Abstract
Plant development is modulated by the convergence of multiple environmental and endogenous signals, and the mechanisms that allow the integration of different signaling pathways is currently being unveiled. A paradigmatic case is the concurrence of brassinosteroid (BR) and gibberellin (GA) signaling in the control of cell expansion during photomorphogenesis, which is supported by physiological observations in several plants but for which no molecular mechanism has been proposed. In this work, we show that the integration of these two signaling pathways occurs through the physical interaction between the DELLA protein GAI, which is a major negative regulator of the GA pathway, and BRASSINAZOLE RESISTANT1 (BZR1), a transcription factor that broadly regulates gene expression in response to BRs. We provide biochemical evidence, both in vitro and in vivo, indicating that GAI inactivates the transcriptional regulatory activity of BZR1 upon their interaction by inhibiting the ability of BZR1 to bind to target promoters. The physiological relevance of this interaction was confirmed by the observation that the dominant gai-1 allele interferes with BR-regulated gene expression, whereas the bzr1-1D allele displays enhanced resistance to DELLA accumulation during hypocotyl elongation. Because DELLA proteins mediate the response to multiple environmental signals, our results provide an initial molecular framework for the integration with BRs of additional pathways that control plant development.
Keywords: cross-regulation, growth
The growth and development of plants is governed by an interconnected web of signaling pathways whose architecture has been shaped during evolution, most likely by constraints imposed by their sessile life habit. The connectivity of the pathways is particularly relevant during hormone signaling (1), which affects almost every aspect of a plant’s life, very often in response to the environment (2). According to the theory of modular biology (3), each hormone-signaling pathway might be considered in principle as an insulated module, because of the extremely high specificity provided by the chemical properties of the hormone. However, current evidence indicates that the degree of interaction between the different pathways is high and that a given hormone frequently modulates the output triggered by the rest (1). The molecular mechanisms that underlie the interactions are beginning to be unveiled and include the regulation of the homeostasis of another hormone and/or the shared participation of certain signaling elements in more than one pathway (1, 4, 5). For example, the negative regulators of the gibberellin (GA) pathway, the DELLA proteins (6), restrain root growth by promoting jasmonate (JA) signaling through inhibition of the JA ZIM-DOMAIN (JAZ) proteins (7) that negatively regulate the JA pathway (8). Conversely, GA metabolism is regulated by auxin, having an impact on hypocotyl elongation and gene expression (9). Moreover, brassinosteroids (BRs) regulate auxin responses. For instance, the negative regulator BRASSINOSTEROID INSENSITIVE2 inactivates the transcription factor AUXIN RESPONSE FACTOR2 that regulates gene expression in response to auxin (10) and thus provides one of the possible molecular mechanisms explaining the synergistic effect that both hormones exert on the expression of many genes (11).
GAs and BRs regulate common physiological responses, e.g., as illustrated by the dwarf phenotype of the GA- and BR-deficient mutants (6, 12). Moreover, both hormones act synergistically to promote hypocotyl elongation of light-grown Arabidopsis seedlings (13), a behavior that, as with BRs and auxin, might be interpreted as an indication of interaction between the two pathways. Similarly, BRs mediate the GA action promoting the skotomorphogenic developmental program in etiolated seedlings (14), suggesting that the two pathways do not always act in parallel. Thus, we hypothesize that the mechanistic link between BRs and GAs involves the molecular interaction between known signaling elements of both pathways.
Results and Discussion
Physiological Responses to GAs Are Largely Dependent on BRs in Etiolated Seedlings.
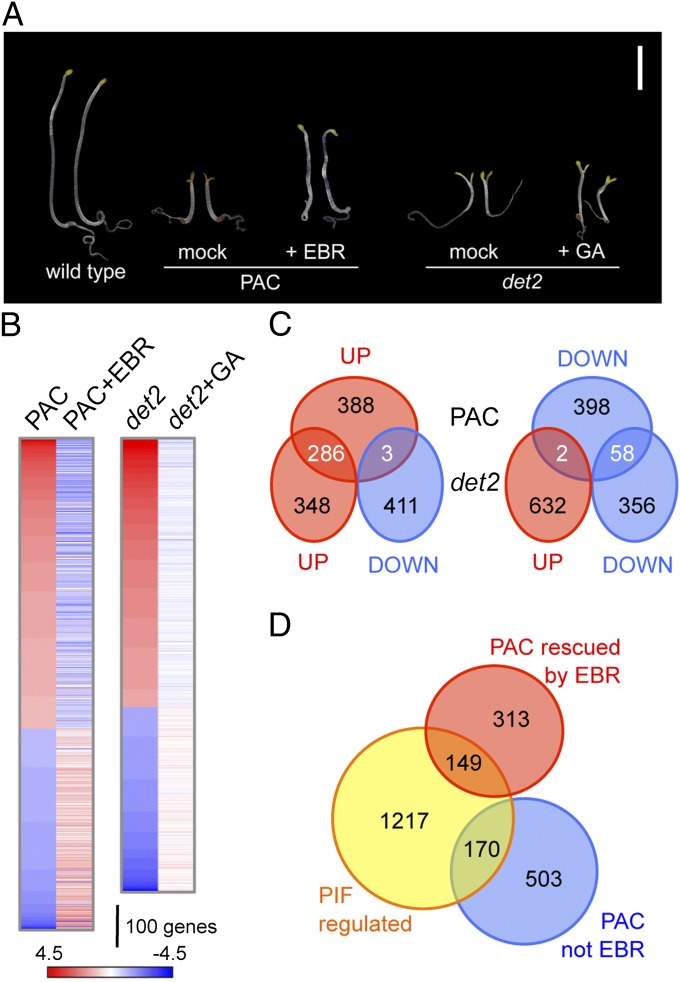
