Abstract
Tripartite motif protein TRIM5α blocks retroviral replication after cell entry, and species-specific differences in its activity are determined by sequence variations within the C-terminal B30.2/PRYSPRY domain. Here we report a high-resolution structure of a TRIM5α PRYSPRY domain, the PRYSPRY of the rhesus monkey TRIM5α that potently restricts HIV infection, and identify features involved in its interaction with the HIV capsid. The extensive capsid-binding interface maps on the structurally divergent face of the protein formed by hypervariable loop segments, confirming that TRIM5α evolution is largely determined by its binding specificity. Interactions with the capsid are mediated by flexible variable loops via a mechanism that parallels antigen recognition by IgM antibodies, a similarity that may help explain some of the unusual functional properties of TRIM5α. Distinctive features of this pathogen-recognition interface, such as structural plasticity conferred by the mobile v1 segment and interaction with multiple epitopes, may allow restriction of divergent retroviruses and increase resistance to capsid mutations.
Retroviral restriction factors are important components of innate immunity defenses that protect higher organisms against retroviral pathogens. The splicing variant alpha of tripartite motif five (TRIM5α) is particularly remarkable because of the potent activity that the TRIM5α of rhesus monkey (rhTRIM5α) displays against HIV-1 (1). TRIM5α is a member of the tripartite motif (TRIM) family of proteins increasingly recognized for their role in innate immunity (2–4). All TRIM proteins share a conserved N-terminal tripartite domain motif consisting of a RING domain, followed by one or two B-box domains and then by a coiled-coil segment. The composition of the C-terminal part of TRIMs varies, and about one half of approximately 100 TRIM proteins in the human genome contain a C-terminal PRYSPRY domain (also known as B30.2 domain), a protein-protein interaction module (2, 3, 5).
Rhesus TRIM5α is a cytoplasmic protein that normally blocks HIV replication after cell entry but prior to completion of reverse transcription (1). Viral determinants of susceptibility to TRIM5α-mediated restriction are located within the capsid protein (6, 7), and the restriction potency correlates with the ability of the cytosolic TRIM5α to cosediment with the assembled viral capsid (8, 9), strongly suggesting that direct interactions of TRIM5α with the viral capsid are required for restriction. The PRYSPRY domain of TRIM5α is believed to form most of the capsid–TRIM5α interface as species-specific sequence variations within the PRYSPRY domain account for differences in the viral specificity of the TRIM5α-mediated restriction (10–12). In fact, the TRIM5α PRYSPRY domains contain some of the most rapidly changing protein segments within primate genomes, an illustration of how the evolutionary antagonism between retroviruses and their primate hosts accelerates remodeling of the host-pathogen interface (13). Most notably, recent evolution of the human TRIM5α PRYSPRY domain resulted in the variant that has poor affinity for the HIV capsid, the vulnerability that contributed to the AIDS pandemic when the simian immunodeficiency virus (SIV) passed from chimpanzees into a human host (1, 8, 9, 14).
TRIM5α binds to the assembled capsid of the mature viral core rather than the monomeric capsid protein, suggesting that TRIM5α may act as a pattern-recognition molecule (4, 8, 9). Remarkably, an EM investigation revealed that the purified tripartite motif of TRIM5α forms hexagonal arrays that match the symmetry of the assembled retroviral capsid (15, 16). This observation suggested a model of TRIM5α–capsid interaction, in which the hexagonal assembly of TRIM5α would juxtapose the PRYSPRY domains with the regularly spaced epitopes on the surface of the assembled capsid, leading to specific, high-affinity binding of TRIM5α to the retroviral core. Mutations that interfere with TRIM5α self-association also disrupt capsid cosedimentation confirming the importance of TRIM5α multimerization and the avidity effect in capsid recognition (15, 17–19). Such multivalent, high-avidity interactions pose significant experimental challenges. The binding of the individual PRYSPRY domains to the capsid surface may be very weak, which may be one of the reasons why direct PRYSPRY–capsid interactions have not yet been demonstrated by biochemical, biophysical, or structural means despite the extensive mutagenesis and evolutionary data suggesting a PRYSPRY–capsid interface.
