Abstract
A wide range of membrane receptors signal through conformational changes, and the resulting protein conformational flexibility often hinders their structural studies. Because the determinants of membrane receptor conformational stability are still poorly understood, identifying a minimal set of perturbations stabilizing a membrane protein in a given conformation remains a major challenge in membrane protein structure determination. We present a novel approach integrating bioinformatics, computational design and experimental techniques that identifies and stabilizes metastable receptor regions. When applied to the beta1-adrenergic receptor, the method generated 13 novel receptor variants stabilized in the intended inactive state among which two exhibit an apparent thermostability higher than WT and M23 (a receptor variant previously stabilized by extensive scanning mutagenesis) by more than 30 °C and 11 °C, respectively. Targeted regions involve nonconserved unsatisfied polar residues or exhibit significant packing defects, features found in all class A G protein-coupled receptor structures. These findings suggest that natural G protein-coupled receptor sequences have evolved to be conformationally metastable through the design of suboptimal polar and van der Waals tertiary interactions. Given sufficiently accurate structural models, our approach should prove useful for designing stabilized variants of many uncharacterized membrane receptors.
Keywords: computational protein design, protein conformational stability, membrane protein modeling, signal transduction, synthetic biology
Membrane proteins represent around 30% of currently sequenced genomes and are critical in the regulation of cell signaling and cell–cell communication (1, 2). When dysfunctional, however, these proteins can be responsible for serious diseases and constitute more than 60% of current drug targets. Despite their abundance and functional importance, membrane proteins are largely underrepresented in the Protein Data Bank, mainly due to technical difficulties in studying these proteins. A large fraction of these proteins such as G protein-coupled receptors (GPCRs) transduces signals across membranes through conformational changes (3–6). The resulting protein conformational heterogeneity and flexibility is a major factor hindering crystallization. An additional obstacle to structural determination is the poor stability of membrane proteins in detergent solution required by traditional crystallization approaches (7, 8).
In recent years, several approaches to stabilize membrane proteins have been explored (9). These methods include development and optimization of solubilizing detergents (10), reconstitution of proteins into lipidic bicelles (11, 12), cleavage of flexible protein regions (13–18), co-crystallization with stabilizing ligands, antibodies, and fusion with soluble proteins (e.g., T4 lysozyme) (13–15, 17, 19–23). Several of these modifications, however, disrupt important functional regions and interactions with either natural ligands or downstream effectors (9).
Mutations stabilizing membrane proteins have also been discovered using various methods. Directed molecular evolution, alanine, leucine scanning, or random mutagenesis approaches have identified stabilizing amino acid substitutions of numerous membrane proteins (7, 24–27). However, these approaches are both very time-consuming and limited in their ability to explore large sequence spaces. Typical libraries of random mutations are limited in size and allow exploring the sequence space for only a fraction of all the residues in large membrane receptors. Alanine scanning mutagenesis may be biased toward selecting surface residues, because single-point mutations in the interior of the receptor will most likely lead to packing defects. Scanning single substitutions at a time is unlikely to select novel stabilizing interaction networks involving multiple residues.
In contrast to water-soluble proteins, membrane proteins exist in a largely hydrophobic environment and, once inserted into the membrane, must complete the folding process in the absence of significant hydrophobic effect. Various studies of membrane protein stability have pointed to van der Waals (VDW) and hydrogen bond interactions as key factors contributing to folding and overall stability (28–30). Polar substitutions are the most prevalent disease-associated transmembrane (TM) mutations (31) and can perturb both membrane protein structure and function through strong nonnative polar interactions. However, attempts to measure the contributions of hydrogen bonds in membrane protein stability have yielded conflicting results. While several studies have reported substantial contribution by hydrogen bonds in the dimerization of TM domains (30, 32, 33), others indicate only modest stabilization by side-chain mediated polar interactions (34, 35). These findings highlight the importance of the structural context in modulating the strength of polar interactions. In principle, by modeling protein structures at atomic resolution, computational structure-based design techniques can predict the energetics of hydrogen bonding and VDW interactions and engineer stabilizing mutations. However, such techniques have only been developed to design simple hydrophobic or cofactor-bound TM peptides (36–38) so far and have never been applied to large polytopic membrane proteins (28, 39–41).
To address these limitations, we have developed a novel computational/experimental approach, which identifies metastable regions and designs stabilizing mutations in large membrane receptors. As a stringent proof of concept, we have applied our method to stabilize the WT beta1-adrenergic receptor (B1AR-WT) and a variant (B1AR-M23) previously thermostabilized by extensive scanning mutagenesis (26). Sequence/structure analysis of all class A GPCRs suggests that GPCR sequences have evolved to occupy metastable conformations.
Results
In this section, we describe the rationale and validation of a novel computational/experimental approach developed to identify and stabilize metastable regions in large multi-pass membrane proteins.
Rationale.
