providing a degree of stability and memory. We conclude that increasing the relative propagation rate contributes to the stability of the domain by reducing the relative scale of the fluctuations, as well as contributes to memory of the domain over a longer time by increasing autocorrelation.
Model Extension: Varying Nucleation Rate.
An extension of the standard model above is to allow the nucleation rate to vary independently of the propagation rate (i.e., allowing nucleation to occur at rate  rather than
rather than  ). To examine this, we extended the standard model by replacing step i with alternative step ia.
). To examine this, we extended the standard model by replacing step i with alternative step ia.
ia) Nucleation: If the target site is unmarked
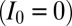 , it is converted to a marked state
, it is converted to a marked state  with probability
with probability 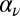 (where
(where  ).
).
Similar to κ, we defined  . By allowing κ and
. By allowing κ and  to vary independently, we found that bounded domains are established across a wide range of nucleation rates (Fig. S2). However, when mark nucleation is much slower than turnover (i.e., near
to vary independently, we found that bounded domains are established across a wide range of nucleation rates (Fig. S2). However, when mark nucleation is much slower than turnover (i.e., near 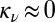 ), domains are not established with high efficiency. Additionally, because nucleation of the mark is a rate-limiting step for establishing a modification domain, the nucleation rate also controls the rate at which the domain is established de novo (Fig. S3). Thus, the primary effects of the nucleation rate are to control the marking density at the target site and the timescale over which inherently bounded domains may be established.
), domains are not established with high efficiency. Additionally, because nucleation of the mark is a rate-limiting step for establishing a modification domain, the nucleation rate also controls the rate at which the domain is established de novo (Fig. S3). Thus, the primary effects of the nucleation rate are to control the marking density at the target site and the timescale over which inherently bounded domains may be established.
Model Extension: Marking Beyond Nearest Neighbors.
Although marks may propagate only through nearest-neighbor contacts in the standard model above, several lines of experimental evidence suggest that discontinuous spreading may also occur (reviewed in ref. 24). To address whether inherently bounded domains may arise through marking at a distance, we have extended the standard model to allow one of two different modes of histone marking beyond nearest-neighbor contacts: a geometric dependence with respect to distance or a power-law dependence with respect to distance. We focus here on the geometric dependence, which may occur, for example, if HP1 oligomers behave as a simple one-stranded equilibrium polymer (25, 26) when extended beyond H3K9me3 marked sites and histone methyltransferases were recruited to the ends of HP1 oligomers. In our implementation, this dependence is treated by replacing step ii with alternative steps iia and iib.
iia) Propagation (left): For each marked nucleosome j on the lattice, integer distance
 is drawn from a geometric distribution with probability
is drawn from a geometric distribution with probability 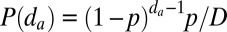 . If the
. If the  position is unmarked
position is unmarked  , it is converted to a marked state
, it is converted to a marked state  with probability
with probability  .
.iib) Propagation (right): Similarly, for each marked nucleosome j on the lattice, integer distance
 is drawn from a geometric distribution with probability
is drawn from a geometric distribution with probability  . If the
. If the  position is unmarked
position is unmarked 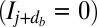 , it is converted to a marked state
, it is converted to a marked state  with probability
with probability  .
.
Note that in both iia and iib, the normalization constant
 |
normalizes probability such that the sum over the entire locus is unity. Here, p describes the probability of marking the nearest-neighbor site, and the probability of mark propagation decays geometrically with increasing distance d. In this model, there are two floating parameters, κ and p. To describe the stability of inherently bounded domains using these two parameters, we constructed heat maps of mean target-site mark density  , specificity S, and the efficiency score E as a function of both κ and p (Fig. 5).
, specificity S, and the efficiency score E as a function of both κ and p (Fig. 5).
Fig. 5.
