Abstract
Supernumerary B chromosomes are optional additions to the basic set of A chromosomes, and occur in all eukaryotic groups. They differ from the basic complement in morphology, pairing behavior, and inheritance and are not required for normal growth and development. The current view is that B chromosomes are parasitic elements comparable to selfish DNA, like transposons. In contrast to transposons, they are autonomously inherited independent of the host genome and have their own mechanisms of mitotic or meiotic drive. Although B chromosomes were first described a century ago, little is known about their origin and molecular makeup. The widely accepted view is that they are derived from fragments of A chromosomes and/or generated in response to interspecific hybridization. Through next-generation sequencing of sorted A and B chromosomes, we show that B chromosomes of rye are rich in gene-derived sequences, allowing us to trace their origin to fragments of A chromosomes, with the largest parts corresponding to rye chromosomes 3R and 7R. Compared with A chromosomes, B chromosomes were also found to accumulate large amounts of specific repeats and insertions of organellar DNA. The origin of rye B chromosomes occurred an estimated ∼1.1–1.3 Mya, overlapping in time with the onset of the genus Secale (1.7 Mya). We propose a comprehensive model of B chromosome evolution, including its origin by recombination of several A chromosomes followed by capturing of additional A-derived and organellar sequences and amplification of B-specific repeats.
Keywords: centromere, genome evolution, promiscuous DNA, non-Mendelian chromosome transmission
Supernumerary B chromosomes are not required for the normal growth and development of organisms and are assumed to represent a specific type of selfish genetic element. B chromosomes do not pair with any of the standard A chromosomes at meiosis, and have irregular modes of inheritance. Because they are dispensable for normal growth, B chromosomes have been considered nonfunctional, with no essential genes. As a result, B chromosomes follow their own species-specific evolutionary pathways. Despite their widespread occurrence in all eukaryotic groups, including insects (1), mammals (2), and plants (3), and their potential as chromosome-based vectors in biotechnology (4), little is known about the origin and molecular composition of these constituents of the genome.
Several scenarios have been proposed for the origin of B chromosomes. The most widely accepted view is that they are derived from the A chromosome complement. Some evidence also suggests that B chromosomes can be spontaneously generated in response to the new genomic conditions after interspecific hybridization. The involvement of sex chromosomes has also been argued for their origin in some species (reviewed in refs. 5–7). Despite the high number of species with B chromosomes, their de novo formation is probably a rare event; the occurrence of similar B chromosome variants within related species suggests that they arose from a single origin.
One of the best-studied plant models for research into B chromosomes is rye (Secale cereale), with a genome comprising seven pairs of A chromosomes (1C ∼7,917 Mbp) and containing between zero and eight B chromosomes, each with 1C ∼580 Mbp. Rye B chromosomes appear to be monophyletic and very stable, being quite similar among rye taxa like S. cereale subsp. segetale, which is very closely related to S. ancestrale (8). This is rather unusual, given that B chromosomes are expected to have an elevated mutation rate compared with the A genome and thus should quickly diverge. At the DNA level, apart from the terminal region of the B chromosome long arm, overall the A and B chromosomes of rye are highly similar (9, 10). The molecular processes that gave rise to Bs during evolution remain unclear, and the characterization of sequences residing on them might shed light on their origin and evolution.
Our analysis provides insight into an enigmatic phenomenon of genome evolution in numerous groups of eukaryotes. We report that B chromosomes of rye are unexpectedly rich in gene-derived sequences, allowing us to trace their origin to parts of the A genome. In addition, compared with A chromosomes, B chromosomes accumulate large amounts of specific repeats and insertions of organellar DNA. We propose a model of the stepwise evolution of B chromosomes after segmental genome duplication followed by the capture of additional A-derived and organellar sequences and amplification of B-specific repeats.
Results
B Chromosomes Are Unexpectedly Rich in A-Derived Genic Sequences.
To identify the origin and evolution of the B chromosome, we performed a comparative sequence analysis of the A and B chromosomes of rye. First, we purified the B chromosome of an isogenic rye line by flow cytometry sorting (Fig. S1) and shotgun sequenced it at 0.9-fold sequence coverage using Roche 454 technology. As a reference, we used the sequence information from all A chromosomes (also purified by flow cytometry sorting) and the genomic DNA of plants both with (+B) and without (0B) B chromosomes (Table S1).
