Abstract
Production of functional proteins requires multiple steps, including gene transcription and posttranslational processing. MicroRNAs (miRNAs) can regulate individual stages of these processes. Despite the importance of the cystic fibrosis transmembrane conductance regulator (CFTR) channel for epithelial anion transport, how its expression is regulated remains uncertain. We discovered that miRNA-138 regulates CFTR expression through its interactions with the transcriptional regulatory protein SIN3A. Treating airway epithelia with an miR-138 mimic increased CFTR mRNA and also enhanced CFTR abundance and transepithelial Cl− permeability independent of elevated mRNA levels. An miR-138 anti-miR had the opposite effects. Importantly, miR-138 altered the expression of many genes encoding proteins that associate with CFTR and may influence its biosynthesis. The most common CFTR mutation, ΔF508, causes protein misfolding, protein degradation, and cystic fibrosis. Remarkably, manipulating the miR-138 regulatory network also improved biosynthesis of CFTR-ΔF508 and restored Cl− transport to cystic fibrosis airway epithelia. This miRNA-regulated network directs gene expression from the chromosome to the cell membrane, indicating that an individual miRNA can control a cellular process more broadly than recognized previously. This discovery also provides therapeutic avenues for restoring CFTR function to cells affected by the most common cystic fibrosis mutation.
Keywords: ATP-binding cassette transporter, epithelial ion transport, protein biosynthesis
MicroRNAs (miRNAs) are an evolutionarily conserved class of small (∼21–24 nt) noncoding RNAs that play key roles in the transcriptional and posttranscriptional regulation of gene expression (1, 2). The effector functions of miRNAs in several facets of pulmonary biology have been identified (3–5), but their actions in airway epithelia are just emerging (6–8). Although there is no established role for miRNAs in the regulation of fluid and electrolyte transport in the airways, this process is vital to homeostasis and is often perturbed in disease states.
CFTR encodes an anion channel that is regulated by ATP hydrolysis and phosphorylation and is expressed in epithelia and other cell types (9, 10). CFTR conducts Cl−, HCO3−, and other anions (11, 12) and through these activities plays a critical role in regulating the volume and composition of airway surface liquid. Mutations in CFTR cause cystic fibrosis (CF) (9, 13), an autosomal recessive disease involving the airways, sweat glands, intestines, pancreas, liver, and reproductive tract. The majority of CF-associated morbidity and mortality arises from progressive pulmonary infection and inflammation (14). The most common CFTR mutation, ΔF508, is present on ∼70% of mutant alleles (13) and causes protein misfolding, degradation, and CF (9, 15). If CFTR-ΔF508 trafficks to the cell membrane, as occurs with low-temperature (16) or chemical chaperone treatment (17), then the mutant protein retains channel function, albeit with reduced residency and open-state probability (18, 19). Because the majority of people with CF have one or two ΔF508 alleles, there is intense interest by academic and industry laboratories in identifying interventions that might restore function to this misprocessed protein.
CFTR is a low-abundance mRNA in airway epithelia (20), and its temporal and spatial expression are tightly regulated (21, 22). Although the CFTR promoter has been studied extensively, its complex regulation remains incompletely understood (23, 24). Because miRNAs play key roles in the transcriptional and posttranscriptional regulation of at least 60% of human genes (1, 2), we hypothesized that they may provide a previously unidentified mechanism for regulating CFTR abundance and thereby its function (20).
Results
MiRNA Profiling Identifies a Candidate Regulator of CFTR.
We profiled global miRNA expression in well-differentiated primary cultures of human airway epithelia by quantitative PCR. Of the 115 miRNAs identified, 31 were highly expressed (Cq <25) (SI Appendix, Table S1). We interrogated these expressed miRNAs for possible direct or indirect interactions with CFTR and found no miRNAs with conserved target sites in the CFTR 3′ UTR. However, Targetscan, Pictar, and Miranda software-based analyses of these 31 miRNAs identified the SIN3A (SIN3 homolog A) gene as a highly conserved candidate miR-138 target. SIN3A is a transcriptional regulator belonging to the Sin3–histone deacetylase core complex (25, 26). Notably, SIN3A protein has conserved motifs that bind to the chromatin insulator protein CCCTC-binding factor (CTCF), a ubiquitously expressed, highly conserved transcriptional repressor that recruits SIN3A and other proteins to the promoters of target genes (27, 28). Importantly, the CFTR locus contains functional CTCF-binding sites (29). We thus hypothesized that miR-138 and SIN3A regulate CFTR.