The analysis of photomorphogenic markers such as hypocotyl elongation and the expression of CAB2 and RbcS genes suggested that BRs act downstream of GAs in etiolated Arabidopsis seedlings (14), a notion that is consistent with the inability of GAs to restore the growth phenotypes of BR-deficient mutants (Fig. 1A) (15, 16). To investigate the extent of this interaction, we examined by microarray analysis the response to GAs and BRs in BR- and GA-deficient backgrounds, respectively. For that purpose, we interrogated the transcriptome of seedlings treated with 1 μM of the GA biosynthesis inhibitor paclobutrazol (PAC) supplemented or not with 1 μM epibrassinolide (EBR) and of seedlings of the BR-deficient det2-1 mutant (17) grown in the presence or absence of 50 μM GA3. As shown in Fig. 1B and in Dataset S1, EBR treatment caused a partial or complete reversion of the expression of 40% of genes (Z score ≥ 1.65; P < 0.05) (18). This reversion was confirmed for a number of selected genes in the GA-deficient mutant ga1-3 (19) (Fig. S1). The impact of the GA treatment on the det2-1 transcriptional profile was smaller, because it reversed the expression of 16% of the genes (Z score ≥ 1.65; P < 0.05) (Fig. 1B and Dataset S1). Importantly, the difference in the effectiveness of the EBR and GA treatments was reflected not only in the total number of differential genes but also in the greater magnitude of the induced changes with EBR treatment (Fig. 1B and Dataset S1). Thus, the high proportion of genes affected similarly in darkness by both GAs and BRs (Fig. 1C) indicates a large overlap in the target sets for the two hormones. However, there is a hierarchy by which GAs require an active BR-signaling pathway to exert their regulation, suggesting that an underlying mechanism involves cross-regulation between signaling elements that globally regulate gene expression in response to these hormones.
Fig. 1.
BRs mediate GA responses in etiolated seedlings. (A) Treatment with 1 μM EBR partially restored hypocotyl elongation of 6-d-old, dark-grown wild-type Col-0 seedlings treated with 1 μM PAC, whereas treatment with 50 μM GA3 did not reverse the growth defects of det2-1 mutants. (Scale bar: 0.5 cm.) (B) Heatmap representing the effect of EBR and GA treatments on global gene expression in GA- and BR-deficient backgrounds, respectively. Shown are the Z scores that correspond to PAC- versus mock-treated wild-type seedlings (PAC); wild-type seedlings treated with PAC and EBR versus PAC-treated seedlings (PAC+EBR); det2-1 mutant versus the wild type (det2); and GA3- versus mock-treated det2-1 mutants (det2+GA). (C) Venn diagram showing overlap among genes differentially regulated in GA- and in BR-deficient seedlings grown in darkness. (D) Venn diagram showing overlap among genes whose expression is restored by EBR in a GA-deficient background and genes misregulated in the pifQ mutant [data taken from Leivar et al. (21)].
The current model for gene regulation by GAs during photomorphogenesis indicates that the regulation occurs, at least in part, through the negative effect that DELLA proteins exert on the basic helix-loop-helix transcription factors of the PHYTOCHROME INTERACTING FACTORs (PIFs) clade (20). Indeed, 28% of PAC-regulated genes also were misregulated in dark-grown quadruple pif mutant seedlings (pifQ) (21, 22) (Fig. 1D and Dataset S1), suggesting that the mechanism based on the DELLA–PIF interaction likely was responsible for this regulation. Remarkably, among the 462 genes that were rescued by EBR treatment in the GA-deficient background, 68% were not misregulated in the pifQ mutant (Fig. 1D and Dataset S1). This finding suggests that DELLA proteins also regulate BR-dependent gene expression through a PIF-independent mechanism.
One possible model explaining the cross-regulation between GA and BR signaling involves the modulation of DELLA proteins by BRs. DELLA proteins are destabilized in response to GAs (23). Thus, we reasoned that BRs might have a negative impact on DELLA stability. To test this idea, we examined by Western blot whether the accumulation of DELLA proteins was affected by BRs. Importantly, levels of the endogenous GAI and RGA proteins were not affected by BR deficiency in the det2 mutant (Fig. S2A). Similarly, neither endogenous GAI and RGA nor transgenic TAP-GAI and GFP-RGA were affected by treatment with 1 μM EBR (Fig. S2 B–D). The same result was observed when we applied EBR to dark-grown RGA::GFP-(rga-Δ17) seedlings that express a dominant, GA-resistant version of RGA (24) (Fig. S2E). These results rule out the possibility that BRs act upstream of DELLA proteins and are supported further by the observation that treatment with 1 μM EBR restored the repression of CAB2 expression in dark-grown 35S::gai-1 seedlings (Fig. S3) (14, 25).
