Abstract
How different organs are formed from small sets of undifferentiated precursor cells is a key question in developmental biology. To understand the molecular mechanisms underlying organ specification in plants, we studied the function of the homeotic selector genes APETALA3 (AP3) and PISTILLATA (PI), which control the formation of petals and stamens during Arabidopsis flower development. To this end, we characterized the activities of the transcription factors that AP3 and PI encode throughout flower development by using perturbation assays as well as transcript profiling and genomewide localization studies, in combination with a floral induction system that allows a stage-specific analysis of flower development by genomic technologies. We discovered considerable spatial and temporal differences in the requirement for AP3/PI activity during flower formation and show that they control different sets of genes at distinct phases of flower development. The genomewide identification of target genes revealed that AP3/PI act as bifunctional transcription factors: they activate genes involved in the control of numerous developmental processes required for organogenesis and repress key regulators of carpel formation. Our results imply considerable changes in the composition and topology of the gene network controlled by AP3/PI during the course of flower development. We discuss our results in light of a model for the mechanism underlying sex-determination in seed plants, in which AP3/PI orthologues might act as a switch between the activation of male and the repression of female development.
Flowers are typically composed of four organ types, which are disposed in four floral whorls. From the outside of the flower to the center, they are sepals, petals, stamens, and carpels (the subunits of the gynoecium). The developmental fate of these different types of organs is specified by a small number of floral organ identity genes. The pivotal role of these genes was uncovered through the analysis of mutants that form flowers with homeotic transformations, i.e., the replacement of one type of organ with another (1–4). Based on the morphological defects of the individual mutants and their genetic interactions, it was proposed that the floral organ identity genes act in a combinatorial manner and have distinct functions during flower development, with the so-called A function genes being required for the formation of sepals and petals, B function genes for petal and stamen development, and C function genes for the formation of stamens and carpels. This well-established ABC model of floral organ identity specification (5) has provided, since its introduction more than 20 y ago, an invaluable framework for the analysis of the genetic mechanisms underlying the formation and evolution of flowers.
Molecular characterization of the floral organ identity genes in different species revealed that they encode transcription factors and belong, with few exceptions, to the family of MADS domain proteins (1–3). The floral organ identity factors were shown to form higher-order complexes together with flower-specific cofactors, which are also MADS domain transcription factors (6, 7). Further insights into the molecular functions of these regulators have come from the recent identification of some of their target genes and interacting proteins (7–11). Despite this progress, our understanding of the developmental mechanisms that mediate the specification and formation of floral organs is still vastly incomplete.
To gain detailed insights into the processes controlled by floral organ identity factors during morphogenesis, we analyzed the activities of the B function regulators APETALA3 (AP3) and PISTILLATA (PI) from Arabidopsis throughout flower development. AP3 and PI are closely related MADS domain proteins that are thought to act as obligate heterodimers (12–14). Only few direct target genes of AP3/PI have been described, which include the A function gene APETALA1 (AP1) (15), NAP (NAC-like, activated by AP3/PI) (16), two GATA transcription factors (17), and three related BANQUO genes (18), which also encode transcriptional regulators. To unravel the gene network underlying B function on a global scale, we used gene perturbation assays, transcript profiling, and genomewide localization studies, in combination with a floral induction system, which allows the analysis of regulatory processes during early flower development by genomic technologies. We supplemented these molecular data with a morphological analysis of the spatial and temporal requirement for AP3/PI activity during flower development. Taken together, our results provide a molecular framework for the control of organ specification by B function.
Results
Genes and Processes Controlled by AP3/PI.