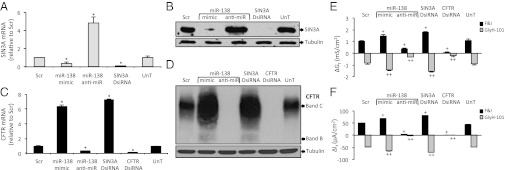
A dual-luciferase reporter assay revealed that miR-138 repressed SIN3A expression in a dose-dependent manner, by binding to its 3′ UTR (SI Appendix, Fig. S1). This effect was site-specific; mutating the two miR-138 binding sites in the SIN3A 3′ UTR relieved the repression in vitro. Transfection of polarized primary cultures of human airway epithelia with an miR-138 mimic reduced, and that of an miR-138 anti-miR increased, SIN3A mRNA and protein levels (Fig. 1 A and B and SI Appendix, Fig. S2). These findings validate SIN3A as an miR-138 target in airway epithelia.
Fig. 1.
miR-138 and SIN3A regulate CFTR expression in airway epithelia. (A) SIN3A mRNA abundance in human primary airway epithelia at 24 h after the indicated interventions (n = 6). Scr, negative control; SIN3A DsiRNA, positive control; UnT, untransfected cells. (B) SIN3A protein abundance in primary airway epithelia at 72 h posttransfection. A representative immunoblot is shown. (C) CFTR mRNA abundance in Calu-3 cells at 24 h after indicated transfections. CFTR DsiRNA, positive control. (D) CFTR protein abundance in Calu-3 cells at 72 h posttransfection (R-769 antibody). (E and F) Changes in conductance (Gt) (E) and transepithelial current (It) (F) with indicated treatments. Basal resistance range, 397–586 ohm*cm2. Error bars indicate mean ± SE. *P < 0.01 relative to Scr; +P < 0.01, ++P < 0.01 relative to ΔGt and ΔIt in Scr-transfected samples on forskolin and IBMX (F&I) and CFTR inhibitor GlyH-101 treatment, respectively.
Mir-138 Regulates CFTR Expression and Function by Relieving SIN3A-Mediated Repression.
To test the hypothesis that miR-138 regulates SIN3A and thereby CFTR expression in airway epithelia, we used the Calu-3 cell line, which expresses CFTR (30). Treatment of Calu-3 cells with an miR-138 mimic or a Dicer-substrate siRNA (DsiRNA) against SIN3A increased CFTR mRNA and protein levels (Fig. 1 C and D and SI Appendix, Fig. S3), whereas the miR-138 anti-miR markedly reduced CFTR mRNA and protein abundance (Fig. 1 C and D and SI Appendix, Fig. S3). CFTR creates an ion permeability, and thus its function can be assessed by measuring transepithelial electrical conductance. The miR-138 mimic and SIN3A DsiRNA treatments increased CFTR-mediated Cl− conductance (Gt) and current (It) in polarized Calu-3 epithelia, whereas the miR-138 anti-miR had the opposite effects (Fig. 1 E and F).
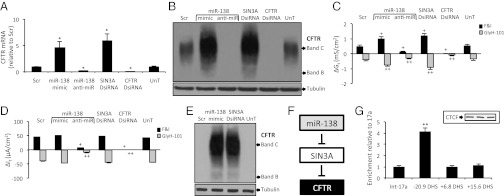
In polarized primary cultures of human airway epithelia, transfection with an miR-138 mimic or SIN3A DsiRNA increased, and that of an miR-138 anti-miR reduced, CFTR mRNA and protein levels (Fig. 2 A and B and SI Appendix, Fig. S4). We note that native tissue and primary cultures of airway epithelial cells in which endogenous CFTR is studied have less band B protein than that detected when recombinant CFTR is expressed (31, 32). Treatment with the miR-138 mimic and the SIN3A DsiRNA increased cAMP-stimulated Gt (Fig. 2C). There was no change in It (Fig. 2D), consistent with the presence of other rate-limiting steps for Cl− secretion in airway epithelia (33). The miR-138 anti-miR reduced both Gt and It responses to cAMP-dependent stimulation (Fig. 2 C and D).