In a reciprocal model, GAs would modulate the activity of BR-signaling elements. The regulation of gene expression in response to BRs relies on a small family of transcription factors that includes BRASSINAZOLE RESISTANT1 (BZR1) and BRI1 EMS SUPPRESSOR1 (BES1) (12). Their activity is regulated negatively by phosphorylation dependent on GSK3-type kinases, whereas BRs promote their dephosphorylation (26, 27). We used dark-grown BZR1::BZR1-CFP (28) and 35S::BES1-GFP (26) seedlings to investigate whether the phosphorylation status of these proteins was affected by GAs. As shown in Fig. S4 A and B, altering GA levels either by a 2-h treatment with 100 μM GA3 or by continuous growth on 0.5 μM PAC did not significantly affect the ability of BRs to modulate the phosphorylation of these two transcription factors.
Taken together, these results suggest that the mechanism for cross-regulation between GAs and BRs could be constituted by DELLA proteins and the BR-dependent transcription factors. This notion is consistent with a model in which DELLA proteins regulate gene expression in etiolated seedlings by inactivating nuclear dephosphorylated BZR1 and BES1 (29, 30), similar to the inactivation that DELLA exert on the PIFs (20) and JAZs (7) upon physical interaction.
DELLA Proteins GAI and RGA Interact Physically with BZR1.
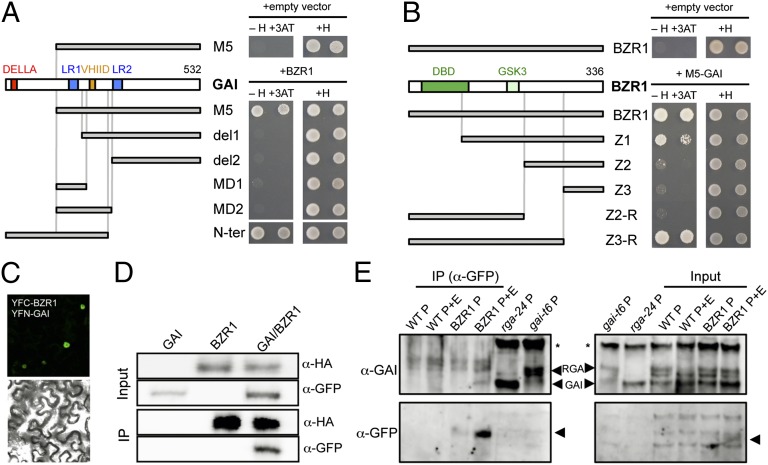
To challenge the model, we first investigated whether GAI interacted physically with BZR1 in a yeast two-hybrid (Y2H) assay (Fig. 2A and Fig. S5A). As bait, we used a truncated version of GAI, M5, that showed reduced autoactivation, as occurs with similar versions of RGA (7, 31). As shown in Fig. 2A, M5 was able to interact strongly with the full-length BZR1. To get clues about the possible molecular outcome of this interaction, we mapped the interacting domains for GAI and BZR1. Deletion of the first leucine heptad repeat (LHR1) (32) was sufficient to prevent interaction of del1 with BZR1, whereas N-ter, which included the entire N-terminal DELLA domain and the LHR1, was able to interact with it (Fig. 2A). These results suggest that the LHR1 is important for the interaction with BZR1, as occurs in the interactions of RGA with PIF4 (31) and with JAZ1 (7). Nonetheless, the LHR1 alone is not sufficient, because small fragments of GAI containing only the LHR1 or the LHR1 and the VHIID motif did not interact (Fig. 2A). A more complex scenario was found when we tested M5 with truncations of BZR1. Two overlapping versions were able to interact: Z3-R, which interacted as strongly as the full-length, and Z1, which showed a weaker interaction (Fig. 2B). This result suggests that the fragment adjacent to the DNA-binding domain (DBD; amino acids 21–104, included in Z3-R) (33), extending from the beginning of Z1 to the beginning of Z3, is sufficient for the interaction and likely cooperates with the DBD to enhance the ability of BZR1 to interact with GAI. To determine whether BZR1 also interacts with other DELLA proteins, we performed Y2H assays using clones expressing a full-length BZR1 and the M5 equivalent version of RGA (Fig. S5B). These assays demonstrate that the ability of BZR1 to interact with DELLA proteins is not restricted to GAI.
Fig. 2.