Local marking increases the robustness of inherently bounded domains. (A) We extended our standard model of marking to include marking at a distance. The probability of marking follows a geometric dependence as described in the main text. Inherently bounded domains are formed most efficiently when marking occurs primarily at the nearest neighbor (near  ). As p is decreased, the efficiency of the domain is decreased and the rates that support domain formation are sharply reduced. (B) Average mark density of the target site depends on the relative propagation rate κ, but does not show a strong dependence on local marking. (C) Specificity of marking shows a strong dependence on local marking. As marking becomes more local (near
). As p is decreased, the efficiency of the domain is decreased and the rates that support domain formation are sharply reduced. (B) Average mark density of the target site depends on the relative propagation rate κ, but does not show a strong dependence on local marking. (C) Specificity of marking shows a strong dependence on local marking. As marking becomes more local (near 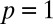 ), specificity is maintained over a broader range of κ. Because of this dependence, domains are established with high target-site mark density and specificity only when marking is primarily local.
), specificity is maintained over a broader range of κ. Because of this dependence, domains are established with high target-site mark density and specificity only when marking is primarily local.
Importantly, the efficiency score E shows that only certain regions of κ and p can support the formation of densely marked domains with high specificity (Fig. 5A). At one extreme, at the limit when 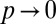 and marking is equally likely everywhere along the locus, inherently bound domains largely do not form under any values of κ because marking is too diffuse. At the other extreme, when
and marking is equally likely everywhere along the locus, inherently bound domains largely do not form under any values of κ because marking is too diffuse. At the other extreme, when  , the behavior of the system is identical to that described in the standard model above with propagation restricted only to nearest neighbors. Here as before, a specific and densely marked domain is established between
, the behavior of the system is identical to that described in the standard model above with propagation restricted only to nearest neighbors. Here as before, a specific and densely marked domain is established between  (Fig. 5A). Between these two extremes, we find that the values of κ that support formation of a densely marked domain with high specificity are gradually broadened as p is increased (i.e., as marking becomes more local).
(Fig. 5A). Between these two extremes, we find that the values of κ that support formation of a densely marked domain with high specificity are gradually broadened as p is increased (i.e., as marking becomes more local).
To sort out the effects of marking at a distance on target-site mark density and specificity, we separately examined the dependence of these parameters on κ and p. Target-site mark density was only weakly dependent on the probability of nearest-neighbor marking, but displayed a strong dependence on the relative propagation rate κ (Fig. 5B). On the other hand, specificity showed a mixed dependence on both κ and p (Fig. 5C): At low values of p, when marking is diffuse over the entire region, a high level of specificity was maintained up to  ; above this value specificity was sharply reduced. As p increased, specificity was maintained over a broader range of up to
; above this value specificity was sharply reduced. As p increased, specificity was maintained over a broader range of up to  as marks remained largely constrained to the narrow area around the target site. Thus, local marking positively contributes to the overall specificity of the domain. These results indicate that, although inherently bounded domains may form when marking is extended beyond nearest-neighbor histones, it is primarily local marking that drives formation of these domains.
as marks remained largely constrained to the narrow area around the target site. Thus, local marking positively contributes to the overall specificity of the domain. These results indicate that, although inherently bounded domains may form when marking is extended beyond nearest-neighbor histones, it is primarily local marking that drives formation of these domains.
Similar results were obtained when using a power-law dependence of marking at a distance (SI Text and Fig. S4). We conclude that, regardless of the mathematical details regarding the distance dependence of marking, inherently bounded domains are established primarily through local contacts. In particular, local marking contributes to the specificity of these domains by constraining histone marks to the vicinity of the target site. Nevertheless, our model provides a mechanism for the appearance of discontinuous marking. In an individual cell, discontinuities transiently arise even when propagation occurs exclusively through local contacts (Fig. 1D). Such discontinuities arise when turnover occurs in an area between two marked regions. Under these circumstances, the mark may continue to be propagated as two separate domains for some time. Additionally, in our simulations, “local” means nearest-neighbor nucleosomes; however, in chromatin, where physical proximity may be maintained through stable looping or other structures, marking may be transmitted across a loop, giving rise to the discontinuities previously described (24).