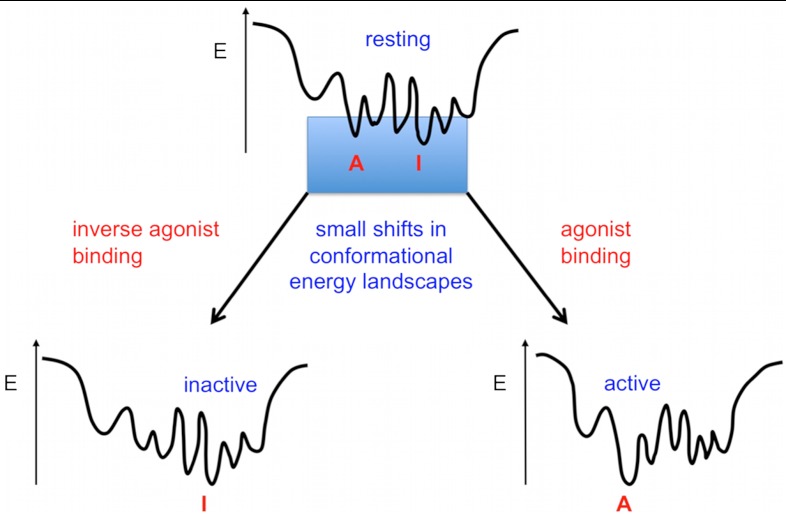
Many membrane receptors function through conformational changes induced by the transient binding of small ligands providing limited binding energy to shift conformational energy landscapes. Therefore, we hypothesize that natural sequences of such receptors have evolved to occupy multiple relatively isoenergetic metastable conformations in the resting state (Fig. 1). If this hypothesis is valid, physical interactions stabilizing each receptor conformation should be suboptimal and substitutions that stabilize specific conformations may be readily identified. The availability of a significant number of GPCR structures allowed us to seek sequence/structure correlations and general molecular determinants of conformational metastability across the class A GPCR family. Our present study focuses on suboptimal hydrogen bonds and van der Waals tertiary interactions, which represent two of the main physical forces stabilizing membrane protein structures (28, 29, 40, 41).
Fig. 1.
Conformational energy landscape of receptor signaling triggered by transient ligand binding. In this hypothetical landscape, a receptor in the resting state occupies an ensemble of conformations close in energy (i.e., metastable). Even relatively modest energy provided by transient ligand binding can lead to shifts in conformational energy landscape that are sufficient to significantly alter the relative population of receptor in distinct conformational states. I and A refer to hypothetical conformations encoding either inactive or active functional states of the receptor.
Prediction of Putative Metastable Regions in B1AR.
Nonconserved unsatisfied polar residues.
To avoid disrupting any polar interactions playing universal structural and functional role in GPCRs, we selected residues exhibiting low sequence conservation in the entire class A GPCR family. Multiple sequence alignments across a representative sample of 270 Class A GPCRs (42) were analyzed to identify nonconserved polar residues in the transmembrane region (i.e., found in less than 20% of the representative Class A GPCRs). The B1AR crystal structure (18) was then analyzed to select among these nonconserved sites unsatisfied polar residues—i.e., not significantly stabilized by hydrogen bonds or electrostatic interactions (SI Methods). Excluding residues that potentially contact ligands, six polar residues satisfying these constraints were found in B1AR (Table S1 and Fig. S1). Of these nonconserved polar residues, two are specific to the beta-adrenergic receptor subfamily (E3.41, T3.42), whereas the others are found across multiple subfamilies but are represented in less than 20% of Class A GPCRs (Figs. S1–S3).
Regions exhibiting packing defects.
The RosettaHoles software (43) and the directional atomic contact analysis method (44) (SI Methods) were used to quantitatively measure packing scores of each residue in the receptor structure. Packing defects were mainly found in three partially buried regions involving the following transmembrane helix (TMH) interfaces: TM2/3 including residue T3.42, TM4/5 including residue V4.56, TM5/6 including residues G6.42 and T6.45, and the cytoplasmic side of TM2/6/7 (Figs. S4 and S5).
Computational Selection of Stabilizing Substitutions.
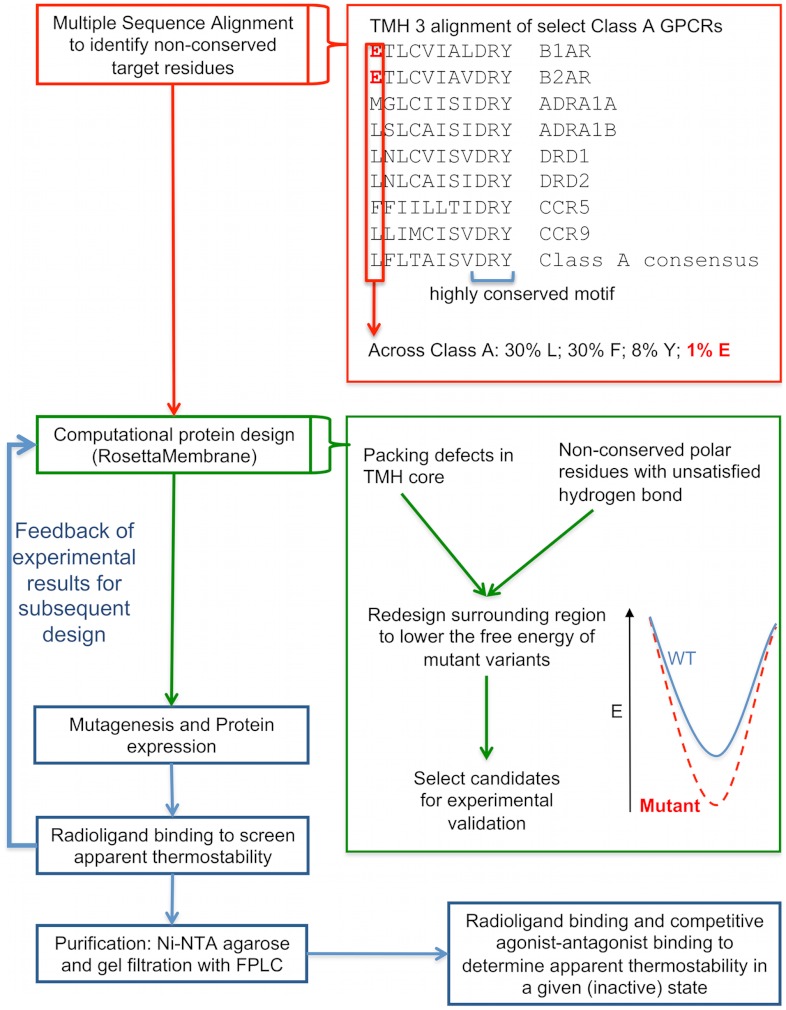
The nonconserved unsatisfied polar and poorly packed metastable sites identified as described above served as starting points for designing conformational stability improvements. The four molecules present in the asymmetric unit of the crystal of the inactive-state B1AR-M23 (18) were used to generate four monomeric starting conformations of the receptor. The design mode of RosettaMembrane (SI Methods) (45) was applied to predict mutations that decrease the energy of the inactive B1AR structure (Fig. 2). Three to eight positions were typically redesigned around each predicted metastable site allowing all possible 20 amino acids at each designable position (SI Methods). Designed solutions were screened for false positives using two criteria: (i) Mutations exhibiting minimal decrease in energy were systematically reverted to wild-type amino acids; (ii) designed solutions exhibiting pathological features (such as disruption of native interactions) and packing defects (43) were discarded (Fig. 2 and SI