GAI interacts physically with BZR1. (A and B) Y2H assays analyzing the interaction between deleted versions of GAI and the full-length BZR1 (A) and between M5 and deleted versions of BZR1 (B). Note that, given the strong autoactivation of N-ter, this GAI derivative was cloned in the prey vector, whereas others were cloned in the bait vector. Drawings on the left illustrate the relevant motifs of each protein and their truncated versions. Two serial dilutions per yeast clone are shown. +H, control medium containing His. −H +3AT, selective medium lacking His and containing 20 mM 3-aminotriazol (3-AT). (C) BiFC analysis in N. benthamiana leaves of GAI and BZR1 fusions to N- and C-terminal fragments of YFP, respectively. (Upper) YFP fluorescence. (Lower) Bright-field image. (D) Coimmunoprecipitation assay showing the interaction between GAI and BZR1 in leaves of N. benthamiana. YFP-GAI and HA-BZR1 were expressed either alone or together in leaves of N. benthamiana. Nuclear proteins were immunoprecipitated with paramagnetic beads coated with anti-HA antibody and were detected by immunoblotting with either anti-HA or anti-GFP (JL-8; Clontech) antibodies. (E) Coimmunoprecipitation assay showing the interaction between GAI and BZR1 in Arabidopsis seedlings. Soluble proteins from wild-type Col-0 and BZR1::BZR1-CFP (BZR1) seedlings grown in darkness for 4 d in MS plates containing 0.5 μM PAC (P) or 0.5 μM PAC and 1 μM EBR (P+E) were used for immunoprecipitations (IP) with anti-GFP (α-GFP) paramagnetic beads. Proteins were immunoblotted and detected consecutively with anti-GAI and anti-GFP (Ab290; Abcam) antibodies. Ten micrograms of soluble proteins were loaded as input. Soluble proteins from the null mutants gai-t6 and rga-24 grown in the dark for 4 d in 0.5 μM PAC plates were used as controls. The asterisk indicates a cross-reacting, nonspecific band that served as a loading control; the arrowheads in the lower blots indicate BZR1-CFP.
The interaction between GAI and BZR1 also was confirmed in planta, as shown by bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaves (Fig. 2C). Nuclear fluorescence caused by the reconstruction of YFP was observed in nuclei of epidermal cells of leaves that coexpressed YFC-BZR1 and YFN-GAI, whereas fluorescence in nuclei of control leaves that coexpressed either YFC-BZR1 and YFN or YFC and YFN-GAI was below detection limits (Fig. S6). Moreover, we tested the physical interaction between the two proteins by coimmunoprecipitation. For that purpose, we first expressed HA-BZR1 and YFP-GAI transiently in leaves of N. benthamiana. As shown in Fig. 2D, YFP-GAI coimmunoprecipitated when HA-BZR1 was pulled down from leaf extracts. Similarly, the endogenous GAI was immunoprecipitated by anti-GFP antibodies from extracts of BZR1::BZR1-CFP seedlings grown in darkness in the presence of PAC and EBR, a situation that favored the accumulation of both GAI and unphosphorylated, nuclear BZR1-CFP (Fig. 2E). As expected, the interaction was not detected when seedlings were grown in the absence of EBR; these seedlings clearly accumulated much less active BZR1-CFP (Fig. 2E). These results further demonstrate that these two proteins interact in vivo.
DELLA Interaction Prevents BZR1 Binding to Target Promoters.
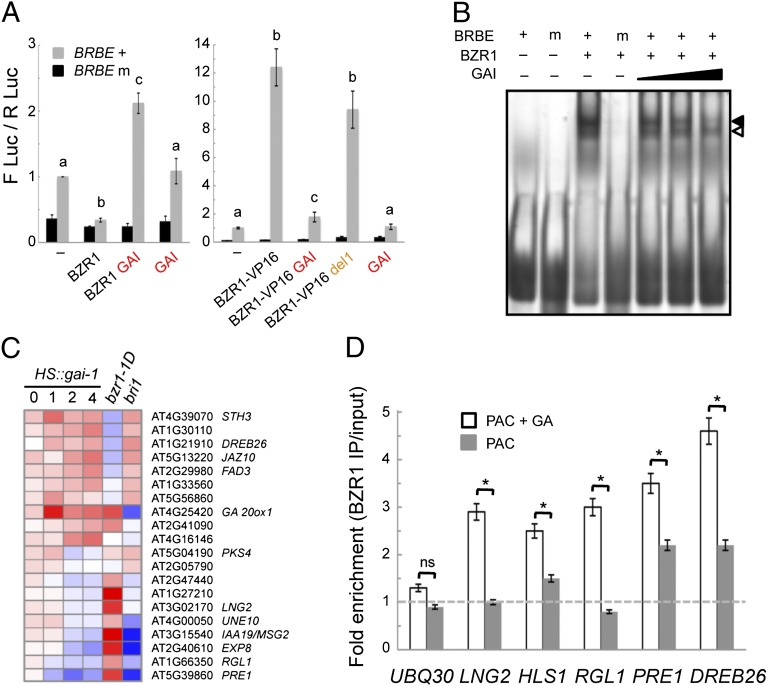
BZR1 acts as a transcriptional regulator, being an activator or repressor depending on the target promoter (34). To examine the effect of the interaction on BZR1 activity, we performed transcriptional assays by transient expression in leaves of N. benthamiana. To do so, we prepared a synthetic promoter consisting of five concatemerized copies of the BZR1-binding site from the CPD gene (33), a minimal 35S promoter, and the viral translational enhancer Ω controlling the expression of the reporter gene LUCIFERASE (LUC) (see Materials and Methods for details). Expression of HA-BZR1 caused a reduction in the LUC activity that was not observed with the mutant version of the promoter (Fig. 3A). This repressor activity also has been observed in the context of another synthetic promoter (33), suggesting that it is an intrinsic feature of BZR1 that may be modulated by coregulator proteins when bound to natural promoters. Remarkably, the repressor activity was largely reversed when YFP-GAI was coexpressed with HA-BZR1, whereas YFP-GAI alone did not affect the LUC activity significantly. To rule out the possibility that GAI was inactivating BZR1 corepressor proteins present in the N. benthamiana cells rather than BZR1 itself, we performed the same assays using a translational fusion of BZR1 to the strong transcriptional activator VP16 (35). YFP-GAI also was able to counteract fully the specific, strong activation of the reporter by HA-VP16-BZR1 (Fig. 3A). On the other hand, coexpression of del1, a version of GAI that failed to interact with BZR1 (Fig. 2A), did not affect its transcriptional activity significantly (Fig. 3A). Taken together, these results support the model that GAI inactivates BZR1 by direct, physical interaction.