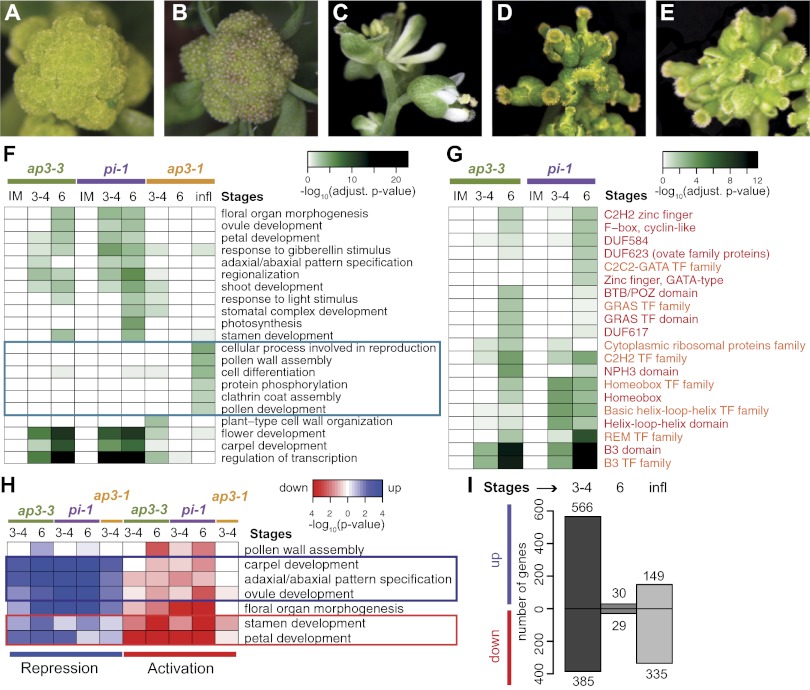
To unravel the transcriptional program underlying B function, we identified genes whose expression depends on AP3/PI activity during early flower development. To this end, we used a floral induction system, which allows the collection of a large number of flowers that are at approximately the same developmental stage (19). Morphological and molecular analyses have shown that flowers produced by this system can serve as a model for the early phase of flower development (19). To perturb B function activity, we introduced the null mutant alleles ap3-3 and pi-1, respectively, into the floral induction system. Activation of flower development in these genetic backgrounds led to the formation of flowers with petal-to-sepal and stamen-to-carpel transformations (Fig. 1 A–E), as expected for B function mutants. We collected mutant flowers at different stages of early flower development (stages according to ref. 20) and compared their gene expression profiles, by whole-genome microarray analysis, with those of corresponding flowers in which B function was not affected. As expected, the number of differentially expressed genes identified in these experiments increased with progressing flower development due to accumulative effects of B function perturbation (SI Appendix, Fig. S1). A search for overrepresented Gene Ontology (GO) terms in the 2,100 differentially expressed genes (Dataset S1) revealed numerous enriched functional categories (Fig. 1F and SI Appendix, Fig. S2). These included several implicated in organogenesis and in particular, in petal and stamen development, in agreement with the known role of the B function genes during flower formation. Additional GO terms that were enriched among the differentially expressed genes included, for example, genes involved in pattern formation, cellular differentiation, and in the response to the phytohormone gibberellin, which has important functions during floral organ development (21). Genes involved in the regulation of transcription were also highly enriched in the dataset. In fact, a gene family enrichment analysis showed that transcription factor families with known roles in flower development were strongly overrepresented among the differentially expressed genes (Fig. 1G). We further found regulators of carpel development to be enriched in the dataset. Notably, a test for coordinated directionality of expression changes within the enriched GO terms revealed that these genes were predominantly up-regulated in very young (i.e., stage ∼3–4) flowers of ap3 and pi mutants (Fig. 1H and SI Appendix, Fig. S2). Because this response occurred shortly after the onset of AP3/PI expression at stage 3 (13, 14), it suggested that these carpel developmental genes are normally repressed by the B function regulators during early flower formation. In agreement with this conjecture is the observation that the gene CRABS CLAW (CRC), which responded strongly in the microarray experiments, is precociously expressed in young floral buds of B function mutants, with its expression expanding from the fourth into the third floral whorl (22) (SI Appendix, Fig. S3).
Fig. 1.
Genes and processes controlled by AP3/PI. (A–E) Floral induction system based on the expression of a fusion between AP1 and the hormone-binding domain of the glucocorticoid receptor in ap1 cauliflower double-mutant plants (19). Inflorescences of a control plant (A) and a plant 6 d after induction of flower formation (B). Mature flowers generated by the floral induction system (C) and after introgression of the ap3-3 (D) and pi-1 (E) alleles. (F) Selected GO terms enriched among genes differentially expressed in flowers of B function mutants. Terms specifically identified in late flower development are marked by a blue box. Benjamini–Hochberg-adjusted P values are shown. A comprehensive view of GO terms is presented in SI Appendix, Fig. S2 and Dataset S3. (G) Gene families and protein domains identified as significantly enriched (or in one case, that of F-box, cyclin-like, as under-represented) in the datasets stemming from the ap3-3 and pi-1 microarray experiments. Benjamini–Hochberg-adjusted P values are shown. Gene families and protein domains are labeled in orange and red, respectively. (TF, transcription factor.) (H) Directionality of gene expression changes within selected GO terms. Terms containing genes predominantly repressed or activated by AP3/PI are marked with blue and red boxes, respectively. A comprehensive view of GO terms is presented in SI Appendix, Fig. S2. (I) Number of genes identified as up- and down-regulated in the ap3-1 microarray experiments.
Functions of AP3/PI in Early- and Late-Stage Flowers.