Methods). This filtering step reduced the number of substitutions for each design, resulting in four solutions with single substitutions (E3.41L; T3.42I; V4.56L; C2.48I), four with pairwise substitutions (E3.41L.S4.53A; T3.37S.E3.41Y; G6.42L.T6.45I; C4.47L.T4.48L), and one triple mutant design (T3.37S.E3.41Y.S4.53A) (Table 1). Subsequent designs sought to combine these mutations leading to two hyperstable variants that consisted of five (E3.41L.S4.53A.V4.56L.G6.42L.T6.45I) and seven (T3.37S.E3.41Y.T3.42I.S4.53A.V4.56L.G6.42L.T6.45I) substitutions compared to WT or M23 (Table 1). Selected variants were validated experimentally by determining their melting temperature (Tm) under saturating concentration of antagonist to assess their apparent conformational stability in the inactive state (Fig. 2).
Fig. 2.
Integrated computational/experimental approach to design stabilized membrane proteins. (Top, red) Multiple sequence alignments (MSA) across class A GPCRs are analyzed to identify potential target sites for stabilization (i.e., nonconserved residues). Top Inset: MSA of Class A GPCRs transmembrane helix 3 (TM3) exhibit a highly conserved motif (DRY) and a nonconserved polar residue specific to beta-adrenergic receptors (E3.41). (Middle, green) Structural analysis is first performed to (i) select unsatisfied among nonconserved polar residues (i.e., nonstabilized by hydrogen bond or electrostatic interactions) and (ii) identify local regions with packing defects. RosettaMembrane is then used to redesign these target regions to identify amino-acid substitutions minimizing the energy of the inactive B1AR structure (middle inset). Designs are subsequently refined and filtered for false positives (SI Methods). (Bottom, blue) Rapid experimental validation using analytical radioligand binding assays determines the apparent melting temperature (Tm) of selected receptor variants. The results are fed back into the design process for subsequent rounds of computational design combining and accommodating locally stabilized regions. The variants resulting in the highest apparent conformational stability are purified to accurately measure their apparent Tm and to assess their conformational stabilization using agonist-antagonist competition binding assays.
Table 1.
Apparent conformational stability of B1AR variants
| B1AR variant | Apparent Tm (°C) | ΔTm vs B1AR-M23 (°C) | ΔTm vs B1AR-WT (°C) |
| WT | 32.4 ± 1.1 | −19.3 | na |
| M23 | 51.7 ± 0.6 | na | +19.3 |
| M23+E3.41L | 58.2 ± 1.2 | +6.5 | +25.8 |
| M23+E3.41L.S4.53A | 59.4 ± 0.5 | +7.7 | +27.0 |
| M23+T3.37S.E3.41Y | 58.6 ± 1.1 | +6.9 | +26.2 |
| M23+T3.37S.E3.41Y.S4.53A | 60.4 ± 1.9 | +8.7 | +28.0 |
| M23+T3.42I | 54.2 ± 1.4 | +2.5 | +21.8 |
| M23+G6.42L.T6.45I | 55.0 ± 0.9 | +3.3 | +22.6 |
| M23+V4.56L | 53.6 ± 1.2 | +1.9 | +21.2 |
| M23+C2.48I | 52.0 ± 1.5 | +0.3 | +19.6 |
| M23+C4.47L.T4.48L | 56.3 ± 0.9 | +4.6 | +23.9 |
| M23+E3.41L.S4.53A.V4.56L | 60.6 ± 1.0 | +8.9 | +28.2 |
| M23+E3.41L.S4.53A.V4.56L.G6.42L.T6.45I (M23-5m) | 62.8 ± 0.8 | +11.1 | +30.4 |
| M23+T3.37S.E3.41Y.T3.42I.S4.53A.V4.56L.G6.42L.T6.45I (M23-7m) | 63.1 ± 0.8 | +11.4 | +30.7 |
| WT+E3.41L.S4.53A.V4.56L.G6.42L.T6.45I (WT-5m) | 40.7 ± 0.5 | na | +8.3 |
| WT+T3.37S.E3.41Y.T3.42I.S4.53A.V4.56L.G6.42L.T6.45I (WT-7m) | 38.0 ± 0.8 | na | +5.6 |
| M23+S2.45A.T3.42I.W4.50L | 46.1 ± 6.5 | −5.6 | na |
| M23+C2.48A | 45.7 ± 0.4 | −6.0 | na |
| M23+C4.47A.T4.48A | 51.1 ± 1.4 | −0.6 | na |
Apparent melting temperatures (Tms) of B1AR variants were determined by monitoring antagonist binding to detergent solubilized receptor incubated at increasing temperatures. Assays were performed as described in SI Methods. Designed variants are categorized (as depicted by horizontal spaces) into (i) initial designs optimizing local regions in B1AR M23 exhibiting unsatisfied, non-conserved polar residues or packing defects, (ii) subsequent combinations of redesigned local regions in B1AR M23 to achieve maximal stabilization, (iii) combinations of redesigned local regions in B1AR WT to achieve maximal stabilization, (iv) designs targeting conserved polar residues (i.e., S2.45, W4.50) along with alanine substitution at surface exposed sites in category 1 (i.e., C2.48, C4.47, T4.48) were negative controls that result in destabilization. Results are reported as mean ± standard error with N = 4.