The arrangement of the HIV capsid protein in the mature retroviral core is well-characterized, and the atomic-resolution model of the entire assembled structure is now available (20); in contrast, structures of the primate TRIM5α PRYSPRY domains have remained elusive, limiting our insight into capsid recognition by TRIM5α. Here we describe the structure of the rhesus TRIM5α PRYSPRY domain determined by a hybrid experimental approach that combines NMR spectroscopy and X-ray crystallography. The structure, NMR titration experiments, and site-directed mutagenesis suggest an extensive capsid–PRYSPRY interface dominated by the highly mobile v1 loop of the PRYSPRY domain. The capsid recognition mechanism, which is reminiscent of antigen recognition by the natural and the early immune response antibodies because it also involves mobile variable loops and high-avidity binding, may facilitate restriction of divergent retroviruses and increase resistance of TRIM5α to capsid mutations.
Results
Structure and Dynamics of the rhTRIM5α PRYSPRY.
Production of the recombinant TRIM5α PRYSPRY domain in bacterial expression systems is impeded by the low total protein expression and by the low soluble-to-insoluble protein ratio. Two features of our protein expression strategy were important for high yields of soluble recombinant protein: (i) codon optimization of TRIM5α sequences and protein expression in the BL21 Rosetta 2 (EMD/Novagen) bacterial strains and (ii) use of the N-terminal solubility enhancement tag derived from the B1 domain of protein G (GB1) (21). Detailed description of protein production and crystallization can be found in SI Experimental Procedures.
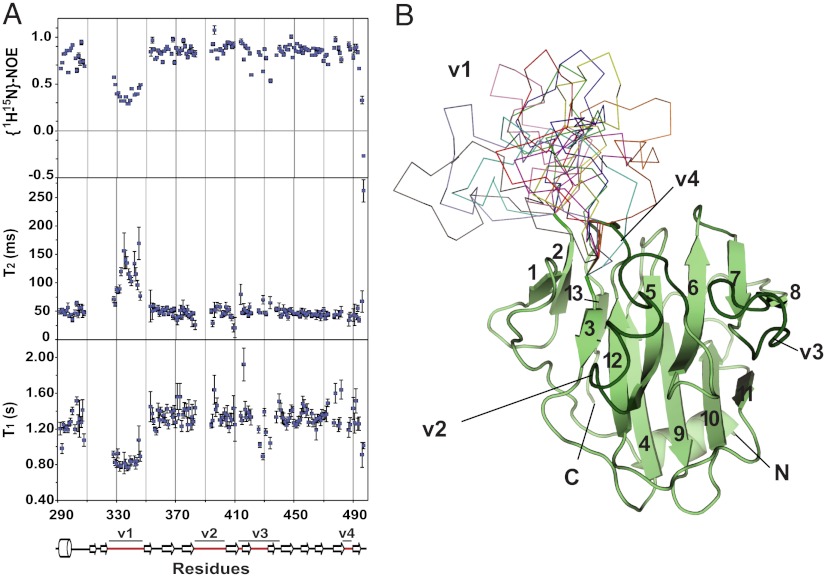
The wild type version of rhTRIM5α PRYSPRY was refractory to crystallization; therefore, we used NMR to identify mobile segments within PRYSPRY that could interfere with protein crystal packing. Measurement of the relaxation parameters of the protein backbone amides revealed that the v1 variable loop of the PRYSPRY domain was indeed highly mobile (Fig. 1A). Analysis of the relaxation data and modeling of the TRIM5α structural core guided production of a v1-deleted rhTRIM5α PRYSPRY construct (designated Δv1 elsewhere in the text). In this construct, the 24-residue v1 segment of the rhPRYSPRY (aa 326–349) was replaced by a two-amino-acid linker, Ala-Gly. The Δv1 PRYSPRY variant produced well-diffracting crystals at multiple crystallization conditions enabling determination of the crystal structure at 1.55 Å resolution. Crystallographic statistics are given in Table 1, and the unusual packing of the protein in the crystal is discussed in Experimental Procedures.
Fig. 1.
PRYSPRY structure and dynamics. (A) The 15N T1, T2 and {15N-1H} NOE measurements of the rhTRIM5α PRYSPRY. (B), The structure of the rhTRIM5α PRYSPRY domain. Ten lowest-energy conformations of the v1 loop calculated using NOE restraints are shown as colored ribbons. The rest of the structure determined by X-ray crystallography is shown as a light green cartoon. The strands forming the beta sandwich PRYSPRY fold are numbered, and the variable loop regions are labeled and highlighted in dark green.
Table 1.