B chromosomes are generally considered nonfunctional, with no essential genes (5–7). Unexpectedly, we found many B sequences with a high homology to the genes of sequenced plant genomes (Fig. S2). Comparison of sequence reads from the rye B chromosome with the estimated size of 580 Mbp (BLASTX ≥70% identity ≥30 amino acids) revealed a total of 4,189, 3,449, and 3,815 homologous nonredundant genes for Brachypodium distachyon, rice (Oryza sativa), and sorghum (Sorghum bicolor), respectively (Table S2). From the comparison of different individual reference datasets, a nonredundant gene count was extracted, comprising at least 4,946 putative B-located genic sequences. In comparison, the short arm of rye A chromosome 1R, with a size of 441 Mbp, is expected to contain ∼2,000 genes (11). However, our analysis does not allow for conclusions regarding the completeness and functionality of the B-located genes.
We made use of the similarity between shared genic sequences of rye A and B chromosomes to determine the mutation frequency and relative age of the B-located sequences. To analyze the differences in SNP frequencies, which should reflect the presence or absence of selective pressure, we compared genic sequences from the As and Bs to rye RNAseq-based contigs (12) by BLASTN and identified SNPs in regions present in all three of the datasets. As expected, the genic sequences of rye A chromosomes revealed a lower SNP frequency (1/72 bp) than their B-located homologs (1/47 bp) compared with the rye RNAseq assemblies (Table S3). This difference is not related to a disparity in the effective population size between the chromosome sets, given that the SNP frequencies of mobile elements were one SNP per 25 bp in the A chromosomes and one SNP per 26 bp in the B chromosomes. Thus, the selection pressure is lower for B-located genes than for A-located genes.
We used sequence alignments of the A and B gene sequences and their homologs in Brachypodium and barley full-length cDNAs in Bayesian phylogenetic analyses to determine the age of origin of the B chromosome The inferred age of 1.1–1.3 million y (My) of rye B chromosomes (Fig. S3) might be overestimated owing to the relaxed selective pressure on B-located genes. Nevertheless, it coincides with the estimated age of 1.7 My for the genus Secale and 0.8 My for the S. strictum/S. vavilovii/S. cereale taxon group, based on a dated rDNA phylogeny of Triticeae (Fig. S4). These ages indicate that rye B chromosomes originated only within the genus Secale and are in accord with the present-day occurrence of B chromosomes in S. cereale alone.
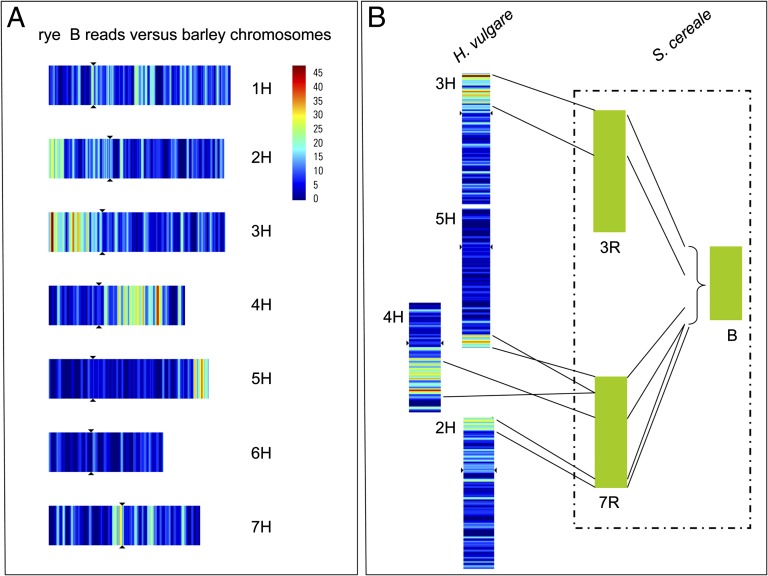
The identification of B-sequence reads with similarity to conserved coding sequences and the close syntenic relationship among grass genomes allowed us to trace the putative chromosomal origin of rye B sequences. Using a stringent filter criterion of ≥30 amino acids/100 bp similarity, we analyzed the rye B reads against the virtual gene map of barley (13) and the assembled genomes of Brachypodium, rice, and sorghum to depict the positional information on the respective chromosomes. Rye B chromosomes apparently contain several prominent blocks of conserved genes corresponding to barley chromosomal regions 2H, 3H, 4H, and 5H, along with thousands of short genic sequences scattered all over the A chromosomes (Fig. 1A). In contrast, reads from the short arm of rye A chromosome 1R (1RS) corresponded mainly to the syntenic barley chromosome 1H (Fig. S5). These results indicate that the randomly scattered pattern observed for rye B sequences is exclusive to B chromosomes and is not shared by A chromosomes. A comparison of the reads with the sequences of Brachypodium and sorghum confirmed the genome-wide scattered distribution of the rye B reads.