Stabilizing Designs Target Nonconserved Polar Residues and Packing Defects.
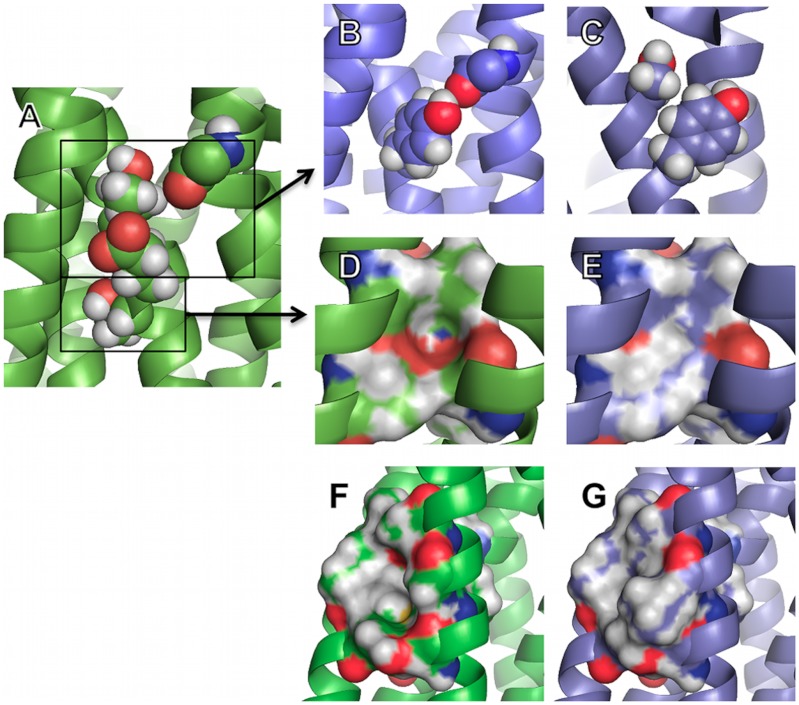
All designs targeting nonconserved polar residues in the receptor core (E3.41L, T3.37S/E3.41Y, T3.42I, G6.42L/T6.45I; Fig. 3) led to a significant increase in stability (2–6 °C increase over B1AR-M23) (Table 1). Conversely, designs that modified conserved polar residues (S2.45A.T3.42I.W4.50L) led to a decrease in apparent Tm (ΔTm = -6 °C versus B1AR-M23) (Table 1). Designs that improved the packing in the receptor core (V4.56L, T3.42I, G6.42L/T6.45I; Fig. 3) also resulted in significant increases in apparent Tm (2–3 °C increase over B1AR-M23). These variants all exhibited an increased local packing score (+0.08, +0.05, and +0.11, respectively) compared to B1AR-M23.
Fig. 3.
Conformational stabilization of B1AR by optimization of polar and van der Waals interhelical interactions. WT and designed structures are colored in green and blue, respectively. (A–E) Unsatisfied polar residues and packing defects at TM2/4/5 interface. (A) E3.41 and T3.37 are nonconserved and unsatisfied polar residues in B1AR WT. (B) Y3.41 improves the packing, eliminates an unsatisfied polar group, and creates a close to optimal hydrogen bond with a backbone unsatisfied carbonyl group on TM4. (C) S3.37, unlike T3.37, accommodates Y3.41 conformation. (D) Surface representation of T3.42 and associated packing defect. (E) I3.42 improves the packing and eliminates an unsatisfied polar group. (F and G) Packing defects at TM5/6 interface. (F) G6.42 and T6.45 create a packing defect between TM5 and TM6 in B1AR WT. (G) The redesigned stabilized region consists of two substitutions: L6.42 and I6.45, which optimize the VDW interactions between TM5 and TM6.
Both Local Regions Buried in the Receptor Core or Exposed at the Receptor Surface Can Be Stabilized.
In addition to designs targeting residues buried in the receptor core, we attempted to stabilize the receptor by targeting surface-exposed positions. Our method identified a few nonconserved unsatisfied surface-exposed cysteine and threonine residues (i.e., C2.48, C4.47, and T4.48). Redesigning local regions around these sites led to two variants: C4.47L.T4.48L significantly stabilized compared to B1AR-M23 (ΔTm = +4 °C) and C2.48I as stable as B1AR-M23 (Table 1). As a control, these sites were also substituted to alanine, and the corresponding mutants were all destabilized compared to B1AR-M23 (C4.47A.T4.48A: ΔTm = -1 °C; C2.48A: ΔTm = -6 °C). These results suggest that RosettaMembrane can also select novel stabilizing physical interactions at the receptor surface. The improved stability observed in C2.48I and C4.47L.T4.48L were predicted to come mainly from (i) an increase in Van der Waals interactions resulting from additional atomic contacts provided by the larger hydrophobic residues substituting the small polar natives (a few atoms at each of the redesigned sites become buried upon mutation) and (ii) an improved solvation in the hydrophobic core of the membrane due to mutations of polar to hydrophobic residues.