Data collection and refinement statistics
| X-ray statistics of the ΔV1 rhSPRY structure (3UV9) | |
| Data collection | |
| Space group | R3 (hexagonal setting) |
| Cell dimensions | |
| a, b, c (Å) | 86.6, 86.6, 77.3 |
| α, β, γ (°) | 90, 90, 120 |
| Wavelength | 0.979 |
| Resolution (Å) | 30–1.55 |
| Rsym* | 0.044 (0.456) |
| I/σI | 31.4 (4.5) |
| Completeness (%) | 98.7 (98.4) |
| Redundancy | 6.4 (6.4) |
| Refinement | |
| Resolution (Å) | 26.9–1.55 |
| No. reflections | 29851 |
| Rwork/Rfree | 0.148/0.196 |
| No. Atoms | |
| Protein | 1462 |
| Solvent | 155 |
| B-factors | |
| Protein | 27.6 |
| Solvent | 39.5 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.949 |
| NMR statistics of the V1 loop structure(2LM3) | |
| Distance constraints for V1 loop | |
| Total number of NOE | 259 |
| Intraresidue | 103 |
| Sequential | 102 |
| Medium-range NOEs (1 < |i-j| < 4) | 49 |
| Long-range NOEs (|i-j| > 5) | 5 |
| Deviations from idealized covalent geometry | |
| Bonds (Å) | 0.0033 ± 0.0001 |
| Angles (°) | 0.5829 ± 0.080 |
| Impropers (°) | 0.4307 ± 0.0137 |
| Ramachandran statistics | |
| Most favored region (%) | 16.7 |
| Additionally allowed region (%) | 38.9 |
| Generously allowed region (%) | 33.3 |
| Disallowed region (%) | 11.1 |
*Values in parentheses are for the highest-resolution shell.
The v1 loop is critical for retroviral restriction. Numerous mutations within the loop are known to disrupt restriction, and conversely, mutations within the v1 of the human TRIM5α can restore HIV restriction with potency approaching that of the rhesus protein (12, 22, 23). Moreover, the composition of the v1 loop has been under strong positive selection during recent primate evolution (13). We analyzed the Nuclear Overhauser Effect (NOE) data from the 15N- and 13C-dispersed NOE–heteronuclear single quantum coherence (HSQC) experiments performed on the wild type version of rhTRIM5α PRYSPRY to calculate v1 conformations consistent with the observed NOE contacts. The pairwise interproton distance restraints obtained from the NOE data limit the number of conformations that the v1 region can possibly adopt. The resulting hybrid X-ray/NMR structure is shown in Fig. 1B. The v1 loop protrudes from the otherwise globular PRYSPRY domain and can adopt multiple divergent conformations without violating the NOE-derived restraints. These findings are consistent with the mobility of the v1 segment apparent from the relaxation measurements.
Structural Evolution of PRYSPRY Domains.
Comparison of the TRIM5α PRYSPRY structure to other known PRYSPRY structures offers insights into the molecular evolution of the primate TRIM5α and the expansion of PRYSPRY-containing TRIM proteins within the genomes of higher organisms. TRIM21 is the closest evolutionary cousin of TRIM5α for which the PRYSPRY structure is known, and the interactions with its binding partner, the invariant Fc segment of IgG, have been biochemically and structurally characterized (24, 25). Structural comparison of the PRYSPRY domains of TRIM5α and TRIM21 reveals that, whereas the core beta sandwich architecture and the packing of the hydrophobic core residues are essentially the same for the two proteins, their surfaces have diverged. The structural divergence between TRIM5α and TRIM21 PRYSPRY is primarily manifested in the distinct backbone conformations within the so-called variable loop regions v1 through v4, which display low sequence homology between different PRYSPRYs. Remarkably, the structural differences are clustered on one face of the protein (Fig. 2A). This structurally divergent protein face forms the binding interface in TRIM21,Gustavus, and SPSB1-4 proteins, the interactions of which with their binding partners have been structurally characterized (24, 26, 27). In primate TRIM5α, the residues that display strong positive selection in recent evolution as well as sequence insertion sites map to the same protein surface (10, 11, 13), offering strong evidence that the TRIM5α PRYSPRY uses the same interface for binding to the retroviral capsids (Fig. 2B). The observation that the structural divergence between closely related PRYSPRY domains is confined to the interaction interface illustrates that the binding specificity of the PRYSPRY domain has conferred enhanced fitness and shaped the evolution of primate TRIM5α, and probably of other PRYSPRY-containing TRIM proteins.
Fig. 2.