Fig. 1.
Multichromosomal origin of the rye B chromosome. (A) Rye B sequence reads mapped onto the barley genome. The heatmap depicts the detected homologous (syntenic) regions in the barley genome. Sequence reads were anchored on barley chromosomes 1H–7H using BLASTN and the best detectable match. Individual chromosomes are numbered. Multiple regions exhibit conserved genes with respect to barley chromosomes 2H, 3H, 4H, and 5H (implied syntenic regions) and multiple small regions on the remaining chromosomes. (B) Syntenic relationship between the A chromosomes of barley and rye. Chromosome 7R corresponds to regions from 2H, 4H, 5H ,and 7H (15). Thus, the B chromosome of rye shows extended similarity to regions of chromosomes 3R and 7R.
B Chromosomes Accumulate Large Amounts of Organellar Sequences.
The rye A and B chromosomes were further compared with respect to the content and frequency of individual classes of repeats. Repeat identification using similarity-based clustering of sequence reads (14) revealed that almost 90% of the rye genome is composed of repetitive DNA, and 70% of the genome is represented by fewer than 60 different repeat families. Although the B chromosomes contained a similar proportion of repeats as the A chromosomes, the two differed significantly in composition owing to an additional massive accumulation of B-specific satellite repeats (Fig. S6 and Table S4). The B-specific satellite repeats were characterized by exceptionally long monomers (0.9–4.0 kb), and their partial similarity to other types of repeats suggests chimeric origins. In addition to satellite repeats, an accumulation of sequences corresponding to the Bianka family of Ty1/copia elements was also observed in the B chromosomes.
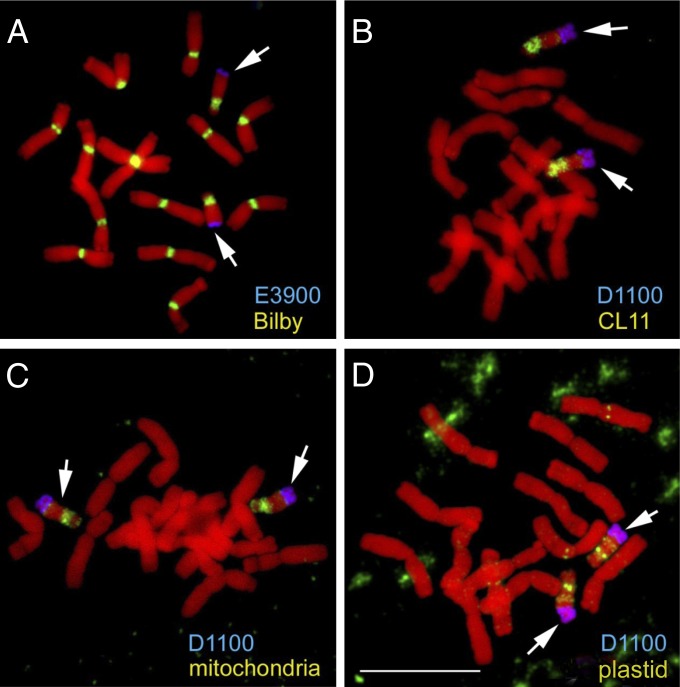
Furthermore, B chromosomes accumulated significant amounts of plastid (NUPT)- and of mitochondrion (NUMT)-derived sequences. All parts of the plastid and mitochondrion genomes were transferred to the B chromosomes, indicating that all sequences are transferable. The higher number of organelle-derived DNA inserts in B chromosomes than in A chromosomes (Fig. S7) and the increased mutation frequency of B-located organellar DNA suggest a reduced selection against organellar DNA in supernumerary chromosomes. We also observed that along with large amounts of mitochondrion-derived DNA, B-enriched high-copy repeats are integrated in the centromeric region (Fig. 2). Therefore, the centromere might facilitate the evolution of the B chromosomes by accumulation and shuffling of sequences. Whether the distinct centromere composition of the B chromosomes plays a role in the B-specific drive mechanism, resulting in non-Mendelian chromosomal segregation behavior, remains to be tested.
Fig. 2.