Expression of floral organ identity genes commences during early floral stages and continues for most of floral organ development (3). This prolonged expression implies that these genes are not only involved in the specification of organ primordia but also in the control of organ maturation during later developmental stages (10, 23). To gain insights into the molecular basis of these different activities, we identified genes whose expression depends on the B function regulators at distinct stages of flower development. To this end, we conducted conditional gene perturbation experiments by introducing the temperature-sensitive AP3 mutant allele ap3-1 (4) into the floral induction system. We found that inactivation of AP3 through a shift from permissive to nonpermissive temperatures led to a strong transcriptional response in young floral buds (stage ∼3–4), whereas, in slightly older buds (stage ∼6), only minor effects on gene expression occurred (Fig. 1I and SI Appendix, Fig. S2). In whole inflorescences of ap3-1 mutant plants, representing flowers predominantly of intermediate and late developmental stages (24), we again detected widespread transcriptional changes after a perturbation of AP3 activity. A GO analysis of the resulting datasets revealed considerable differences between gene groups identified as enriched at early and late stages of development (Fig. 1F and SI Appendix, Fig. S2). Furthermore, genes responding early showed, as expected, a significant overlap (P < 0.0001, Pearson χ2 test) with genes identified as differentially expressed in the null mutant experiments, whereas genes identified at late stages did not. The latter genes included many with known roles in microsporogenesis (i.e., pollen formation; Fig. 1F and SI Appendix, Fig. S2), a process that commences at intermediate stages of flower development and is completed shortly before anthesis. These results together provide molecular evidence for an involvement of AP3/PI in the control of different processes during petal and stamen organogenesis and indicate that they regulate largely unique sets of genes at distinct phases of flower development.
Spatial and Temporal Requirement for B Function.
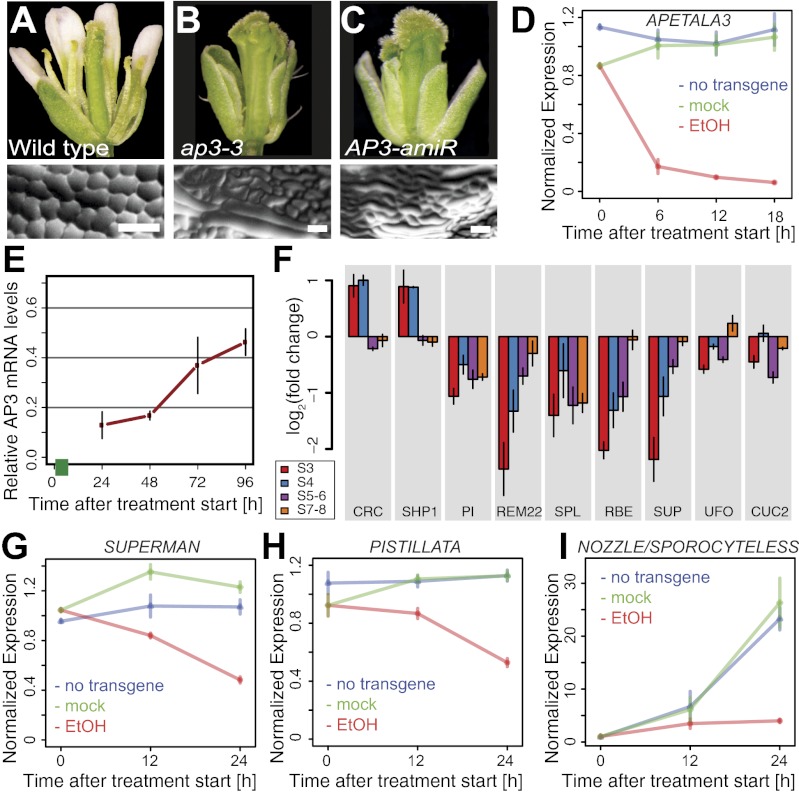
We next investigated whether the strong reduction in the number of differentially expressed genes detected in the ap3-1 microarray experiments around stage 6 was indicative of a temporarily reduced role of the B function regulators in flower development. To this end, we generated functional artificial microRNAs (amiRNAs) (25) against AP3 and PI (Fig. 2 and SI Appendix, Fig. S4 and Table S1). We then expressed these amiRNAs from an ethanol-inducible promoter system (denoted 35S:AlcR/pAlcA) (26) in WT plants to allow a controlled perturbation of AP3 and PI function. A detailed characterization of the AP3 amiRNA line showed that a pulsed induction of amiRNA expression led to a prompt and substantial (Fig. 2D), but time-limited (Fig. 2E), decrease of AP3 mRNA levels, followed by a marked reduction in the levels of the corresponding protein (SI Appendix, Fig. S4). Moreover, this reduction in AP3 activity led to changes in the expression of genes known or suspected to be transcriptionally regulated by AP3 during early flower development (Fig. 2 G–I), indicating that the gene knockdown was successful.
Fig. 2.