Stabilizing Mutations Can Be Combined To Generate Hyperstable B1AR Variants.
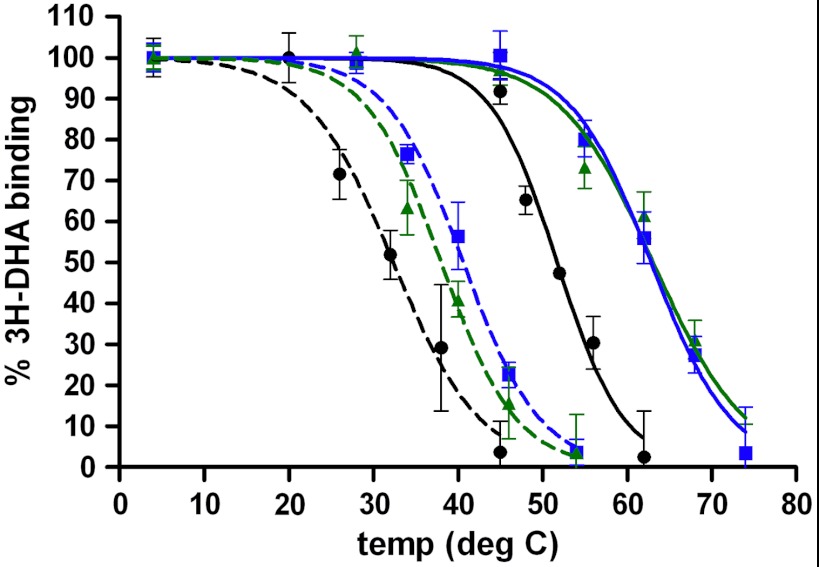
Designs targeting individual metastable sites and successfully cross-validated experimentally were combined to generate two hyperstable variants. M23-7m (M23+T3.37S.E3.41Y.T3.42I.S4.53A.V4.56L.G6.42L.T6.45I) showed a large increase in apparent thermostability (Tm = 63.1 ± 0.8 °C) compared to B1AR-M23 (ΔTm = +11.4 °C) and B1AR-WT (ΔTm = +30.7 °C) (Table 1 and Fig. 4). Additionally, M23-5m (M23+E3.41L.S4.53A.V4.56L.G6.42L.T6.45I) displayed similar increase in apparent thermostability (Tm = 62.8 ± 0.8 °C) (Table 1 and Fig. 4).
Fig. 4.
Hyperstabilized B1AR variants. Melting curves of B1AR variants were obtained by measuring tritiated antagonist (3H-DHA) binding to detergent solubilized receptor incubated at increasing temperatures. Assays were performed as described in SI Methods. When designed in WT B1AR (dotted lines), 5 m (blue square) and 7 m (green triangle) variants exhibited Tm increases of 8 and 6 °C over B1AR-WT (black circle), respectively. When designed in the M23 background (solid lines), 5 m (blue square), and 7 m (green triangle) variants both exhibited Tm increases of 11 °C over B1AR-M23 (black circle, solid line) and 31 °C over B1AR-WT (black circle, dotted line). Error bars show the standard errors N = 4.
Designed B1AR-M23 Variants are Stabilized in the Intended Inactive Conformation.
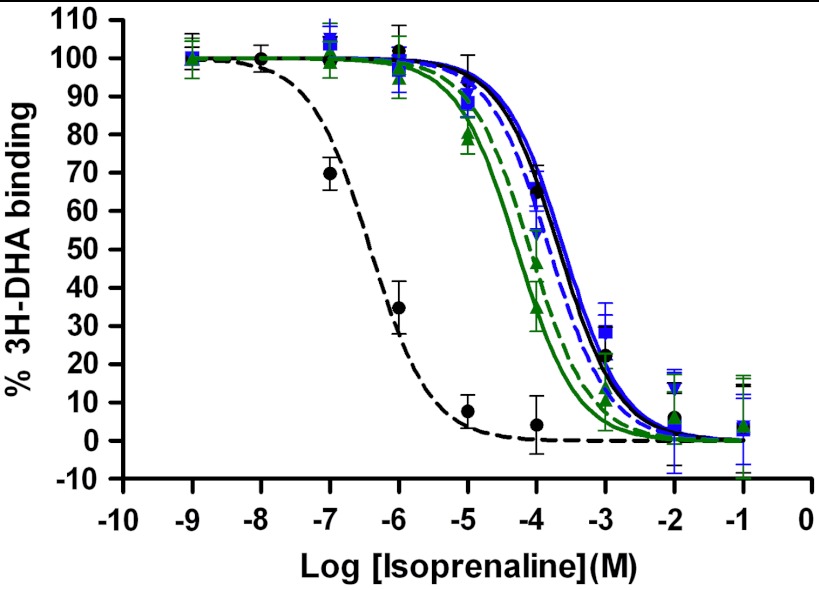
Competition binding assays using increasing concentrations of the agonist isoprenaline against [3H]-DHA revealed increased EC50 values for the B1AR variants (Fig. 5). Compared to WT (EC50 = 389 nM), M23-7m and M23-5m (EC50 = 52 μM, 251 μM) preferentially bind the DHA antagonist, indicating that the variants were stabilized in the intended inactive conformation.