Fig. 3.
Interaction with GAI inactivates BZR1. (A) Effect of GAI on the transcriptional activity of BZR1. The reporter construct with wild-type (BRBE +) or the mutated (BRBE m) BZR1-binding sites was transiently expressed in leaves of N. benthamiana by agroinfiltration, either alone (−) or with the effectors HA-BZR1 (BZR1), HA-VP16-BZR1 (BZR1-VP16), YFP-GAI (GAI), HA-VP16-BZR1 plus YFP-GAI (BZR1-VP16 GAI), or HA-VP16-BZR1 plus YFP-del1 (BZR1-VP16 del1). The y-axis represents the ratio between the activities of firefly LUC and the control Renilla LUC. Values were normalized with respect to the ratio obtained for the wild-type reporter alone. Three biological repeats were performed; error bars represent SEM. Statistically significant differences (P < 0.001) according to one-way ANOVA test are indicated by different letters. (B) EMSA showing the effect of GAI on the DNA-binding ability of BZR1. The black arrowhead indicates the band whose intensity was most strongly affected by GAI. (C) Heatmap representing the expression of genes that are putative common targets for GAI and BZR1 based on meta-analysis (see text for description). Color indicates fold expression change at different times (in hours) after HS::gai-1 induction or in bzr1-1D bri1 versus bri1 or in bri1 versus wild type. (D) ChIP of BZR1-CFP followed by quantitative PCR of selected target genes in 4-d-old seedlings grown in darkness in 1 μM PAC with or without 10 μM GA3. UBQ30 is a control gene whose promoter is not bound by BZR1. Values represent the fold enrichment of BZR1-bound DNA in immunoprecipitated samples relative to the total input DNA. Three biological repeats were performed; error bars represent SEM. *P < 0.01 (Student’s t test); ns, no significance.
The Y2H results were compatible with the possibility that GAI prevents the DNA-binding activity of BZR1, thereby providing an explanation for the inhibitory effect of GAI on BZR1 transcriptional activity (Fig. 3A), as occurs with PIFs (31, 36, 37). To test this idea, we examined the DNA-binding ability of BZR1 in the presence or absence of DELLA proteins. First, we used in vitro tests, performing EMSAs. As shown in Fig. 3B, His-BZR1 bound a probe containing the BZR1-binding site of the CPD promoter, whereas binding was not detectable with the mutated probe (33). Importantly, the binding decreased with increasing amounts of His-GAI (Fig. 3B). These results provide biochemical evidence of the inhibitory effect that the interaction with GAI has on the DNA-binding ability of BZR1. To investigate if this inhibitory effect is significant in plants, we examined the ability of BZR1 to bind its natural target promoters in vivo. To select targets in which DELLA-BZR1 interaction could be relevant, we searched for genes whose expression was regulated directly by GAI in etiolated seedlings (38) that had differential expression in a bzr1-1D–mutant background (34) and whose promoters were bound by BZR1 in vivo (Fig. 3C) (34). Interestingly, 15 of the 20 genes matching all criteria displayed opposite regulation by GAI and BZR1. As shown in Fig. 3D, the binding of BZR1-CFP to the promoter region of five of these genes was reduced in dark-grown BZR1::BZR1-CFP seedlings treated with PAC, as compared with seedlings treated simultaneously with PAC and GA3, although neither the amount nor the phosphorylation status of BZR1-CFP was affected by treatments (Fig. S4C). These results strongly suggest that GAI exerts a negative effect on the DNA-binding activity of BZR1 in vivo by sequestering the protein into an inactive complex, a situation that parallels the mechanism by which DELLA proteins inactivate PIFs (31, 36, 37).
BZR1 Mediates Cellular Responses to GAs.