Fig. 2.
miR-138 and SIN3A regulate CFTR expression in primary cultures of human airway epithelia and cells with no CFTR expression. (A) CFTR mRNA abundance in primary airway epithelia at 24 h after interventions (n = 6). (B) CFTR protein abundance from primary airway epithelia at 72 h posttransfection; R-769 antibody, representative immunoblot. (C and D) Changes in conductance (Gt) (C) and transepithelial current (It) (D) with indicated treatments. Each bar represents six primary airway epithelial cell cultures each from three donors, pretransfected with the indicated reagents. Basal resistance range, 415–672 ohm*cm2. (E) CFTR protein abundance in HeLa cells; R-769 antibody. (F) Schematic representing miR-138– and SIN3A-mediated regulation of CFTR expression. (G) Fold enrichment of SIN3A, assessed by quantitative PCR after ChIP. Data are normalized to CFTR intron 17a DHS. (Inset) CTCF immunoblot of lysates from three airway epithelia donors. Error bars indicate mean ± SE; *P < 0.01 relative to Scr; **P < 0.01 relative to intron 17a; +P < 0.01 and ++P < 0.01 relative to ΔGt and ΔIt in Scr-transfected samples on F&I and GlyH-101 treatment, respectively.
SIN3A Is a Transcriptional Repressor of CFTR Expression.
The foregoing data show that miR-138 and SIN3A regulate CFTR expression in epithelia that normally express CFTR. To learn whether they also can control CFTR expression in cells that do not produce CFTR, we studied HeLa and HEK293T cells. The miR-138 mimic and SIN3A DsiRNA markedly increased CFTR mRNA and protein expression (Fig. 2E and SI Appendix, Figs. S5 and S6). Transfected HeLa cells also exhibited cAMP-dependent anion permeability, as assessed by iodide efflux (SI Appendix, Fig. S7). These results implicate SIN3A as a potent regulator of CFTR expression, and further support the notion that miR-138 regulates CFTR expression by repressing SIN3A (Fig. 2F).
To assess whether SIN3A-mediated CFTR repression involves CTCF-mediated recruitment of SIN3A to the CFTR promoter (34), we performed ChIP in primary human airway epithelial cells with a SIN3A antibody. Because SIN3A-mediated transcriptional repression involves recruitment to the promoter of target genes (35), we specifically assessed SIN3A enrichment at the CFTR promoter. CTCF has been demonstrated to bind within the −20.9 kb DNase I hypersensitive site (DHS) (i.e, distance from the transcriptional start site) (29) at the CFTR promoter. In addition to this site, we also assessed SIN3A enrichment at the +6.8 kb DHS (i.e., distance from the transcriptional stop site), which has been shown to bind CTCF in a tissue-specific manner (29, 36). Indeed, the −20.9 kb DHS was enriched for SIN3A compared with two control regions, CFTR intron 17a and +15.6 kb DHS (Fig. 2G). The demonstration by ChIP that CTCF and SIN3A interact at the −20.9 kb DHS in primary human airway epithelial cells provides insight into the repressor functions of this important regulatory region, as well as the nature of protein interactions at the CFTR promoter.
MiR-138 and SIN3A Regulate Genes Influencing CFTR Protein Maturation.