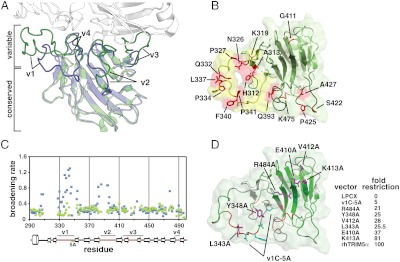
The capsid-binding surface of PRYSPRY. (A) rhTRIM5α PRYSPRY (green) superimposed on the structure of TRIM21 PRYSPRY (blue) in complex with the IgG Fc chain (white). Structurally divergent variable loops (v1 through v4) are highlighted and labeled. (B) The view of the predicted capsid-interacting surface of the rhTRIM5α PRYSPRY. Residues under positive evolutionary selection are colored red and labeled (13). The mobile surface formed by the v1 loop is colored yellow, whereas v2, v3, and v4 segments are highlighted in dark green. (C) The broadening rates of the 15N TROSY HSQC signals plotted for all assigned residues of the wild-type rhTRIM5α PRYSPRY (blue) and the v1C-5A PRYSPRY mutant (green). (D) The view of the capsid-binding surface illustrating the results of NMR titrations (Fig. 2C) and mutagenesis. Backbone segments perturbed by HIV CA-NTD titrations are colored in red, unaffected in green, and unassigned in gray. Residues tested by point mutagenesis are shown in magenta, whereas the segment changed to five alanines in the v1C-5A mutant is shown in cyan. HIV-1 restriction activity measurements for mutations located on the variable face of the rhTRIM5α PRYSPRY are listed in the table.
Mapping of the PRYSPRY–Capsid Interface.
To further characterize the surface involved in the PRYSPRY–capsid interactions we performed NMR titrations of the 15N-labeled rhPRYSPRY with the unlabeled N-terminal domain of the HIV capsid (CA-NTD). The NMR spectra revealed that the addition of CA-NTD to PRYSPRY produced broadening/weakening of the PRYSPRY NMR signals as a function of the CA-NTD concentration (SI Text). The rates of signal broadening were not uniform, but rather were most pronounced for the signals of the v1 and v2 segments and for the residues in the N-terminal segment preceding the v1 loop (Fig. 2C and D). The broadening of the NMR signals is indicative of the conformational changes in the v1 and v2 segments of the PRYSPRY structure upon CA-NTD binding. The fact that the broadening rather than peak shifting is observed is characteristic of the intermediate exchange regime on the NMR timescale, when the exchange rates between the bound and the unbound forms are comparable to frequency differences between NMR signals of the two states.
To test whether the PRYSPRY–capsid interaction observed by NMR is physiologically relevant we performed the same titration with the v1C-5A mutant of the rhesus PRYSPRY, which contains a five-alanine substitution of the NFNYC (aa 345–349) segment in the C-terminal portion of the mobile v1 loop. Point mutations of surface PRYSPRY residues result only in partial reduction of the restriction activity, therefore we chose the v1C-5A mutant because it is severely impaired in HIV restriction, but is nevertheless structurally intact (SI Text). The characteristic enhanced broadening of the NMR signals observed for the wild-type rhesus PRYSPRY is not present in v1C-5A mutant (Fig. 2C), suggesting that the PRYSPRY–capsid interaction observed by NMR correlates with the restriction activity of TRIM5α.
The binding between CA-NTD and PRYSPRY is very weak, because even when the two proteins are mixed at 200 μM each the binding is not close to saturation. By extending the NMR titration experiments to higher CA-NTD concentrations we can estimate that the dissociation constant of the PRYSPRY/CA-NTD binding is approximately 410 ± 90 μM (SI Text). This remarkably weak interaction of the isolated PRYSPRY domain with CA-NTD confirms the importance of the avidity effect in TRIM5α recognition of the mature viral cores. The fact that the binding occurs in the intermediate NMR exchange regime (see above) even for such weak interaction indicates that the kon rate for the binding is slow, consistent with the observation that the binding is accompanied by significant conformational changes.
We do not observe NMR signals for some backbone segments at the binding interface, raising the possibility that the residues not visible by NMR may still be important for capsid binding. Indeed, site-directed mutagenesis revealed that features of the structurally variable face of the PRYSPRY domain outside of the v1 and v2 loops are important for restriction (Fig. 2D). For example, alanine substitution of residue E410 in the v3 region, which is important for MLV restriction by the human TRIM5α (28), and the neighboring V412, displayed as much attenuation of HIV-1 restriction by rhTRIM5α as did the point mutations in the v1 loop. Mutation of R484 in the v4 loop reduced restriction potency from 100-fold to 21-fold. Thus, although the v1 and v2 loops undergo the most dramatic and observable conformational changes upon capsid binding they are not the sole determinants of HIV restriction on the variable face of TRIM5α PRYSPRY (Fig. 2D). Together these results suggest an extended interaction interface with multiple epitopes akin to the one observed in the TRIM21-IgG Fc complex.