If the negative interaction of GAI with BZR1 is physiologically relevant, this regulation would have to meet at least two conditions: (i) DELLA-dependent differential binding of BZR1 to target promoters would have to result in significant changes in the expression of those genes; and (ii) the output of certain morphological traits associated to DELLA activity should be dependent on the relative levels of both proteins.
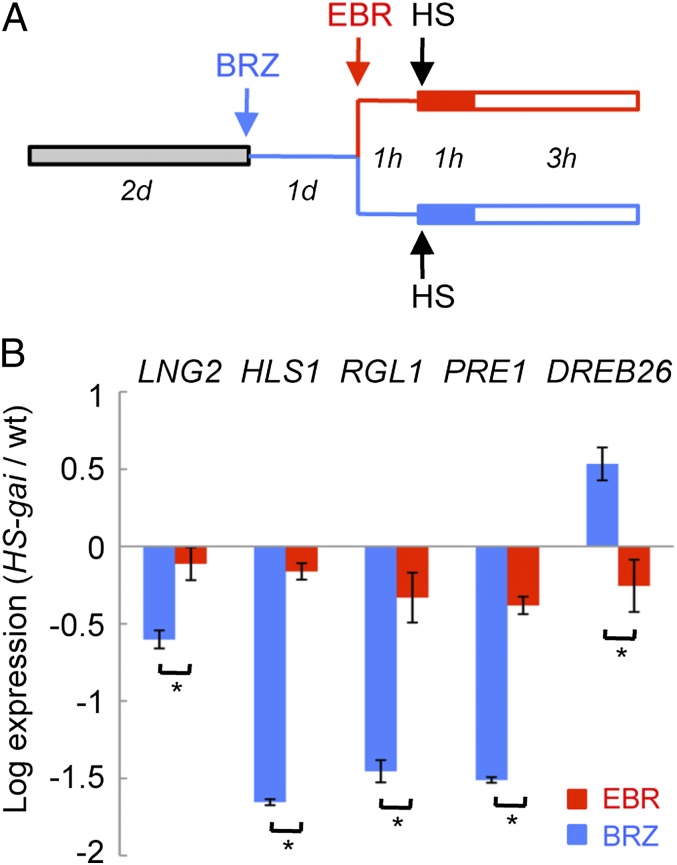
To test these criteria, we first examined the early, immediate consequences of the activation of the BR pathway on the activity of gai-1 in etiolated HS::gai-1 seedlings, comparing the ability of gai-1 to induce or repress the expression of DELLA- and BZR1 target genes in dark-grown seedlings treated with brassinazole (BRZ) or with BRZ and EBR (Fig. 4A). As shown in Fig. 4B, an EBR treatment that induced full dephosphorylation of BZR1-CFP in BRZ-treated seedlings (Fig. S4A) alleviated the molecular phenotype caused by the accumulation of gai-1 on the target genes; this result is consistent with the proposal that the regulation of these genes by GAI is mediated by BZR1. Consistent with these results, repression of the CPD gene in response to exogenous EBR was largely abolished in dark-grown gai-1–mutant seedlings (Fig. S7A).
Fig. 4.
Activation of the BR pathway counteracts the activity of gai-1. (A) Scheme depicting the experimental design. Two-day-old dark-grown, wild-type Col-0 and HS::gai-1 seedlings were transferred to plates containing 3 μM BRZ for an additional day. Half the seedlings were mock-treated with 3 μM BRZ (blue), and half were treated with 1 μM EBR + 3 μM BRZ (red) starting 1 h before heat shock (HS) for 1 h at 37 °C. Samples were harvested 4 h after the beginning of the heat shock. (B) Changes in expression induced by HS::gai-1 in selected target genes expressed relative to EF1α. Three biological repeats were performed; error bars represent SEM. *P < 0.01 (Student’s t test).