Discussion
Despite abundant experimental data regarding the genomic distributions of histone marks, there are surprisingly few physical models that predict the spatial distribution and dynamics of these marks. One earlier model of H3K9me3 marking at the yeast mating-type locus rationalizes how memory of the mark may be maintained over cell generations (27); however, the spatial distribution anticipated by that model is inconsistent with the genomic distribution of noncentromeric H3K9me3 marks in mammalian cells (20). We speculate that this discrepancy may reflect important differences in the way that H3K9me3 marks are propagated in yeast and mammals within nonrepetitive coding regions.
In our previous work, we showed that our standard model of inherently bounded histone marking—presented in detail here—is consistent with the local distribution of H3K9me3 marking at >99% of noncentromeric H3K9me3 domains throughout the genome in mouse embryonic stem cells (20). Additionally, when the accumulation of mark density predicted by the model is fitted to experimental results, we obtained specific values for  and
and  , both rates on the order of 0.1–0.2 h−1. The values of
, both rates on the order of 0.1–0.2 h−1. The values of  that we obtained in mouse embryonic fibroblasts and embryonic stem cells fall between two different experimental measures of histone turnover in live cells (20, 28, 29). On the basis of the reasonableness of these values, the degree of similarity in the shapes of H3K9me3 enrichment profiles, and the striking predictive power throughout the genome, we propose that most mammalian H3K9me3 domains arise through the general mechanisms described by our model. The simple processes of nucleation, local propagation, and histone turnover are sufficient to drive the formation of dynamic H3K9me3 domains that are peaked in their mean genomic profiles and are of the appropriate length scales. Importantly, our model predicts that inherently bounded domains have continuously decaying H3K9me3 profiles when averaged over an ensemble of cells, rather than a plateau with sharp edges that would arise through a well-positioned boundary or insulator element. Although boundary elements have been described in several specific instances (30), we propose that the majority of mammalian H3K9me3 domains may not require such elements because the propagation of the domains is inherently limited by the rate of mark and/or histone turnover. In this context, turnover of the mark may occur through a variety of mechanisms. We expect that enzymatic demethylation and displacement of core histones are the primary contributors to mark turnover, although the dynamics described by our model are general and consistent with these and other mechanisms.
that we obtained in mouse embryonic fibroblasts and embryonic stem cells fall between two different experimental measures of histone turnover in live cells (20, 28, 29). On the basis of the reasonableness of these values, the degree of similarity in the shapes of H3K9me3 enrichment profiles, and the striking predictive power throughout the genome, we propose that most mammalian H3K9me3 domains arise through the general mechanisms described by our model. The simple processes of nucleation, local propagation, and histone turnover are sufficient to drive the formation of dynamic H3K9me3 domains that are peaked in their mean genomic profiles and are of the appropriate length scales. Importantly, our model predicts that inherently bounded domains have continuously decaying H3K9me3 profiles when averaged over an ensemble of cells, rather than a plateau with sharp edges that would arise through a well-positioned boundary or insulator element. Although boundary elements have been described in several specific instances (30), we propose that the majority of mammalian H3K9me3 domains may not require such elements because the propagation of the domains is inherently limited by the rate of mark and/or histone turnover. In this context, turnover of the mark may occur through a variety of mechanisms. We expect that enzymatic demethylation and displacement of core histones are the primary contributors to mark turnover, although the dynamics described by our model are general and consistent with these and other mechanisms.
Multiple lines of experimental evidence suggest an ongoing competition between enzymatic methylation and demethylation throughout the genome. The H3K4me3 and H3K9me3 demethylase Lsd1 appears at both active and silenced genes and plays an important role during differentiation by decommissioning enhancer sites that have accumulated these marks (31). In addition, depletion of the H3K9-specific demethylases Jmjd1a and Jmjd2c leads to accumulation of H3K9me3 and differentiation of ES cells (32). Both of these results indicate that demethylases are continually opposing the continued placement of H3K9me3. Thus, H3K9me3 marks are governed not simply by the enzymatic placement of the mark, but instead by the continual steady-state balance between the placement and removal of the marks. Reduction of demethylase activity pushes the balance toward propagation of the mark, resulting in increased accumulation of H3K9me3. Importantly, this steady-state balancing act is integral to the maintenance of cell identity and pluripotency.