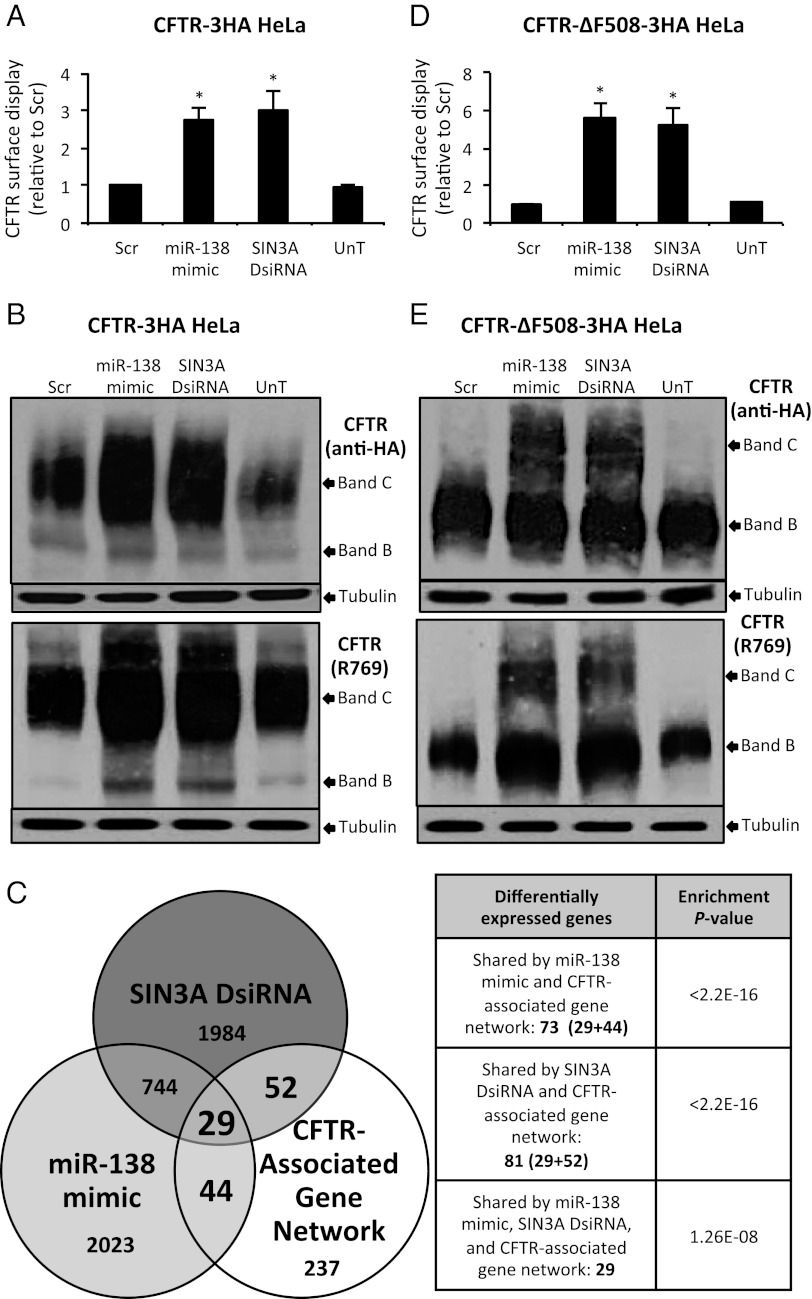
To explore whether miR-138 and SIN3A might have posttranscriptional effects on protein biosynthesis in addition to their direct transcriptional regulation of CFTR, we performed additional experiments using HeLa cells stably expressing HA-tagged WT CFTR under control of the CMV promoter (18). Cell-based ELISA using an HA antibody revealed an increase in HA-tagged CFTR at the cell surface after treatment with the miR-138 mimic or SIN3A DsiRNA (Fig. 3A and SI Appendix, Fig. S8A), with no changes in transgene mRNA abundance (SI Appendix, Fig. S8B). This result was further supported by immunoblot analysis (Fig. 3B and SI Appendix, Fig. S8 C and D). These data indicate that miR-138 has important posttranscriptional effects on CFTR biosynthesis.