Discussion
Here we describe the high-resolution structure of the rhesus TRIM5α PRYSPRY domain and map its interactions with the HIV capsid, offering insight into the structural basis of retroviral capsid binding. We find that capsid recognition by the PRYSPRY domain displays three distinctive features: (i) the capsid-binding interface of the PRYSPRY domain is located on the structurally divergent face of the protein formed by hypervariable loops designated v1 through v4, (ii) the binding surface is dominated by a mobile v1 segment that undergoes considerable structural rearrangements upon capsid binding , and (iii) the interaction of the isolated PRYSPRY domain with the capsid is weak and likely to involve multiple capsid epitopes.
The structural features of the TRIM5α PRYSPRY domain reveal striking similarities between capsid recognition by TRIM5α and antigen recognition by germ line antibodies. Germ line or near germ line amino acid sequences of the variable domains are frequently observed in the natural and early IgM antibodies (29), which are commonly polyreactive (capable of binding structurally unrelated epitopes), bind antigens relatively weakly, and display high-avidity binding owing to the presence of 10 antigen-binding sites in a single pentameric IgM macromolecule. These properties make IgM distinct from the affinity-matured IgG antibodies that bind their antigens with very high affinity and specificity. Structural and biophysical studies of antigen binding by the germ line versus affinity-matured antibodies revealed that the antigen-binding loops of the germ line antibodies and the antibodies of the early immune response are very mobile and undergo significant structural rearrangements upon antigen binding, whereas in the affinity-matured antibodies amino acid changes acquired via somatic hypermutation and clonal selection lock these loops in well-defined conformations and thereby transform antigen binding into a high-affinity, lock-and-key mode of interaction (30–32). The structural plasticity of the antigen binding surface explains the relatively low antigen affinity of germ line antibodies and is essential for their polyreactivity, a distinctive property viewed as critical for diverse biological functions of IgM (29).
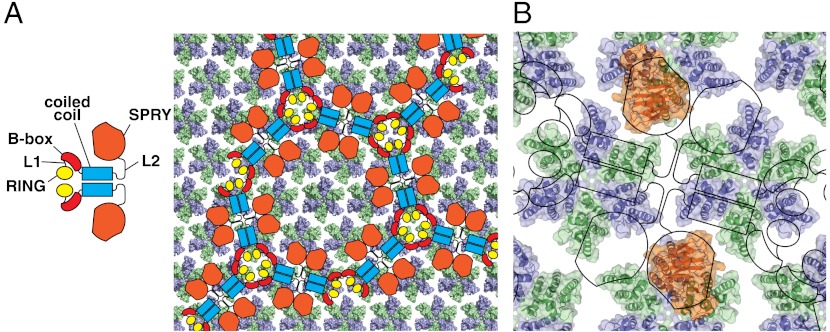
The long and mobile v1 loop that protrudes from the otherwise globular structure is a characteristic feature of the TRIM5α PRYSPRY. The mobility of v1 in TRIM5α and the associated loss of conformational entropy upon capsid binding almost certainly contribute to the weak affinity of the isolated PRYSPRY domain for the capsid. In contrast, structural and biophysical characterization of TRIM21-IgG interactions revealed that in the TRIM21 PRYSPRY the v1 loop is rather rigid, flanking an extensive preformed binding interface and resulting in a significantly tighter interaction with its target, the Fc chain of IgG (25). Such differences in v1 mobility resemble differences between germ line and affinity-matured antibodies and most likely reflect different biological functions of TRIM5α and TRIM21. TRIM21 is thought to function as a cytosolic IgG receptor (33) that binds to the structurally invariant Fc segment, whereas TRIM5α binds directly to a viral structural element; thus, the mobility of the v1 region may be beneficial in evolutionary terms because it would allow TRIM5α to adapt to mutations on the surface of the capsid. The relatively low affinity of the individual PRYSPRY modules for the capsid resulting from v1 mobility is compensated for by the avidity effect produced by higher-order multimerization of TRIM5α into hexagonal arrays that match the symmetry of the assembled retroviral capsids (15) (Fig. 3A).