FISH of rye mitotic metaphase chromosomes with the centromeric retrotransposons Bilby (A), the B-specific pericentromeric Ty1/copia repeat CL11 (B), mitochondrial DNA (C), and plastid DNA (D). B chromosome-specific satellite repeats E3900 and D1100 were used for identifications of the Bs. The Bs are indicated by arrows. (Scale bar: 10 μm.)
Discussion
We have described a unique comprehensive model of B chromosome evolution based on comparative sequence analysis of the A and B chromosomes of rye. Considering the similar age of the genus Secale and the age of its B chromosomes, it is tempting to speculate that B chromosomes originated as a by-product of chromosome rearrangement events. This hypothesis is supported by the notion that the rye genome underwent a series of rearrangements after its split from the wheat and barley lineages and as such is an exception to otherwise pronounced genome colinearity in Triticeae (15). Thus, chromosomes 3R of rye and 3H of barley are mainly conserved and syntenic to each other, whereas rye 7R shares conserved synteny with barley chromosomal regions 2H, 4H, and 5H (Fig. S8). Based on the comparison of the rye B-specific sequence reads to the linear genome model of barley (13), we conclude that the rye B chromosomes originated primarily from the rye chromosomal regions 3RS and 7R after multiple chromosomal rearrangements (Fig. 1B). A multichromosomal origin of B-chromosome sequences is further supported by the many short sequences that are similar to other regions of the A chromosomes. A comparable amalgamation of diverse A-derived sequences has been previously postulated for the B chromosomes of maize (16) and Brachyscome dichromosomatica (17). The intron-containing gene reads found among the B-sequence reads, corresponding to regions outside of the 3RS and 7R regions, might represent insertions into the B chromosomes that occurred during double-strand break repair (18) or results from hitchhiking genomic fragments with transposable elements, as demonstrated for noncollinear genes of Triticeae (19).
The most unexpected result of our analysis is the discovery that B chromosomes are rich in gene fragments that represent copies of A chromosome genes. Although our analysis does not allow us to draw any conclusions regarding the completeness and functionality of the B-located genes, preliminary analyses indicate that B-located sequences are transcribed only weakly (20). Considering the coexistence of sequence-identical A- and B-derived transcripts, it is likely that “dosage compensation” occurs in rye, with an equal expression regardless of the copy number of the respective gene. An efficient dosage compensation mechanism might explain the weak phenotype caused by B chromosomes.
What mechanism could account for the accumulation of organellar DNA in B chromosomes of rye? Transfer of organellar DNA to the nucleus is very frequent (21), but most of the “promiscuous” DNA is also rapidly lost again via a counterbalancing removal process (22). If this expulsion mechanism is impaired in B chromosomes, then the high turnover rates that prevent such sequences on the A chromosomes from accumulating and degrading would be absent and allow for sequence decay. Thus, the dynamic equilibrium between frequent integration and rapid elimination of organellar DNA could be imbalanced for B chromosomes. We also observed that the large amounts of mitochondrion-derived DNA integrated preferentially in the B pericentromeric region. Pericentromeric regions generally contain few functional genes, and this low gene density may facilitate the repeated integration of the organelle-derived DNA (23). Alternatively, consistent with the rapid evolution of centromeres (24) after sequence integration, subsequent amplification of these sequences might have occurred within this region. Future analyses of other B-bearing species are needed to address the question whether organelle-to-nucleus DNA transfer is an important mechanism that drives the evolution of B chromosomes.