Fig. 5.
Designed B1AR variants are stabilized in the intended inactive state. Competition binding assays of adrenergic receptor agonist (isoprenaline) against antagonist (tritiated dihydroalprenolol) were performed as described in SI Methods. The 5 m (blue square) and 7 m (green triangle) combined mutant designs in both the WT (dotted lines) and M23 (solid lines) backgrounds exhibited significant isoprenaline EC50 increases over B1AR-WT (black circle, dotted line, EC50 = 389 nM), with EC50 shifts similar to that of B1AR-M23 (black circle, solid line, EC50 = 214 μM). The EC50 values for the mutant designs are WT-5m (154 μM), WT-7m (81 μM), M23-5m (250 μM), M23-7m (52 μM). Error bars show the standard errors N = 4.
Stabilization of B1AR-WT in the Inactive State.
In addition to further stabilizing the B1AR-M23 variant, we tested the ability of our technique to identify mutations that preferentially stabilize the receptor inactive state starting from the structurally uncharacterized WT sequence. According to its relatively high affinity for agonist (low EC50 of 389 nM) and constitutive activity (46), the WT receptor, unlike the M23 variant, has a significant propensity to occupy the active state. We first modeled the WT receptor from the M23 X-ray structure and performed in silico mutagenesis using the design mode of RosettaMembrane described in this study. Designed stabilizing mutations were identified among which several were similar to those selected in the M23 background. We selected and tested two variants, WT-5m and WT-7m (where 5 m and 7 m correspond to the same mutations than in the M23-5m and M23-7m variants). WT-5m and WT-7m variants both exhibited the intended increase in apparent thermostability (ΔTm of 8.3 °C and 5.6 °C compared to WT; Fig. 4) and the intended loss in agonist affinity (EC50 of 154 μM and 81 μM versus 389 nM for WT; Fig. 5). The new variants designed in the WT background indicate that the method can not only stabilize a native receptor sequence but also modulate its pharmacological properties.
Prediction of Experimentally Selected Stabilizing Mutations in GPCRs.
In addition to designing novel stabilizing mutations, the method could also predict the thermostabilizing effects of 70% (14 among 20) of the substitutions identified by scanning mutagenesis to generate the B1AR-M23 (26) and the adenosine A2 (A2AR) Rant receptor variants (47) (SI Text and Fig. S6).
Metastable Motifs are Predicted in all Structurally Characterized Class A GPCRs but are Found in Regions Specific to Each Member or Subfamily.
Sequence bioinformatics [i.e., evolutionary trace (48)] and structural analysis were performed on Class A GPCRs for which structures are available (bovine rhodopsin, squid rhodopsin, B1AR, B2AR, A2AR, CXCR4, D3DR, H1HR, and M2AR) (SI Methods).
As shown in Table S1, around 30% of all polar residues present in the TM regions and not involved in ligand binding are conserved in less than 20% class A GPCRs (SI Methods). Among these, around 90% (an average of 6 residues per GPCR) are unsatisfied (i.e., stabilized by less than 0.2 kcal/mol in hydrogen bond energy). Evolutionary trace analysis (48) revealed that a majority of these residues have specific locations and encode distinct physical interactions in each GPCR subfamily (SI Methods and Figs. S1–S3). Partially buried locations of these nonconserved polar residues cluster at the following TM interfaces: bovine rhodopsin (TM1/7, TM3/4/5), squid rhodopsin (TM2/3/4, TM1/2/7, TM6/7), B1AR (TM3/4/5, TM5/6), B2AR (TM3/4/5, TM5/6, TM6/7), A2AR (TM2/3/4, TM3/5, TM1/7), CXCR4 (TM2/3/4, TM5/6, TM2/7), D3DR (TM2/3/4, TM3/5, TM5/6), HH1R (TM2/3/4 TM3/5, TM5/6, TM1/7), M2AR (TM2/3/4, TM3/4/5, TM5/6, TM6/7, TM1/2/7).
In class A GPCR structures, packing defects were found in diversely located partially buried regions involving the following TMH interfaces (Figs. S4 and S5): bovine rhodopsin (TM2/3/4, TM3/4/5, TM2/6, TM1/7, TM6/7), squid rhodopsin (TM2/4, TM3/4/5, TM5/6, TM2/6/7), B2AR (TM3/4, TM4/5, TM6/7, TM2/6/7), A2AR (TM2/3/4, TM3/4/5, TM3/6/7), CXCR4 (TM2/3, TM4/5, TM5/6, TM6/7, TM3/6), D3DR (TM3/4/5, TM3/5/6), HH1R (TM2/3/4, TM3/4/5, TM3/5/6, TM6/7), M2AR (TM2/3/4, TM3/5/6, TM6/7).
This analysis indicates that polar and packing determinants for metastability are distributed in distinct local regions among class A GPCRs.