Effects of an inducible AP3 knockdown. (A–E) Characterization of a line expressing an ethanol-inducible amiRNA against AP3. (A) WT flower. (B) ap3-3 mutant flower. (C) Flower of a 35S:AlcR/pAlcA:AP3-amiRNA plant in which amiRNA expression had been induced by two 24-h treatments (at an interval of 4 d) with ethanol vapor. (D) Knockdown of AP3 mRNA levels in the floral induction system upon ethanol treatment. Plants were treated with ethanol vapor 3 d after induction of flower formation, or were mock-treated, for different time periods (as indicated). (E) Recovery of AP3 mRNA accumulation after a pulsed induction of AP3-amiRNA expression in the floral induction system. Plants were exposed to ethanol vapor for 6 h (green bar) 2 d after the induction of flower development. Following an 18-h recovery period, tissue was collected at different time points (as indicated), and AP3 mRNA levels relative to those in mock-treated flowers were determined. (F) Expression of selected genes (SI Appendix, Table S3, includes full gene names) in flowers of different stages (s) after an amiRNA-mediated AP3 knockdown. AP3 mRNA levels in these flowers were compared with corresponding flowers with unimpaired AP3 activity. (G–I) Transcriptional response of selected genes known or suspected to act downstream of the B function regulators after induction of AP3-amiRNA expression in the floral induction system. Plants were treated with ethanol (EtOH) vapor 3 d after induction of flower formation or mock-treated for different time periods (as indicated). In D and G–I, AP3 mRNA levels were normalized against the mean expression in flowers of ethanol and mock-treated plants carrying the 35S:AlcR/pAlcA:AP3-amiRNA transgene, as well as in flowers of control plants lacking that transgene, at the 0 h time point. Bars indicate SEM of four (D) or three (E–I) independent quantitative RT-PCR experiments.
By using the inducible amiRNA lines, we assessed floral phenotypes at time of anthesis (stage 13) after a pulsed knockdown of B function gene activity in whole inflorescences (Fig. 3). For this analysis, we mainly focused on AP3, but obtained similar results for PI (SI Appendix, Fig. S5). In agreement with the results of previous studies (4, 27), we found considerable differences in the morphological aberrations resulting from a loss of B function, which depended on the stage of development at which gene activities were perturbed (Fig. 3 and SI Appendix, Fig. S8). In flowers in which AP3/PI had been knocked down at intermediate stages (stage ∼8–10), petal-to-sepal transformations occurred gradually in consecutive buds. In contrast, stamens in these flowers retained their identity but became increasingly underdeveloped and did not dehisce pollen. A complete transformation of stamens into carpel-like organs was observed in flowers that were at very early stages (stage ∼2–4) of development during amiRNA induction (Fig. 3). These results indicate that the specification of stamen identity occurs significantly earlier than that of petals (SI Appendix, Fig. S5N). Notably, flowers with stamen-to-carpel transformations were directly preceded by flowers with an almost WT appearance (Fig. 3). In these mostly normal flowers, the induction of amiRNA expression had occurred around the stage for which we observed only a small number of differentially expressed genes in the ap3-1 microarray experiments. Thus, the requirement of B function appears to be temporarily reduced upon completion of the earliest stages of flower development. To test this idea further, we monitored the expression of selected response genes after an amiRNA-mediated perturbation of AP3 function at different developmental stages (Fig. 2F). In agreement with the ap3-1 microarray data, we found that their responsiveness decreased significantly as flower development reached more intermediate stages.