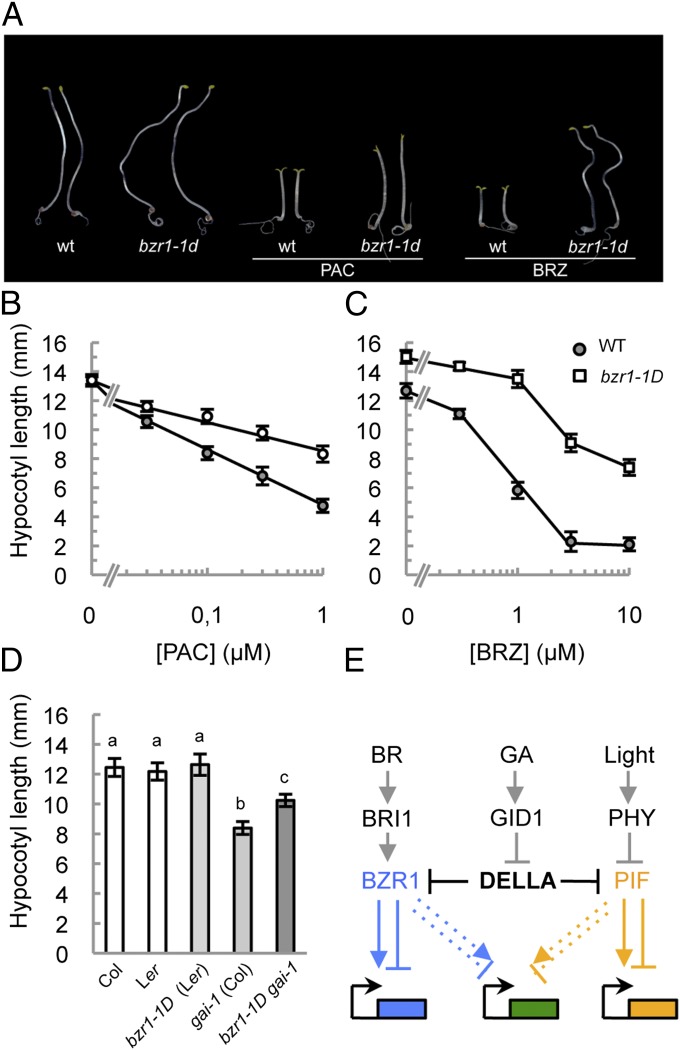
Next, we examined the sensitivity of dominant bzr1-1D–mutant (28) seedlings to increasing doses of PAC, using hypocotyl length as a diagnostic tool. Importantly, the mutant seedlings displayed resistance to PAC-dependent growth inhibition as compared with the wild-type seedlings (Fig. 5 A and B), although the resistance was lower than that shown for BRZ (Fig. 5C) (28). To confirm these results genetically, we examined the hypocotyl length in F1 seedlings from the cross between bzr1-1D and gai-1. As shown in Fig. 5D, the presence of the dominant bzr1-1D allele alleviated the growth restraint caused by gai-1 as compared with the heterozygous bzr1-1D and gai-1 control seedlings. These results indicate that BZR1 has a positive role in regulating hypocotyl growth in response to GA in dark-grown seedlings. Moreover, GAs and BRs synergistically promote the hypocotyl elongation of light-grown Arabidopsis seedlings (13). To evaluate whether the interaction of GAI and BZR1 underlies this synergism, we analyzed the hypocotyl growth phenotype of light-grown bzr1-D in GA dose–response curves. As would be expected if our model also operates to control hypocotyl elongation in the light, the mutant was hypersensitive to applied GA3 (Fig. S7B).
Fig. 5.
BZR1 mediates the growth response of hypocotyls to GAs. (A) Seven-day-old wild-type Col-0 and bzr1-1D seedlings were grown in darkness in control medium or in medium supplemented with either 1 μM PAC or 3 μM BRZ. Two representative seedlings per genotype are shown. (B and C) Hypocotyl growth of 7-d-old, dark-grown wild-type Col-0 and bzr1-1D seedlings in response to different concentrations of PAC (0.03, 0.1, 0.3, and 1 μM) (B) and BRZ (0.3, 1, 3, and 10 μM) (C). Error bars in B and C indicate SD (n > 15 seedlings). Experiments were repeated twice with similar results; results from one representative experiment are shown. (D) Hypocotyl length of F1 seedlings from the cross between the dominant mutants gai-1 and bzr1-1D, along with seedlings from the corresponding control crosses. Statistically significant (P < 0.001) differences according to one-way ANOVA test are indicated by different letters. Error bars represent SD (n > 20). (E) Model depicting the molecular mechanism for the integration of GA, BR, and light signaling. DELLA proteins modulate gene expression in the context of photomorphogenesis through interaction with at least PIFs and BZR1 for distinct sets of genes. Our results also suggest that there is a third group of targets whose expression is dependent on both BZR1 and PIFs (Fig. 1D). GID1, GIBBERELLIN INSENSITIVE DWARF1; PHY, phytochrome.
Concluding Remarks
Our results support a molecular mechanism for the integration of the GA- and BR-signaling pathways based on the inactivation of BZR1 upon interaction with DELLA proteins. The concurrence of DELLA proteins, BZR1, and PIFs establishes a likely framework for interaction among GAs, BRs, and light in the control of photomorphogenic development (Fig. 5E) and allows the plant to funnel the flow of information to regulate the expression of different specific sets of genes coordinately. Two further important implications can be drawn from the model. First, the mechanism might operate at other stages of development; for instance during germination, as suggested by the observation that BRs promote germination of GA-deficient seeds (39). Second, other DELLA proteins (6) and the BZR1 paralogs BES1 and BEH1–4 (26, 40) might establish equivalent interactions. This added complexity would increase the regulatory possibilities of the mechanism considerably, in a combinatorial manner, because of the probable differences in binding affinities between protein partners and depending on the cellular context. In summary, our model provides a mechanistic framework that will help us understand how these two major hormone pathways regulate common developmental processes during the entire life cycle of the plant.
Materials and Methods
Plant Material and Growth Assays.
Arabidopsis thaliana accession Col-0 was used as wild type. All mutants and transgenic lines have been described elsewhere and are listed in SI Materials and Methods along with details of seedlings growth assays.
Microarray Analysis.
Details of microarray analysis are provided in SI Materials and Methods. The microarray experiment is deposited in the GEO database under the accession number GSE32889.
Y2H Assays.