Fig. 3.
miR-138 regulates CFTR processing. (A) Surface display, as detected by ELISA, of epitope-tagged CFTR in CFTR-3HA HeLa cells transfected with indicated reagents. (B) CFTR protein abundance in CFTR-3HA HeLa cells at 24 h posttransfection. (Upper) Anti-HA antibody. (Lower) R769 antibody. (C) Schematic showing regions of intersection of SIN3A DsiRNA, miRNA-mimic, and CFTR-associated genes data sets; P < 0.05 (SI Appendix, Tables S2–S4). (D) Surface display of epitope-tagged CFTR in CFTR-ΔF508-3HA HeLa cells transfected with indicated reagents. (E) CFTR protein abundance in CFTR-ΔF508-3HA HeLa cells at 24 h posttransfection. (Upper) Anti-HA antibody. (Lower) R769 antibody. Error bars indicate mean ± SE; *P < 0.01 relative to Scr.
Subsequent global mRNA transcript profiling in Calu-3 epithelia treated with the miR-138 mimic or SIN3A DsiRNA identified a common set of 773 genes whose expression changed in response to these interventions (Fig. 3C). Intersecting these gene sets with a curated list of 362 genes with protein products known to associate with CFTR (i.e., CFTR-associated gene network; SI Appendix, Table S2) revealed that 34.5% (125 of 362) were in the CFTR-associated gene network, a significant enrichment over random expectations (Fig. 3C and SI Appendix, Table S3). These 125 genes function in several cellular compartments and many positively influence CFTR protein expression or stability (SI Appendix, Table S4). Pathway and Gene Ontology analysis of the 773 differentially expressed genes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (37) revealed significant enrichment of gene sets in pathways that include chaperones, unfolded protein response, protein ubiquitination and proteosomal catabolic processes, negative regulation of apoptosis, and heat shock (SI Appendix, Table S5). These findings further support the conclusion that miR-138 enhances CFTR biogenesis and influences the expression of genes at multiple steps along its biosynthetic pathway.
Manipulating the miR-138–SIN3A Network Rescues Misprocessed ΔF508 CFTR.
The most common CFTR mutant, ΔF508, generates a protein with an altered structure that is unstable, mislocalized, and rapidly degraded via endoplasmic reticulum-associated degradation (15). Interventions that improve biosynthetic processing, such as low temperature (16), chemical chaperones (17), and small molecules (38, 39), can partially restore CFTR-ΔF508 anion channel function; however, overexpression of the ΔF508 cDNA in heterologous cells or primary airway epithelia does not restore CFTR-dependent anion conductance (40). Because miR-138 increased the biosynthesis of WT CFTR (Fig. 3 A and B), we hypothesized that it also might improve the biosynthesis of CFTR-ΔF508.
We transfected HeLa cells stably expressing HA-tagged CFTR-ΔF508 cDNA under the control of the CMV promoter (18) with the miR-138 mimic or SIN3A DsiRNA. Suprisingly, we found that mutant CFTR, as detected by ELISA using an HA-specific antibody, reached the cell surface (Fig. 3D and SI Appendix, Fig. S9A) with no change in transgene mRNA abundance (SI Appendix, Fig. S9B). Immunoblot analysis with an HA-antibody detecting only the transgene CFTR-ΔF508 protein product demonstrated that both interventions increased the abundance of the mature, fully glycosylated CFTR band C (Fig. 3E and SI Appendix, Fig. S9 C and D).