Fig. 3.
Pulsed perturbation of AP3 gene activity reveals multiple roles during flower development. Flowers formed consecutively by an inflorescence of a plant carrying a 35S:AlcR/pAlcA:AP3-amiRNA transgene are shown in a composite image. The plant was treated with a single 24-h pulse of ethanol vapor, and flowers were photographed at time of anthesis (stage 13). Approximate stages at the time of AP3 perturbation are indicated (labels mark the beginning of each stage). Red arrows indicate the abrupt transformation of stamens into carpels in consecutive flowers.
Genomewide Localization of AP3/PI Binding Sites.
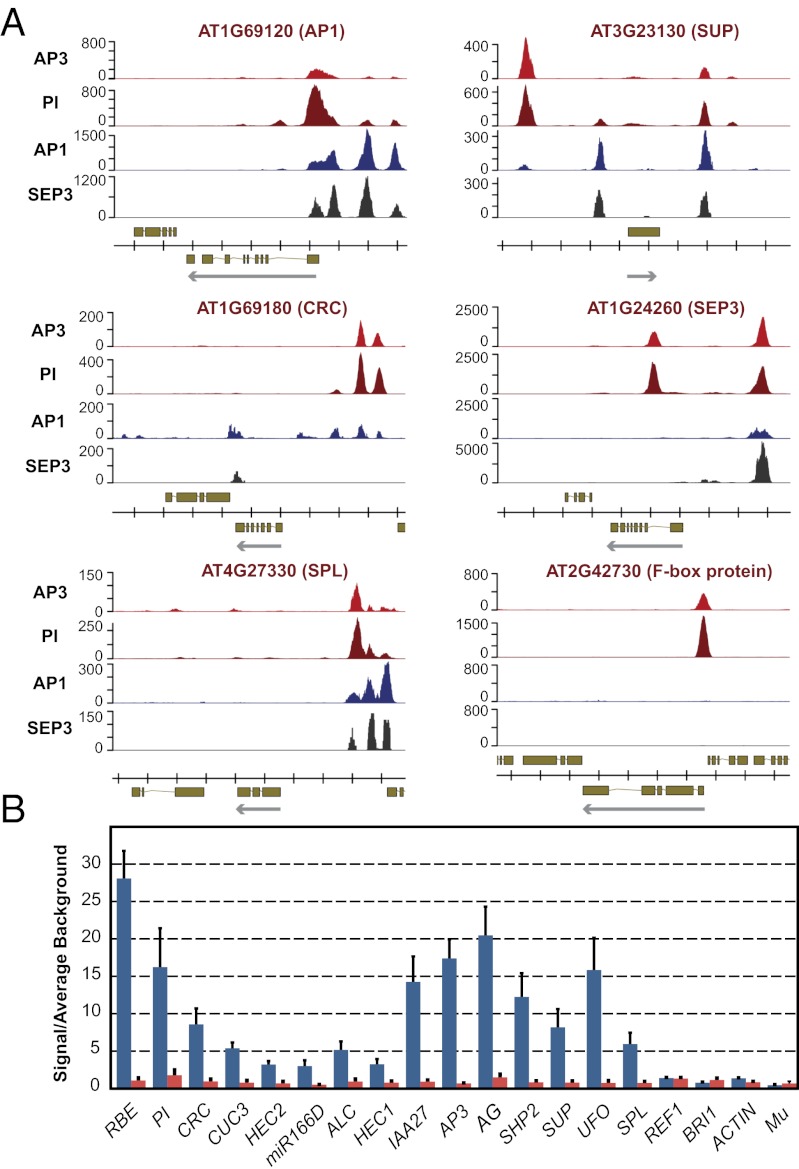
The microarray experiments described here led to the identification of genes whose expression depends on AP3/PI activity. To distinguish between direct and indirect target genes, we identified the binding sites of AP3 and PI on a genomewide scale. To this end, we generated constructs that mediate the expression of AP3 and PI, respectively, fused to GFP under the regulatory regions of the endogenous genes (SI Appendix, Fig. S6). When expressed in ap3 and pi plants, these fusion proteins led to a full rescue of the mutant phenotypes. Subsequently, we crossed these lines into the floral induction system and collected floral buds at stage ∼5. With this material, we conducted ChIP assays with GFP-specific antibodies followed by next-generation sequencing (ChIP-Seq). Analysis of the ChIP-Seq data revealed more than 1,500 high-confidence binding sites for AP3 and PI in the Arabidopsis genome (Fig. 4 and Dataset S2). To validate the binding data, we tested and confirmed selected binding sites in independent ChIP experiments followed by quantitative real-time PCR analysis (Fig. 4B). We also studied the effect of transfer DNA insertions within binding sites located in the vicinity of the floral regulators UNUSUAL FLORAL ORGANS (UFO) (28) and RABBIT EARS (RBE) (29), which respond transcriptionally to the perturbation of B function (Fig. 2F). We found that these insertions led to phenotypes resembling those of the corresponding loss-of-function mutants (SI Appendix, Fig. S7), implying functionality of the binding sites.
Fig. 4.
AP3/PI bind the promoter regions of known developmental regulators (A) ChIP-Seq results for selected target genes. Traces for AP1 (11) and SEP3 (30) are shown for comparison. Genes found in the genomic regions analyzed, and their exon-intron structure and the direction of transcription (arrows) are indicated at the bottom of each panel. AP1 had been previously identified as a target gene of AP3/PI (15), whereas SUPERMAN (SUP) and SPOROCYTELESS (SPL) were likely direct targets (10, 37). Several AP3/PI binding sites lie within evolutionary conserved regions (38) of the CRC promoter. Data for SEP3 and for the F-box protein-coding gene AT2G42730 are shown to exemplify marked differences in the binding patterns of the MADS domain proteins. (B) Validation of ChIP-Seq results for selected binding sites using quantitative PCR. ChIP was carried out by using genomic DNA from inflorescence tissue of 35S:AP1-GR ap1-1 cal-1 (red bars) and 35S:AP1-GR pPI:PI-GFP ap1-1 cal-1 pi-1 plants (blue bars), respectively, which was collected 4 d after dexamethasone treatment. GFP-specific antibodies different from those used for ChIP-Seq were used to ensure independence of the experiments. Data were normalized by using the mean signals of four reference genomic regions: REF1 (11), ACTIN, BRI1, and Mu transposon (15). Bars indicate SEs from the analysis of three independent biological sets of samples.