Construction of vectors, interaction tests, and detection of fusion proteins are described in SI Materials and Methods. Primers used for cloning are listed in Dataset S2.
BiFC, Coimmunoprecipitation Assays, and Protein Analysis.
Details of BiFC and coimmunoprecipitation assays and details of the protein extraction and Western blot analysis from whole seedlings are described in SI Materials and Methods. Primers used for cloning are listed in Dataset S2.
Reporter Construct and Transcriptional Assays.
The synthetic promoter consisted of five concatemerized copies of the wild-type (GCAGAAACCCCCCGTGTGCCCACTCTCCCC) or mutant (GCAGAAACCCCAAAAAAACCCACTCTCCCC) BZR1-binding site (underlined) of the CPD promoter (33), upstream of the minimal 35S promoter and the Ω translational enhancer, as described for the TCS cytokinin reporter (41). Details of the reporter constructs are provided in SI Materials and Methods. Transient expression in leaves of N. benthamiana was as described (37). In the infiltration mixture, the ratio of Agrobacterium-carrying reporter and effector constructs was 1:4. Firefly and the control Renilla LUC activities were assayed from leaf extracts with the Dual-Glo Luciferase Assay System (Promega) and quantified with a GloMax 96 Microplate Luminometer (Promega).
EMSA.
Details regarding the different constructs and conditions used for EMSAs are provided in SI Materials and Methods.
ChIP and PCR Amplification.
Wild-type Col-0 and BZR1::BZR1-CFP seedlings were used for ChIP analysis. Details are given in SI Materials and Methods. Quantitative PCR oligonucleotides used for amplifications are listed in Dataset S2.
RNA Extraction, Northern Blot, and Quantitative RT-PCR.
Details of total RNA extraction and analysis are given in SI Materials and Methods. Primers used are listed in Dataset S2.
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre, Tai-ping Sun, Zhi-Yong Wang, Yanhai Yin, Ana Caño-Delgado, Luís López-Molina, and François Parcy for providing seeds or reagents; Laura García-Cárcel and Gastón Pizzio for help in the early stages of this work; and Salomé Prat for fruitful discussions, sharing unpublished results, and careful reading of the manuscript. J.G.-B. holds a Consejo Superior de Investigaciones Científicas Fellowship of the Joint Admissions Exercise Predoctoral Program. E.G.M. is recipient of a postdoctoral “Juan de la Cierva” contract from the Spanish Ministry of Science and Innovation. A.L. was supported in part by a fellowship of the Fondo per gli Investimenti della Ricerca di Base Progetto Giovani of the Italian Ministry of Education, University, and Research. Work in the authors’ laboratory was funded by Spanish Ministry of Science and Innovation Grants BIO2007-60923, BIO2010-15071, and CSD2007-00057 and by Generalitat Valenciana Grants ACOMP/2010/190 and PROMETEO/2010/020. Rothamsted Research is funded by the Biotechnology and Biological Sciences Research Council (BBSRC) of the United Kingdom.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray analysis reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE32889).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119992109/-/DCSupplemental.
References
- 1.Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–R373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Alabadí D, Blázquez MA, Carbonell J, Ferrándiz C, Pérez-Amador MA. Instructive roles for hormones in plant development. Int J Dev Biol. 2009;53:1597–1608. doi: 10.1387/ijdb.072423da. [DOI] [PubMed] [Google Scholar]
- 3.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761)(Suppl):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 4.Kuppusamy KT, Walcher CL, Nemhauser JL. Cross-regulatory mechanisms in hormone signaling. Plant Mol Biol. 2009;69:375–381. doi: 10.1007/s11103-008-9389-2. [DOI] [PubMed] [Google Scholar]
- 5.Jaillais Y, Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 7.Hou X, Lee LY, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca S, Chico JM, Solano R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr Opin Plant Biol. 2009;12:539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Frigerio M, et al. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 2006;142:553–563. doi: 10.1104/pp.106.084871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clouse SD. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, et al. Physiological roles of brassinosteroids in early growth of Arabidopsis: Brassinosteroids have a synergistic relationship with gibberellin as well as auxin in light-grown hypocotyl elongation. J Plant Growth Regul. 2003;22:259–271. [Google Scholar]
- 14.Alabadí D, Gil J, Blázquez MA, García-Martínez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004;134:1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 16.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 17.Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koornneef M, Van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- 20.Davière JM, de Lucas M, Prat S. Transcriptional factor interaction: A central step in DELLA function. Curr Opin Genet Dev. 2008;18:295–303. doi: 10.1016/j.gde.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Leivar P, et al. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin J, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silverstone AL, et al. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dill A, Jung HS, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alabadí D, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53:324–335. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- 26.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 27.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 29.Ryu H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gampala SS, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 32.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 33.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 36.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallego-Bartolomé J, et al. Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 2011;67:622–634. doi: 10.1111/j.1365-313X.2011.04621.x. [DOI] [PubMed] [Google Scholar]
- 38.Gallego-Bartolomé J, Alabadí D, Blázquez MA. DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE. 2011;6:e23918. doi: 10.1371/journal.pone.0023918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 41.Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.