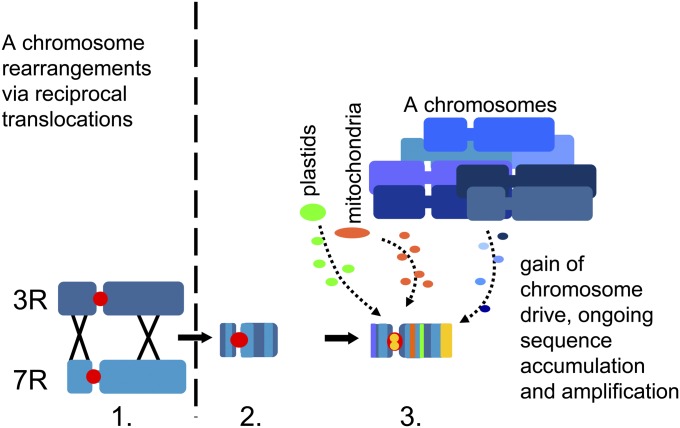
Based on our findings, we propose a multistep model for the origin of a selfish chromosome (Fig. 3). Initially, a proto-B chromosome was formed by segmental or whole-genome duplication, subsequent chromosome translocations, unbalanced segregation of a small translocation chromosome, and subsequent sequence insertions. The recombination with donor A chromosomes became restricted, likely owing to multiple rearrangements involving different A chromosomes, which no longer allowed extended pairing with the originally homologous A regions. This restriction of recombination can be considered the starting point for the independent evolution of B chromosomes. The presence of fast-evolving repetitive sequences, along with reduced selective pressure on gene integrity, could predispose a nascent B chromosome to undergo further rapid structural modifications required to establish a drive mechanism. Because an increased gene dosage may affect gene expression, the expression of paralogues on B chromosomes might have been reprogrammed (potentially through epigenetic mechanisms) early during the evolution of the B chromosomes. Thus, proto-B genes might have first been suppressed by silencing mechanisms and then degenerated owing to mutations and the insertion of sequences derived from other A-chromosomal regions and organellar genomes, except for those coding and/or noncoding sequences providing drive and an advantage for the maintenance of B chromosomes. Our model predicts that B chromosomes occur primarily in taxa with elevated levels of chromosomal rearrangements and phylogenetic groups with unstable chromosome numbers.
Fig. 3.
Model of the stepwise evolution of the rye B chromosome after segmental genome duplication. (1) Reciprocal translocation of duplicated fragments of the 3R and 7R chromosomes and unbalanced segregation of a small translocation chromosome results in (2) decay of meiotic A–B pairing and the formation of a proto-B. (3) The accumulation of organellar and A chromosome-derived DNA fragments, amplification of B-specific repeats, erosion and inactivation of A-derived genes (Muller's ratchet), and gain of chromosome drive resulted in the B chromosome.
Materials and Methods
Purification of Mitotic Chromosomes and 454 Sequencing.
A and B chromosomes of rye (S. cereale) inbred line 7415 (25) were isolated by flow cytometry sorting and shotgun sequenced by Roche 454 (11, 13).
Analysis of Repetitive DNA and Organellar DNA Insertions.
The content of the repetitive DNA per sequence read was identified by Vmatch (http://www.vmatch.de) against the MIPS-REdat Poaceae v8.6.1 repeat library. The clustering analysis of sequence reads was performed as described previously (14). The A and B sequence reads were compared (BLASTN) against the plastid and mitochondrial genomes of wheat (AB042240 and AP008982).
Identification of Gene Reads and Comparative Genomics.
Gene numbers were estimated by BLAST comparisons with the repeat-filtered reads against the proteins/coding sequences of B. distachyon, rice (O. sativa), and sorghum (S. bicolor) and against EST collections. Rye 1RS (EMBL-EBI European Bioinformatics Institute, http://www.ebi.ac.uk, NCBI accession no. SRX019678) and B chromosome datasets were compared with reference genomes (BLASTX) as described previously (13).
Detection of SNPs and Dating of Rye B Chromosome Origin.
Genic 454 shotgun reads from A and B chromosomes were mapped against matching sequences from the rye transcriptome dataset using BWA (26). An SNP-based comparison of A and B chromosomes was performed to date the age of the rye B. The regions that contained high-quality SNPs in A and B were mapped onto the corresponding barley full-length cDNAs (26, 27) and Brachypodium reference genome version 1.2 using BLASTN. The datasets were phylogenetically analyzed with Bayesian inference in MrBayes 3.1.2 (28) and dated in BEAST 1.6.1 (29). More detailed descriptions of methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank I. Schubert, R. N. Jones, M. Puertas, J. Timmis, D. Weigel, J. Birchler, and B. Steuernagel for fruitful discussions; S. König for excellent technical support of the 454 sequencing of the rye A and B chromosomes; K. Burg for sequence information for 1RS; and J. Číhalíková, R. Šperková, and Z. Dubská for their help with chromosome sorting. This work was supported by the German Research Foundation Grant HO 1779/10-1/14-1; German Federal Ministry of Education and Research Grant FKZ 0315063B (Tritex and Gabi Rye Express Projects 0315954C and 0315063C); Czech Science Foundation Grant P501/12/G090; Ministry of Education, Youth and Sports of the Czech Republic Grant OC10037; European Regional Development Fund Operational Programme Research and Development for Innovations Grant ED0007/01/01; and Academy of Sciences of the Czech Republic Grant AVOZ50510513.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive database, http://www.ebi.ac.uk/ena/ (accession no. ERP001061).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204237109/-/DCSupplemental.