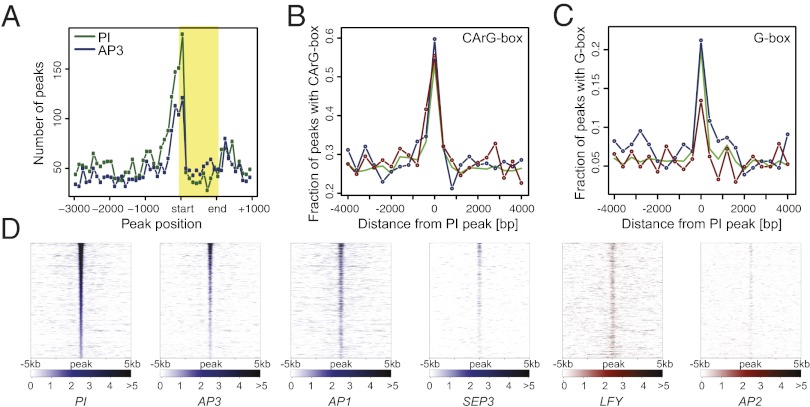
Further support for the validity of the ChIP-Seq data came from the analysis of the locations of binding sites in the Arabidopsis genome. We observed that many of these sites were located in close proximity to transcription start sites (Fig. 5A), in agreement with what has been found for other transcription factors in Arabidopsis (11, 30, 31). We also identified, through de novo predictions, overrepresented DNA sequence motifs in the regions bound by AP3/PI. We found CArG box-like sequences (consensus, 5′-CC(A/T)6GG-3′), the canonical binding sites of MADS domain proteins (12), to be highly enriched (Fig. 5B). In this analysis, we also identified several other sequence motifs as overrepresented (SI Appendix, Fig. S8). Although we currently do not know whether they are functionally relevant in the context of AP3/PI-dependent gene regulation, we noted that G-boxes (consensus, 5′-CACGTG-3′), which serve as binding sites for basic leucine-zipper and basic helix–loop–helix transcription factors (32), are significantly overrepresented (P < 0.05) at binding sites in promoters of genes that are repressed by the B function regulators compared with those of activated genes (Fig. 5C).
Fig. 5.
Genomewide binding data for AP3/PI. (A) Distribution of binding sites around transcription start sites. Plot showing the number and relative distribution of ChIP-Seq peaks in the proximity (from 3,000 bp upstream to 1,000 bp downstream) of transcribed regions of potential target genes. Binding site positions within transcribed regions (shaded box) were normalized relative to their length. Distribution of CArG (B) and G-boxes (C) within 400-bp windows around PI binding sites in the promoters of up-regulated (blue line) or down-regulated (red line) genes, or of all genes in the dataset (green line). (D) Comparison of binding data for selected floral regulators. Heat maps show binding data from different ChIP-Seq experiments for genomic regions surrounding 1,852 high-confidence PI binding sites (data for MADS domain proteins are shown in blue, and data for LFY and AP2 in red). Normalized Poisson enrichment scores are depicted in a color-code scale. All data were sorted (from top to bottom) according to descending PI peak height.
The ChIP-Seq experiments described earlier revealed that AP3/PI associate with many sites in the Arabidopsis genome. In agreement with the idea that AP3/PI act as obligate heterodimers, we found that the genomewide binding patterns for AP3 and PI were very highly correlated (Fig. 5D and SI Appendix, Fig. S8) and, in many cases, indistinguishable (Fig. 4A). We next compared the binding data for AP3/PI to those recently published for several floral regulators, which included the MADS domain proteins AP1 and SEPALLATA3 (SEP3) (11, 30), as well as the structurally unrelated transcription factors LEAFY (LFY) (31, 33) and AP2 (8). We detected a high degree of correlation in the case of AP1, SEP3, and LFY, but not for AP2 (Fig. 5D and SI Appendix, Fig. S8). These results are in agreement with the finding that AP1 and SEP3 can interact with the B function regulators in the transcriptional complexes that control petal (AP1 and SEP3) and stamen (SEP3) development, respectively (7), and the observation that LFY binds to many genes also targeted by AP1 and SEP3 (31). Despite the considerable degree of overlap between the different datasets, we also detected clear binding pattern disparities (e.g., Fig. 4A). Whether they are the result of different experimental setups or reflect bona fide differences in the binding specificities of the transcription factors remains to be elucidated.
Target Genes of the Bifunctional Transcription Factors AP3/PI.
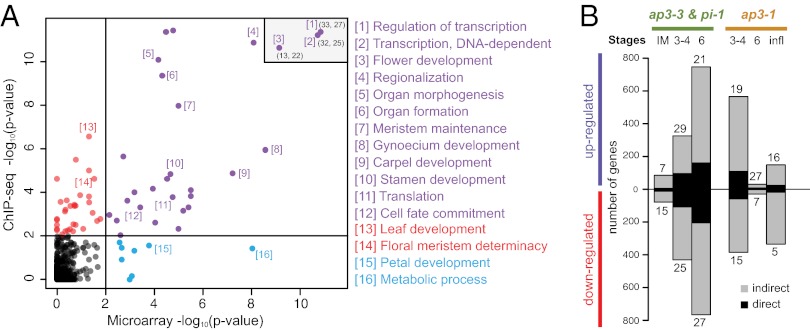
A GO term analysis of the genes bound by AP3 and PI (defined as genes with at least one binding site in the region from 3 kb upstream to 1 kb downstream of the transcribed sequence) revealed as enriched many of the same terms as the analysis of the ap3/pi microarray data described earlier (Fig. 6A). These include, for example, genes involved in stamen development, pattern formation (i.e., “regionalization”) and in the regulation of transcription. In total, 469 of the 2,100 genes that were detected as differentially expressed in the null mutant microarray experiments (∼22%; Dataset S1) are bound by AP3/PI, implying that they are under direct control of the B function regulators. This list of high-confidence target genes contains many known floral regulators, including genes involved in processes such as organ boundary formation and in the response to different hormones (SI Appendix, Table S2). In agreement with the results from our microarray experiments, which suggested that regulators of carpel formation are under transcriptional control of AP3/PI during early flower development, we found binding sites in the putative promoters of many of the corresponding genes (Fig. 4A and SI Appendix, Fig. S3). An analysis of the directionality of gene expression changes of the high-confidence targets genes revealed a similar number of genes that were up- and down-regulated upon B function perturbation (Fig. 6B). Thus, AP3/PI appear to act as bifunctional transcription factors in the control of flower development.