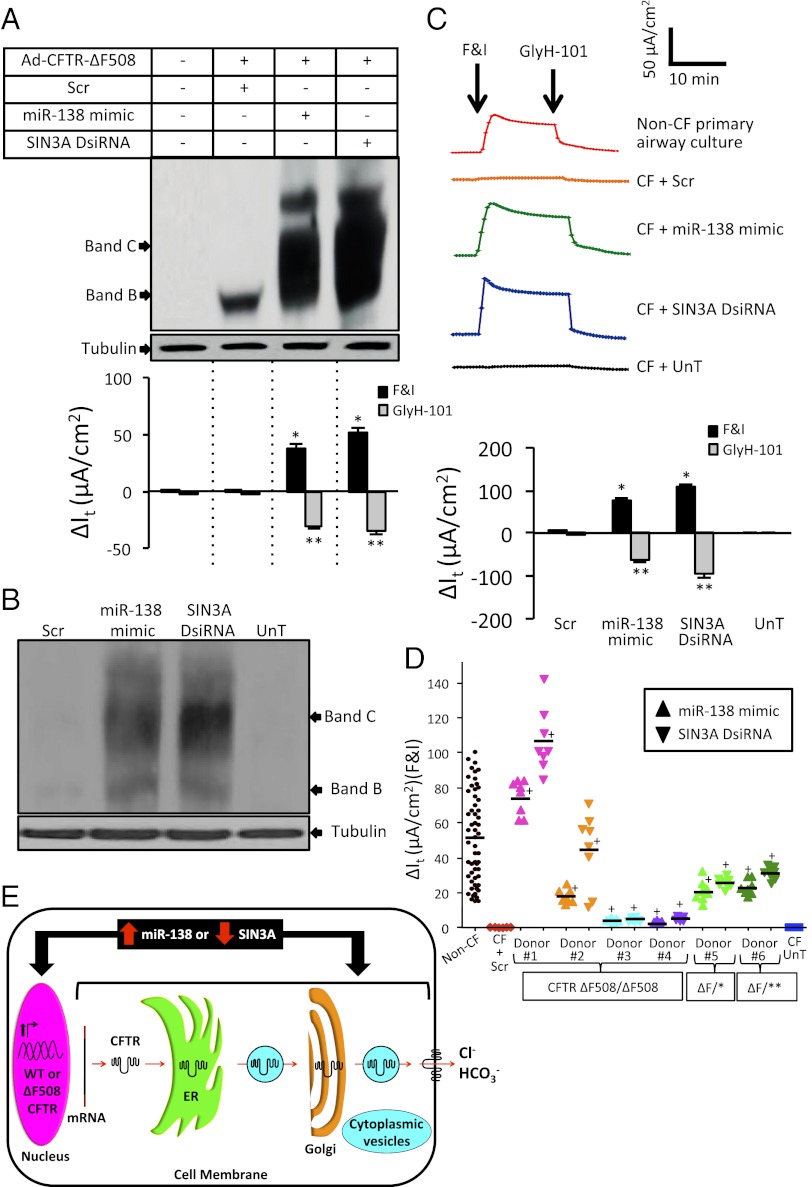
We also expressed a recombinant CMV promoter-driven CFTR-ΔF508 cDNA in primary human CFTR null airway epithelia (CFTR Q493X/S912X) using an adenovirus (Ad) vector (41). In this setting, CFTR-mediated Cl− current was restored only in epithelia pretreated with the miR-138 mimic or SIN3A DsiRNA (Fig. 4A and SI Appendix, Fig. S10). These results further indicate that miR-138– and SIN3A-regulated genes influence the posttranscriptional processing of CFTR.
Fig. 4.
SIN3A inhibition yields partial rescue of Cl− transport in CF epithelia. (A) (Upper) CFTR protein abundance from airway epithelia (CFTR Q493X/S912X, 24-1 antibody) after indicated treatments. (Lower) Change in It values after F&I stimulation and GyH-101 inhibition (one donor, three replicates). Basal resistance range, 279–360 ohm*cm2. (B) Representative CFTR immunoblot from primary epithelia (CFTR ΔF508/ΔF508) at 72 h posttransfection; R-769 antibody, donor 2 in D. (C) Responses of CFTR ΔF508/ΔF508 epithelia to indicated interventions (donor 1). (Upper) It tracings of responses to F&I, followed by GlyH-101 treatment (epithelia pretreated with amiloride and DIDS). (Lower) Summary of change in It in response to F&I, followed by GlyH-101 treatment (one donor, eight replicates). Basal resistance range, 488–691 ohm*cm2. Error bars indicate mean ± SE. *P < 0.01, **P < 0.01 relative to ΔIt in Scr-transfected samples after F&I and GlyH-101 treatments, respectively; +P < 0.01 relative to Scr. (D) Changes in It values after F&I treatment of six primary CF airway epithelia cultures transfected with indicated reagents. Six untreated or Scr-treated CF samples served as negative controls; eight non-CF samples served as WT controls. ΔF/* denotes ΔF508/3659delC; ΔF/** denotes ΔF508/R1162X. Horizontal bars indicate means. Basal resistance range, 295–819 ohm*cm2. (E) Working model of steps in CFTR transcription and protein biosynthesis pathway in which miR-138–regulated gene products influence WT and CFTR-ΔF508 (Fig. 3C and SI Appendix, Tables S2–S5).