Fig. 3.
Possible arrangement of the TRIM5α PRYSPRY on the surface of the HIV-1 mature core. (A) Domain structure of TRIM5α and one of the several possible relative orientations of the hexagonal arrays formed by TRIM5α and the HIV capsid (adapted from ref. (15)). HIV capsid assembly is derived from PDB ID codes 3DIK (47) and 3H47 (48). Only the N-terminal domains of the capsid that form the outer surface of the core are shown. Capsid C-terminal domains are omitted for clarity because they face the interior and are not accessible to interact with the PRYSPRY. Capsid monomers within the hexamers are shown in alternating colors (green and blue). (B) The structure of the rhTRIM5α PRYSPRY (orange) with the variable interaction face oriented toward the surface of the assembled capsid. The figure is only meant to illustrate the relative sizes of the PRYSPRY, CA-NTD, and the hexagonal arrays formed by the TRIM5α and the HIV capsid.
In a functional parallel to IgM polyreactivity, primate TRIM5α proteins are remarkable for their ability to restrict divergent retroviruses that share few similarities in the amino acid composition of their capsids. Rhesus TRIM5α PRYSPRY, for example, displays potent restriction activity against HIV-1, but is also moderately active against MLV (34). Conformational plasticity of the PRYSPRY domain is the most likely explanation of the broad specificity displayed by some TRIM5α variants, analogous to the mechanism of the IgM polyreactivity.
Similarities to IgM should be taken into consideration when using the high-resolution PRYSPRY structure reported here to interpret extensive mutagenesis data available for TRIM5α and to design future studies. Just as in the case of antigen–antibody interactions, TRIM5α mutations that affect binding are not necessarily altering direct contacts with the ligand, but rather may act by limiting the conformational space accessible to the v1 variable loop segment. For example, v1 dynamics may help explain how point mutations within the v1 of the human TRIM5α can alter its viral specificity profile or restore its activity against HIV (12, 22, 23, 35, 36).
It is informative to consider the relative sizes of the protein assemblies and binding interfaces involved in the TRIM5α–capsid interaction. Fig. 3 shows one possible arrangement of TRIM5α at the surface of the HIV mature core proposed in the EM study of TRIM5α hexagonal assemblies (15). We still lack the intermolecular PRYSPRY–capsid distance restraints that would allow identification of the PRYSPRY binding sites, so the placement of the PRYSRPY domains on the capsid surface is completely arbitrary and many different arrangements could be envisioned. The figure, however, illustrates two important points. First, the placement of the PRYSPRY domains is relatively sparse and TRIM5α uses only a small fraction of the capsid monomers in the core for the binding (Fig. 3A). Second, the variable face of PRYSPRY, oriented toward the capsid surface in Fig. 3B is larger than the solvent-exposed surface of the capsid N-terminal domain monomer and is likely to recognize epitopes spanning more than one capsid monomer within the hexamer. Such an extended interaction surface with multiple epitopes is consistent with numerous functional studies (28, 35, 37) and with the analysis of the MLV mutants that escape restriction by TRIM5α (38), which showed that point mutations that compromise restriction are located almost 30 Å apart on the capsid surface.
In summary, our structural and biophysical results reveal that the PRYSPRY domain of TRIM5α displays distinctive features of an immune recognition module that has evolved to bind pathogen-associated molecular patterns displayed on the surface of the retroviral capsid. Dynamics of the v1 segment located on the structurally variable capsid-binding surface of the domain and parallels to IgM–antigen interactions suggest that mutations that affect the mobility of this variable loop may have strong effects on the affinity and specificity of capsid binding. Conformational plasticity of the binding interface, weak capsid affinity of the isolated PRYSPRY domains and interaction with multiple epitopes are important factors to be considered in the studies of capsid recognition by TRIM5α.
Experimental Procedures
Protein Production and Purification.
Detailed description of protein production and crystallization can be found in SI Experimental Procedures.
X-ray Crystallography.
The structure of rhPRYSPRY was determined by the molecular replacement method implemented in PHASER (39), using Protein Data Bank (PDB) ID code 2WL1 (40) as the search model. Coordinates for the model were refined using PHENIX (41) including simulated annealing with torsion angle dynamics and individual anisotropic displacement parameter refinement, and alternated with manual rebuilding using COOT (42). Data collection and refinement statistics are shown in Table 1.