Fig. 6.
Correlation between GO terms enriched in the microarray and ChIP-Seq datasets. (A) Plot showing the correlation between GO terms identified in null mutant microarray (combined data from ap3-3 and pi-1; 5-d time point) and ChIP-Seq (PI) experiments. Selected terms are labeled (Dataset S4 provides a full list). For three highly significant terms, rounded coordinates (x;y) are listed in the upper right corner. (B) Proportion of direct targets among genes identified as up- or down-regulated in the ap3-3 and pi-1 experiments and in the ap3-1 experiment.
Discussion
The genomewide identification of AP3/PI binding sites and target genes revealed that these transcription factors control the expression of more than 460 genes, which can be assigned to different functional categories involved in a multitude of cellular and developmental processes commonly associated with organogenesis. This implies that AP3/PI directly participate in the regulation of many of the pathways and genes that are required for petal and stamen formation. Our results also show that AP3/PI control different processes during early and late flower development and indicate considerable differences in the spatial and temporal requirement of B function. Through conditional gene perturbation experiments we found that, in Arabidopsis, the fate of stamens is specified much earlier than that of petal primordia, which appear to remain uncommitted until intermediate stages of flower development. This finding is in agreement with what has been reported previously for flowers of Antirrhinum majus (snapdragon) (27), a species that is distantly related to Arabidopsis. Unexpectedly, morphological and molecular data from our experiments support the idea of a temporary reduction in the requirement for B function after stamen primordia have been specified. Thus, the control of stamen formation by B function appears to be discrete, whereas that of petal development is more continuous.
It has been suggested that AP3/PI regulate the expression of only a few transcription factor-coding genes and thus act relatively directly in the control of petal and stamen morphogenesis (34). However, we detected differential expression for a large number of such genes in young ap3 and pi mutant flowers (∼13% of differentially expressed genes; ∼26% of high-confidence targets). In contrast, in more mature flowers, which are represented here through the analysis of gene expression in ap3-1 inflorescences, the number of differentially expressed genes that code for transcription factors was much reduced compared with early stages and similar to their genomewide distribution of ∼6% (35). These results imply that, during early developmental stages, AP3 and PI mediate floral organ formation to a considerable extent by controlling the expression of other transcriptional regulators, whereas, during late stages, AP3/PI may act more directly in the control of organogenesis or, alternatively, may function through only a few downstream factors. Together with our discovery that AP3/PI regulate different sets of genes at distinct phases of flower formation, these findings suggest that the composition and the topology of the gene network underlying AP3/PI function changes considerably over developmental time.
It has been proposed that a key event in the evolution of seed plants was the establishment of partially nonoverlapping expression domains of B and C function genes that originated from an ancient duplication event. Hence, sex determination could be mediated by a simple switch-like mechanism in which the presence or absence of B function results in the formation of male or female structures, respectively (36). Our results show that, in Arabidopsis, the B function regulators AP3/PI are bifunctional transcription factors that activate or repress downstream targets, thus acting as a context-dependent transcriptional switch. In some cases, this appears to happen in concert with C function, for example, in the activation of the key regulator of microsporogenesis, SPL (Figs. 2I and 4A) (10). On the contrary, we found that AP3/PI suppress genes, such as those involved in carpel development (e.g., CRC), which are otherwise activated by C function (9), implying opposing regulatory roles. Based on these results, we propose that the molecular mechanism for sex-determination in seed plants depends on the interplay of synergistic and antagonistic interactions between B and C function regulators.
Materials and Methods
Previously published Arabidopsis strains used in this study included the following: 35S:AP1-GR ap1-1 cal-1 (19), ap3-1 (4), ap3-3 (14), and pi-1 (4). Plants were grown on a soil:vermiculite:perlite (3:1:1) mixture at 20 °C under constant illumination with cool white fluorescent light unless indicated otherwise. Cloning strategies and methods used for PCR-based genotyping of mutant alleles and transgenes are described in SI Appendix. Plant transformations, quantitative RT-PCR assays, and in situ hybridizations were carried out as outlined in SI Appendix. Transcript profiling experiments were done by using custom microarrays (Agilent Technologies) and a previously described protocol (11). ChIP-Seq analysis was performed, with minor modifications, as described previously (11). Analysis of microarray and ChIP-Seq data were done using custom R scripts and functions provided by the Bioconductor project. Detailed experimental and data analysis procedures are provided in SI Appendix.