References
- 1.Wilson EB. The supernumerary chromosomes of Hemiptera. Science. 1907;26:870. [Google Scholar]
- 2.Hayman DL, Martin PG. Supernumerary chromosomes in the marsupial Schoinobates volans (Kerr) Aust J Biol Sci. 1965;18:1081–1082. doi: 10.1071/bi9651081. [DOI] [PubMed] [Google Scholar]
- 3.Longley AE. Supernumerary chromosomes in Zea mays. J Agric Res. 1927;35:769–784. [Google Scholar]
- 4.Yu W, Lamb JC, Han F, Birchler JA. Telomere-mediated chromosomal truncation in maize. Proc Natl Acad Sci USA. 2006;103:17331–17336. doi: 10.1073/pnas.0605750103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camacho JPM, Sharbel TF, Beukeboom LW. B-chromosome evolution. Philos Trans R Soc Lond B Biol Sci. 2000;355:163–178. doi: 10.1098/rstb.2000.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones N, Houben A. B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci. 2003;8:417–423. doi: 10.1016/S1360-1385(03)00187-0. [DOI] [PubMed] [Google Scholar]
- 7.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Belknap Press of Harvard Univ Press; 2006. p. 602. [Google Scholar]
- 8.Niwa K, Sakamoto S. Origin of B chromosomes in cultivated rye. Genome. 1995;38:307–312. doi: 10.1139/g95-038. [DOI] [PubMed] [Google Scholar]
- 9.Timmis JN, Ingle J, Sinclair J, Jones RN. Genomic quality of rye B chromosomes. J Exp Bot. 1975;26:367–378. [Google Scholar]
- 10.Tsujimoto H, Niwa K. DNA structure of the B chromosome of rye revealed by in situ hybridization using repetitive sequences. Jpn J Genet. 1992;67:233–241. [Google Scholar]
- 11.Kubaláková M, et al. Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome. 2003;46:893–905. doi: 10.1139/g03-054. [DOI] [PubMed] [Google Scholar]
- 12.Haseneyer G, et al. From RNA-seq to large-scale genotyping: Genomics resources for rye (Secale cereale L.) BMC Plant Biol. 2011;11:131. doi: 10.1186/1471-2229-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer KF, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23:1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novák P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devos KM, et al. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet. 1993;85:673–680. doi: 10.1007/BF00225004. [DOI] [PubMed] [Google Scholar]
- 16.Peng SF, Lin YP, Lin BY. Characterization of AFLP sequences from regions of maize B chromosome defined by 12 B-10L translocations. Genetics. 2005;169:375–388. doi: 10.1534/genetics.104.032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houben A, Verlin D, Leach CR, Timmis JN. The genomic complexity of micro B chromosomes of Brachycome dichromosomatica. Chromosoma. 2001;110:451–459. doi: 10.1007/s00412-001-0173-1. [DOI] [PubMed] [Google Scholar]
- 18.Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wicker T, et al. Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell. 2011;23:1706–1718. doi: 10.1105/tpc.111.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carchilan M, Kumke K, Mikolajewski S, Houben A. Rye B chromosomes are weakly transcribed and might alter the transcriptional activity of A chromosome sequences. Chromosoma. 2009;118:607–616. doi: 10.1007/s00412-009-0222-8. [DOI] [PubMed] [Google Scholar]
- 21.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 22.Sheppard AE, Timmis JN. Instability of plastid DNA in the nuclear genome. PLoS Genet. 2009;5:e1000323. doi: 10.1371/journal.pgen.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuo M, Ito Y, Yamauchi R, Obokata J. The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast-nuclear DNA flux. Plant Cell. 2005;17:665–675. doi: 10.1105/tpc.104.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall AE, Keith KC, Hall SE, Copenhaver GP, Preuss D. The rapidly evolving field of plant centromeres. Curr Opin Plant Biol. 2004;7:108–114. doi: 10.1016/j.pbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Jimenez MM, Romera F, Puertas MJ, Jones RN. B-chromosomes in inbred lines of rye (Secale cereale L), 1: Vigor and fertility. Genetica. 1994;92:149–154. [Google Scholar]
- 26.Matsumoto T, et al. Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 2011;156:20–28. doi: 10.1104/pp.110.171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato K, et al. Development of 5006 full-length CDNAs in barley: A tool for accessing cereal genomics resources. DNA Res. 2009;16:81–89. doi: 10.1093/dnares/dsn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 29.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.