TRIM5α Δv1 PRYSPRY is assembled into a domain-swapped trimer in the crystal (SI Text). The N-terminal segments (aa 292–325) are cyclically swapped in the monomers within the trimer. Apparently, the v1 deletion and the high protein concentration at crystallization conditions lead to the formation of the domain-swapped trimer, a stabilized structure that promoted crystal growth. Oligomerization by swapping of substructural elements can be functionally significant, especially for proteins occurring in the intrinsically crowded multicopy environments such as viral shells, protein filaments, flagella, etc. (43, 44), but many domain-swapped oligomers observed in crystals are simply crystallization peculiarities not linked to the function of the native proteins. We believe the latter to be the case for the Δv1 PRYSPRY crystals, as the wild type and the Δv1 PRYSPRY constructs are predominantly monomeric in our NMR samples, and the observed domain-swapped trimerization obstructs protein surfaces expected to contact the viral capsid. The coordinate file of the unswapped rhesus Δv1 PRYSPRY monomer with the B factors preserved is available at http://ivanovlab.uthscsa.edu/trim5alpha/. The structure of the monomer is not perturbed by domain-swapped oligomerization with the exception of the short linker region connecting the swapped element to the rest of the structure (in our case the Ala-Gly linker used to substitute the v1 loop) (SI Text).
NMR Spectroscopy.
The 15N T1 and T2 relaxation parameters and [1H-15N]-heteronuclear NOE values were obtained from standard experiments (45) performed at 298 K on a 700 MHz spectrometer using an 15N-labeled approximately 0.3 mM rhPRYSPRY sample.
NOE-based distance restraints for the variable loop v1 were extracted from 700-MHz three-dimensional NOESY-[1H,15N]-HSQC and 700-MHz three-dimensional NOESY-[1H,13C]-HSQC spectra. No long-range NOEs were observed between the v1 and the protein outside of v1. The final structures of the rhPRYSPRY were calculated using XPLOR-NIH. Simulated annealing with NOE-derived distance restraints was applied to the variable loop v1 only (residues 325–349) whereas positions of the rest of the atoms in protein were held fixed at the coordinates determined by X-ray crystallography. Structural statistics shown listed in Table 1 are typical of a disordered loop region in a protein structure determined by NMR spectroscopy.
NMR titration of 15N-labeled rhPRYSPRY ( approximately 0.19 mM) with unlabeled CA-NTD was conducted at 298 K on a Bruker Avance 700 MHz spectrometer by recording a series of 1H-15N Transverse Relaxation Optimized Spectroscopy, TROSY-HSQC spectra containing molar ratios from 1∶0 up to 1∶2.5. For each cross-peak the intensity was evaluated using NMRView and plotted as a function of the protein concentration ratio. Broadening rates R for each peak were determined by fitting signal intensity plots above with simple exponential decays I(c) = Ioe-Rc, where c is the protein concentration ratio (CA-NTD/PRYSPRY).
The Kd value of the interaction was estimated from a similar titration but extended to higher CA-NTD concentrations (1∶10 [PRYSPRY]/[CA-NTD] ratio) (SI Text). The details of the fitting procedure are described in SI Experimental Procedures.
All NMR parameters determined for each residue and plotted in Fig. 1A and Fig. 2C are listed in SI Text.
Restriction Assays.
Retroviral vectors encoding wild-type or mutant rhTRIM5α proteins were created using the pLPCX vector, as previously described (46). Cf2Th canine thymocytes were transduced with the different constructs and selected in 5 μg/mL of puromycin (Sigma). Recombinant HIV-1 expressing GFP was prepared as described (35). All recombinant viruses were pseudotyped with the vesicular stomatitis virus G glycoprotein. For infections, 3 × 104 Cf2Th cells seeded in 24-well plates were incubated at 37 °C with virus for 24 h. Cells were washed and returned to culture for 48 h, and then subjected to FACS analysis with a FACScan (Becton Dickinson). HIV-1 viral stocks were titrated by serial dilution on Cf2Th cells to determine the concentration of infectious viruses.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Oleg Tsodikov and the staff of Life Sciences Collaborative Access Team (LS-CAT) sector at the Advanced Photon Source at the Argonne National Laboratory for assistance with collection of the X-ray diffraction data. Support for the NMR Core Facility and the X-ray Crystallography Core Laboratory is provided by University of Texas Health Science Center Executive Research Committee and the Cancer Therapy Research Center (P30 Cancer Center Support Grant from the National Cancer Institute CA054174). This research was supported in part by National Institutes of Health (NIH) Grants R21 AI068548 (to D.I.), R21 AI084612 (to D.I.), R01 AI087390 (to F.D.-G.), and the Robert A. Welch Foundation grant AQ-1399 (to P.J.H.). The Scholar Award from the Cancer Prevention and Research Institute of Texas (CPRIT) (D.I.) and the NIH K99/R00 Pathway to Independence Award (F.D.-G.) are gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structures factors have been deposited in the Protein Data Bank (PDB), www.pdb.org (PDB ID codes 3UV9 and 2LM3). The chemical shift data have been deposited in the Biological Magnetic Resonance Data Bank (BMRB code: 18097).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203536109/-/DCSupplemental.