Discussion
We have developed a novel integrated computational/experimental method to uncover the atom-level determinants governing membrane receptor conformational stability and to rationally design conformationally stabilized receptor variants (Fig. 2). To stringently test our method, we targeted B1AR, a GPCR for which the M23 variant had been previously thermostabilized by extensive scanning mutagenesis (26). We identified several putative metastable regions characterized by nonconserved sequence motifs establishing suboptimal polar and VDW tertiary interactions (Fig. 3). Mutations were designed at these sites to generate 13 novel B1AR variants stabilized in the intended inactive conformation (Table 1). By combining multiple locally stabilized regions, two hyperstabilized variants were generated exhibiting an apparent melting temperature (Tm) higher than that of WT and M23 by up to 30.7 °C and 11.4 °C, respectively (Fig. 4 and Table 1). Mutations designed in the WT receptor background induced an up to 396-fold loss in agonist affinity, indicating that the method can modulate the pharmacological properties of WT receptor sequences (Fig. 5). In addition to designing novel stabilizing mutations, the method was able to predict the thermostabilizing effects of 70% of the substitutions previously identified by experimental scanning mutagenesis to generate the stabilized B1AR-M23 (26) and A2AR Rant receptor variants (47) (Fig. S6). Bioinformatics and structural analysis on all available class A GPCR sequences and structures revealed the recurrent presence and subfamily/member specific location of metastable motifs.
A number of empirical and time-consuming approaches have been explored in the past to stabilize membrane receptors. However, because of the lack of rational and limited ability to explore large sequence spaces, these studies have provided limited insights into the determinants of conformational stability and moderate stabilization of membrane receptors. We report here an integrated computational/experimental approach to uncover the sequence/structure/energetic relationships governing the conformational stability of large membrane receptors and to rationally design hyperstable receptors (Fig. 2). Our computational design technique allows the exploration of large sequence spaces and the selection of multiple substitutions at a time, as highlighted by the identification of novel stabilizing physical interaction networks in the receptor core (e.g., T3.37S.E3.41Y.S4.53A, G6.42L.T6.45I, E3.41L.S4.53A; Table 1). The energy function models membrane protein atom-level interactions explicitly and lipid–protein interactions implicitly allowing for the fast and accurate selection of stabilizing motifs at specific location in the membrane. As highlighted by the E3.41Y substitution, polar residues are selected only if they can form optimized polar interactions compensating for the substantial desolvation cost of burying these residues in the hydrophobic core of the membrane. As a striking validation of our predictions for metastable motifs, all local regions bearing partially buried nonconserved unsatisfied polar residues or exhibiting packing defects were stabilized with a minimal number of substitutions. All substitutions in these regions predicted to be stabilizing were successfully cross-validated experimentally (Table 1). As a negative control, targeting conserved polar residues (e.g., S2.45, W4.50) strongly destabilized the receptor and led to uncooperative unfolding (Table 1). Stabilizing the receptor through the redesign of surface-exposed regions proved also quite successful. Attempts focused on exposed nonconserved and unsatisfied cysteine or threonine residues that led to receptor variants either significantly stabilized or as stable as the B1AR-M23 variant (Table 1). By contrast, simple alanine substitutions of these residues performed as a control led to significant decrease in stability compared to B1AR-M23 (Table 1). These results indicate that RosettaMembrane can design stabilizing physical interactions not only in the core but also at the surface of the receptor. Overall, the ability to generate several B1AR variants stabilized in the inactive state by up to 30.7 °C compared to WT strongly suggest that naturally evolved GPCR sequences are far from optimal for conformational stability.
In absence of hydrophobic effect in the lipid membrane, the forces stabilizing membrane proteins are dominated by short-range van der Waals, hydrogen-bond, and longer range electrostatic interactions. In an elegant study (34), Bowie and coworkers found that the contribution of eight hydrogen bonds to the stability of Bacteriorhodopsin range from 0.2 to 1.8 kcal/mol with an average of 0.6 kcal/mol. While this study suggests that polar interactions are not always highly optimized in membrane proteins, Bacteriorhodopsin is a relatively stable prokaryotic membrane protein (49), for which substantial conformational changes upon function have not been consistently observed (50), and the targeted residues were conserved and functionally important. In our study, we instead sought to uncover general determinants governing the conformational regulation of eukaryotic membrane receptor signaling. We specifically focused on non-conserved sequence/structure motifs unlikely to be involved in important conserved structural or functional properties. Strikingly, around 90% of non-conserved polar residues in structurally characterized GPCRs were found unsatisfied (stabilized by less than 0.2 kcal/mol) (Table S1). Numerous packing defects were also found in all GPCR structures. These findings strongly suggest that the design of suboptimal tertiary VDW and polar interactions is a general mechanism encoding metastability in naturally evolved GPCRs. Combination of evolutionary-trace and structural analysis reveal that many metastable motifs are conserved at the subfamily level and distributed in local regions that are specific to each receptor member or subfamily (Figs. S1–S5). These observations suggest the existence of a functional selectivity in the distribution of metastable motifs. As class A GPCRs can be activated by a large diversity of molecules, ranging from small ligands to small proteins, the need for conformational metastability to direct effective signaling may differ depending on the binding energy provided by each specific molecule. By promoting local flexibility, metastable regions may trigger conformational changes and allosteric paths specific to each member or subfamily, thereby encoding functional selectivity. A predictive model of how such metastable motifs affect the conformational energy landscape and signaling of a given receptor will require the modeling of the receptor structure and predicting its energy in diverse conformational states. Accordingly, reengineering the conformational regulation of receptor function will likely involve the simultaneous modeling of such conformational states within a multistate design framework.
Our ability to generate several variants significantly more stabilized than the M23 variant identified by extensive scanning mutagenesis demonstrates the power of our structure-based design technique in identifying novel stabilizing interactions in large membrane receptors. By targeting nonconserved metastable sites only and filtering out any false positives, our approach selects a minimal number of stabilizing mutations and is unlikely to disrupt major functional determinants of the receptors, which constitute a major advantage over empirical approaches. Many applications such as the design of receptors with reprogrammed signaling properties, the thermostabilization of membrane-embedded scaffolds or molecular sensors will benefit from our rational design technique. As the success of structure-based design approaches partly relies on the accuracy of starting structural models, combining novel improved homology modeling techniques of membrane protein structures (51, 52) and design techniques may allow for rationally engineering stabilized variants of many uncharacterized receptors.