Supplementary Material
Acknowledgments
The authors thank J. L. Riechmann for sharing the design of the Agilent microarrays, T. Kavanagh and M. Ramaswami for sharing equipment, J. Muiño for advice on data analysis, and J. L. Riechmann and J. Goodrich for comments on the manuscript. This study was supported by Science Foundation Ireland Grants 09/SIRG/B1600 (to E.G.), 07/IN.1/B851 (to F.W.), and 10/IN.1/B2971 (to F.W.). A.R. was supported by a scholarship from the Irish Research Council for Science, Engineering and Technology. E.G. was supported by a long-term fellowship from the European Molecular Biology Organization.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray and ChIP-Seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38363).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207075109/-/DCSupplemental.
References
- 1.Lohmann JU, Weigel D. Building beauty: The genetic control of floral patterning. Dev Cell. 2002;2:135–142. doi: 10.1016/s1534-5807(02)00122-3. [DOI] [PubMed] [Google Scholar]
- 2.Jack T. Molecular and genetic mechanisms of floral control. Plant Cell. 2004;16(Suppl):S1–S17. doi: 10.1105/tpc.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krizek BA, Fletcher JC. Molecular mechanisms of flower development: An armchair guide. Nat Rev Genet. 2005;6:688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- 4.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coen ES, Meyerowitz EM. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 6.Goto K, Kyozuka J, Bowman JL. Turning floral organs into leaves, leaves into floral organs. Curr Opin Genet Dev. 2001;11:449–456. doi: 10.1016/s0959-437x(00)00216-1. [DOI] [PubMed] [Google Scholar]
- 7.Smaczniak C, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yant L, et al. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell. 2010;22:2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, et al. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;430:356–360. doi: 10.1038/nature02733. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann K, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 12.Riechmann JL, Krizek BA, Meyerowitz EM. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA. 1996;93:4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- 14.Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 15.Sundström JF, Nakayama N, Glimelius K, Irish VF. Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis. Plant J. 2006;46:593–600. doi: 10.1111/j.1365-313X.2006.02720.x. [DOI] [PubMed] [Google Scholar]
- 16.Sablowski RW, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 17.Mara CD, Irish VF. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 2008;147:707–718. doi: 10.1104/pp.107.115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mara CD, Huang T, Irish VF. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell. 2010;22:690–702. doi: 10.1105/tpc.109.065946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2006;2:e117. doi: 10.1371/journal.pgen.0020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plackett AR, Thomas SG, Wilson ZA, Hedden P. Gibberellin control of stamen development: A fertile field. Trends Plant Sci. 2011;16:568–578. doi: 10.1016/j.tplants.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999;126:2387–2396. doi: 10.1242/dev.126.11.2387. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell. 2007;19:3516–3529. doi: 10.1105/tpc.107.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell. 2004;16:1314–1326. doi: 10.1105/tpc.021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deveaux Y, et al. The ethanol switch: A tool for tissue-specific gene induction during plant development. Plant J. 2003;36:918–930. doi: 10.1046/j.1365-313x.2003.01922.x. [DOI] [PubMed] [Google Scholar]
- 27.Zachgo S, et al. Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development. 1995;121:2861–2875. doi: 10.1242/dev.121.9.2861. [DOI] [PubMed] [Google Scholar]
- 28.Lee I, Wolfe DS, Nilsson O, Weigel D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol. 1997;7:95–104. doi: 10.1016/s0960-9822(06)00053-4. [DOI] [PubMed] [Google Scholar]
- 29.Takeda S, Matsumoto N, Okada K. RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development. 2004;131:425–434. doi: 10.1242/dev.00938. [DOI] [PubMed] [Google Scholar]
- 30.Kaufmann K, et al. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7:e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter CM, et al. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell. 2011;20:430–443. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Sibéril Y, Doireau P, Gantet P. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur J Biochem. 2001;268:5655–5666. doi: 10.1046/j.0014-2956.2001.02552.x. [DOI] [PubMed] [Google Scholar]
- 33.Moyroud E, et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–1306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zik M, Irish VF. Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell. 2003;15:207–222. doi: 10.1105/tpc.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riechmann JL, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 36.Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot (Lond) 2007;100:603–619. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai H, Krizek BA, Jacobsen SE, Meyerowitz EM. Regulation of SUP expression identifies multiple regulators involved in Arabidopsis floral meristem development. Plant Cell. 2000;12:1607–1618. doi: 10.1105/tpc.12.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JY, et al. Activation of CRABS CLAW in the Nectaries and Carpels of Arabidopsis. Plant Cell. 2005;17:25–36. doi: 10.1105/tpc.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.