References
- 1.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Ozato K, Shin DM, Chang TH, Morse HC., III TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23:46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011;7:e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owens CM, Yang PC, Gottlinger H, Sodroski J. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowan S, et al. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song BW, et al. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits line age-specific length and sequence variation in primates. J Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 15.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci USA. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz-Griffero F. Caging the beast: TRIM5alpha binding to the HIV-1 core. Viruses. 2011;3:423–428. doi: 10.3390/v3050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz-Griffero F, et al. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J Virol. 2007;81:10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz-Griffero F, et al. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J Virol. 2009;83:10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P, Lugovskoy AA, Wagner G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR. 2001;20:11–14. doi: 10.1023/a:1011258906244. [DOI] [PubMed] [Google Scholar]
- 22.Pham QT, Bouchard A, Grutter MG, Berthoux L. Generation of human TRIM5alpha mutants with high HIV-1 restriction activity. Gene Ther. 2010;17:859–871. doi: 10.1038/gt.2010.40. [DOI] [PubMed] [Google Scholar]
- 23.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human TRIM5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 24.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci USA. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filippakopoulos P, et al. Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4. J Mol Biol. 2010;401:389–402. doi: 10.1016/j.jmb.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo JS, Suh HY, Park SY, Oh BH. Structural basis for protein recognition by B30.2/SPRY domains. Mol Cell. 2006;24:967–976. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Perron MJ, Stremlau M, Sodroski J. Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J Virol. 2006;80:5631–5636. doi: 10.1128/JVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Notkins AL. Polyreactivity of antibody molecules. Trends Immunol. 2004;25:174–179. doi: 10.1016/j.it.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 31.Yin J, Beuscher AEt, Andryski SE, Stevens RC, Schultz PG. Structural plasticity and the evolution of antibody affinity and specificity. J Mol Biol. 2003;330:651–656. doi: 10.1016/s0022-2836(03)00631-4. [DOI] [PubMed] [Google Scholar]
- 32.Manivel V, Sahoo NC, Salunke DM, Rao KV. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity. 2000;13:611–620. doi: 10.1016/s1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 33.Mallery DL, et al. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci USA. 2010;107:19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perron MJ, et al. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz-Griffero F, et al. A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology. 2008;378:233–242. doi: 10.1016/j.virol.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maillard PV, Reynard S, Serhan F, Turelli P, Trono D. Interfering residues narrow the spectrum of MLV restriction by human TRIM5alpha. PLoS Pathog. 2007;3:e200. doi: 10.1371/journal.ppat.0030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diaz-Griffero F, et al. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 2007;369:400–410. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohkura S, et al. Novel escape mutants suggest an extensive TRIM5alpha binding site spanning the entire outer surface of the murine leukemia virus capsid protein. PLoS Pathog. 2011;7:e1002011. doi: 10.1371/journal.ppat.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinert C, Grutter C, Roschitzki-Voser H, Mittl PR, Grutter MG. The crystal structure of human pyrin b30.2 domain: Implications for mutations associated with familial Mediterranean fever. J Mol Biol. 2009;394:226–236. doi: 10.1016/j.jmb.2009.08.059. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Gronenborn AM. Protein acrobatics in pairs—dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remaut H, Waksman G. Protein-protein interaction through beta-strand addition. Trends Biochem Sci. 2006;31:436–444. doi: 10.1016/j.tibs.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Palmer AG., III NMR probes of molecular dynamics: Overview and comparison with other techniques. Annu Rev Biophys Biomol Struct. 2001;30:129–155. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- 46.Lienlaf M, et al. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): Structure of the RING domain of TRIM5alpha. J Virol. 2011;85:8725–8737. doi: 10.1128/JVI.00497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 48.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.