Materials and Methods
Detailed description of materials and methods is given in SI Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the members of the Barth lab for insightful discussions during this study and critical comments on the manuscript. K.M.C. was partially funded by a training fellowship from the Keck Center of the Gulf Coast Consortia, on the Houston Area Molecular Biophysics Program, National Institute of General Medical Sciences T32GM008280.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205512109/-/DCSupplemental.
References
- 1.Boyd D, Schierle C, Beckwith J. How many membrane proteins are there? Protein Sci. 1998;7:201–205. doi: 10.1002/pro.5560070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choe HW, Park JH, Kim YJ, Ernst OP. Transmembrane signaling by GPCRs: Insight from rhodopsin and opsin structures. Neuropharmacology. 2011;60:52–57. doi: 10.1016/j.neuropharm.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 4.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz TW, Hubbell WL. Structural biology: A moving story of receptors. Nature. 2008;455:473–474. doi: 10.1038/455473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowie JU. Stabilizing membrane proteins. Curr Opin Struct Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 8.Warne T, Serrano-Vega MJ, Tate CG, Schertler GF. Development and crystallization of a minimal thermostabilised G protein-coupled receptor. Protein Expr Purif. 2009;65:204–213. doi: 10.1016/j.pep.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Bill RM, et al. Overcoming barriers to membrane protein structure determination. Nat Biotechnol. 2011;29:335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 10.Yu SM, et al. An improved tripod amphiphile for membrane protein solubilization. Protein Sci. 2000;9:2518–2527. doi: 10.1110/ps.9.12.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faham S, Bowie JU. Bicelle crystallization: A new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 12.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson MA, et al. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 17.Tate CG, Schertler GF. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenbaum DM, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 23.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deber CM, et al. Val → Ala mutations selectively alter helix-helix packing in the transmembrane segment of phage M13 coat protein. Proc Natl Acad Sci USA. 1993;90:11648–11652. doi: 10.1073/pnas.90.24.11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar CA, et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci USA. 2008;105:14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Bowie JU. Building a thermostable membrane protein. J Biol Chem. 2000;275:6975–6979. doi: 10.1074/jbc.275.10.6975. [DOI] [PubMed] [Google Scholar]
- 28.Barth P. Modulating membrane protein stability and association by design. Curr Opin Struct Biol. 2007;17:460–466. doi: 10.1016/j.sbi.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 30.Senes A, Ubarretxena-Belandia I, Engelman DM. The Calpha—H…O hydrogen bond: A determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore DT, Berger BW, DeGrado WF. Protein–protein interactions in the membrane: Sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 33.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 34.Joh NH, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453:1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.North B, et al. Characterization of a membrane protein folding motif, the Ser zipper, using designed peptides. J Mol Biol. 2006;359:930–939. doi: 10.1016/j.jmb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Yin H, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 37.Korendovych IV, et al. De novo design and molecular assembly of a transmembrane diporphyrin-binding protein complex. J Am Chem Soc. 2010;132:15516–15518. doi: 10.1021/ja107487b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordova JM, Noack PL, Hilcove SA, Lear JD, Ghirlanda G. Design of a functional membrane protein by engineering a heme-binding site in glycophorin A. J Am Chem Soc. 2007;129:512–518. doi: 10.1021/ja057495i. [DOI] [PubMed] [Google Scholar]
- 39.Ghirlanda G. Design of membrane proteins: Toward functional systems. Curr Opin Chem Biol. 2009;13:643–651. doi: 10.1016/j.cbpa.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Aguilar JM, Saven JG. Computational design of membrane proteins. Structure. 2012;20:5–14. doi: 10.1016/j.str.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senes A. Computational design of membrane proteins. Curr Opin Struct Biol. 2011;21:460–466. doi: 10.1016/j.sbi.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Mirzadegan T, Benko G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry. 2003;42:2759–2767. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheffler W, Baker D. RosettaHoles: Rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18:229–239. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vriend G, Sander C. Quality control of protein models: Directional atomic contact analysis. J Appl Crystallogr. 1993;26:47–60. [Google Scholar]
- 45.Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engelhardt S, Grimmer Y, Fan GH, Lohse MJ. Constitutive activity of the human beta(1)-adrenergic receptor in beta(1)-receptor transgenic mice. Mol Pharmacol. 2001;60:712–717. [PubMed] [Google Scholar]
- 47.Magnani F, Shibata Y, Serrano-Vega MJ, Tate CG. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc Natl Acad Sci USA. 2008;105:10744–10749. doi: 10.1073/pnas.0804396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez GJ, Yao R, Lichtarge O, Wensel TG. Evolution-guided discovery and recoding of allosteric pathway specificity determinants in psychoactive bioamine receptors. Proc Natl Acad Sci USA. 2010;107:7787–7792. doi: 10.1073/pnas.0914877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris NJ, Booth PJ. Folding and stability of membrane transport proteins in vitro. Biochim Biophys Acta. 2012;1818:1055–1066. doi: 10.1016/j.bbamem.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Hirai T, Subramaniam S. Protein conformational changes in the bacteriorhodopsin photocycle: Comparison of findings from electron and X-ray crystallographic analyses. PLoS One. 2009;4:e5769. doi: 10.1371/journal.pone.0005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michino M, et al. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19:1108–1126. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.