This balance is reflected in our model by the relative propagation rate κ. We find that when propagation of a histone mark is limited to local cis-spreading, formation of a robust, bounded steady-state histone modification domain is optimized when κ is between 1.0 and 1.5. In fact, this is the range of κ that also best fits experimental data from mouse embryonic stem cells and fibroblasts (20). This relatively narrow range suggests that such inherently bounded domains arise only when the rates of marking  and turnover
and turnover  are coupled. It should not be surprising that these processes have comparable rates: No histone modification domains would ever arise if turnover were much faster than marking, but on the other hand, the genome would quickly saturate with histone modifications if marking were much faster than turnover. Similarly in position-effect variegation of the Drosophila eye, the addition or loss of a single allele of a euchromatic or heterochromatic factor can shift the balance from a variegated phenotype to either a fully expressed or a silenced state at the white locus (33). This exquisite sensitivity indicates that heterochromatin dynamics are finely tuned to the dosage/activity balance of chromatin factors. On the basis of this argument, we speculate that the factors responsible for
are coupled. It should not be surprising that these processes have comparable rates: No histone modification domains would ever arise if turnover were much faster than marking, but on the other hand, the genome would quickly saturate with histone modifications if marking were much faster than turnover. Similarly in position-effect variegation of the Drosophila eye, the addition or loss of a single allele of a euchromatic or heterochromatic factor can shift the balance from a variegated phenotype to either a fully expressed or a silenced state at the white locus (33). This exquisite sensitivity indicates that heterochromatin dynamics are finely tuned to the dosage/activity balance of chromatin factors. On the basis of this argument, we speculate that the factors responsible for 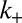 and
and  likely coevolved such that κ is maintained in this range.
likely coevolved such that κ is maintained in this range.
This general model abstracts over much of the complexity that is already known or that remains an outstanding question about H3K9me3 biology. In particular, our model does not address the mechanisms by which these marks are transmitted during or after replication, although we have previously shown that straightforward variations of the model can provide the means for epigenetic memory of the mark (20). Other potential mechanisms for epigenetic memory include factors that promote continued recruitment of marking factors to the target site, such as antisense RNA-based targeting of H3K9me3 silencing factors (34, 35).
Our model of inherently bounded histone modification domains presents a challenge for direct observation of the chromatin states in individual cells over time. In vivo, the accumulation and removal of marks like H3K9me3 are relatively slow (20), indicating that single-cell assays should require observations over long periods of time with temporal resolution on the order of hours. As single-cell chromatin assays mature, quantitative models like the one presented here will provide a biophysical framework for studying chromatin regulation.
Supplementary Material
Acknowledgments
We thank members of the G.R.C. laboratory, in particular O. Bell and N. Hathaway, for advice and helpful discussions. Research in the G.R.C. laboratory is supported by National Institutes of Health Grants HD55391 and NS046789 and the Howard Hughes Medical Institute. C.H. is supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development Fellowship F32HD072627.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211172109/-/DCSupplemental.
References
- 1.Peters AH, et al. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 2.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsui T, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 5.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 6.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 8.Canzio D, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verschure PJ, et al. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritsch L, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 12.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 13.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 15.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 16.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence RJ, Volpe TA. Msc1 links dynamic Swi6/HP1 binding to cell fate determination. Proc Natl Acad Sci USA. 2009;106:1163–1168. doi: 10.1073/pnas.0811161106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 22.Schotta G, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Box GEP, Jenkins GM, Reinsel GC. Time Series Analysis: Forecasting and Control. 3rd Ed. Upper Saddle River, NJ: Prentice-Hall; 1994. [Google Scholar]
- 24.Talbert PB, Henikoff S. Spreading of silent chromatin: Inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 25.Hill T. An Introduction to Statistical Thermodynamics. New York: Dover; 1960. [Google Scholar]
- 26.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 27.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 28.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zee BM, Levin RS, DiMaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess-Beusse B, et al. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whyte WA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert A, et al. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu SG, et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.