Next, we tested whether manipulating the miR-138–SIN3A network rescued CFTR-ΔF508–mediated Cl− transport in CF primary airway epithelial cells. Expression of the miR-138 mimic or SIN3A DsiRNA increased CFTR-ΔF508 mRNA and protein even in primary cultures of CF airway epithelia (Fig. 4B and SI Appendix, Figs. S11 and S12). Both interventions also restored CFTR-ΔF508–mediated Cl− transport in these epithelia (Fig. 4C and SI Appendix, Fig. S12 B and C). Significant restoration of CFTR-ΔF508–mediated Cl− transport to varying levels was observed in primary CF epithelia from multiple human donors (Fig. 4D), and similar results were obtained in a cell line homozygous for the ΔF508 mutation (SI Appendix, Fig. S13). Measurement of lactate dehydrogenase release over a 2-wk period from Calu-3 and CFBE cells transfected with oligonucleotides used in this study (SI Appendix, Fig. S14) revealed no cytotoxicity.
Discussion
Here we show that relieving transcriptional repression through the expression of an miR-138 mimic or knockdown of SIN3A profoundly influenced CFTR expression and anion channel activity in airway epithelia, as well as in cells with no basal CFTR expression. Thus, miR-138, acting via SIN3A and other target genes, orchestrates a cellular program that influences WT and mutant CFTR, increasing the biogenesis and cell surface delivery of both (Fig. 4E and SI Appendix, Tables S3–S5). Through these interactions, the fate of the CFTR-ΔF508 protein in airway epithelia was redirected from proteosomal degradation to a functional anion channel. These findings provide insight into the regulation of CFTR expression and identify a gene network with therapeutic promise.
The present study reveals a previously unrecognized role for SIN3A in the repression of CFTR expression. Despite more than 20 y of study (42), the mechanisms governing CFTR transcription remain incompletely understood. The CFTR promoter has features of a “housekeeping” gene (43, 44), and several transactivating factors and regulatory elements have been characterized (43, 45–47). Given that SIN3A-mediated transcriptional silencing involves associated histone deacetylases (35), identifying an interaction between SIN3A and CTCF on the CFTR promoter at the −20.9 kb DHS improves our understanding of how CFTR is transcriptionally regulated. However, although SIN3A recruitment by CTCF has been reported previously (28), other DNA-binding proteins also could recruit SIN3A to other sites on the CFTR promoter, thereby regulating CFTR transcription. A ChIP-seq approach could be used to further identify SIN3A recruitment sites on the CFTR promoter. The reciprocal effects of the miR-138 anti-miR in decreasing CFTR mRNA, protein, and transepithelial Cl− permeability emphasize the role of miR-138 in regulating CFTR expression.
We have identified a genomic signature associated with the rescue of CFTR-ΔF508 (Fig. 3C and SI Appendix, Tables S3–S5). Strengths of this study include our use of multiple cell models, including both primary airway epithelia and cells that do not normally express CFTR, to test the effect of miR-138 and SIN3A on CFTR biosynthesis. Our key finding that the miR-138–SIN3A network rescues CFTR-ΔF508 biosynthesis was also replicated in primary airway epithelia from multiple CF donors. The current work also has limitations, primarily the need for additional studies to better understand the mechanisms through which miR-138 influences CFTR expression and biosynthesis. We do not yet know which miR-138– and SIN3A-regulated genes are ultimately responsible for directing CFTR-ΔF508 processing in airway epithelia. However, SIN3A inhibition alone was sufficient to achieve this result, suggesting that genes directly regulated by SIN3A, or the downstream targets of SIN3A-regulated genes, are key to the effect. A ChIP-seq approach could be used to further identify SIN3A-regulated genes in airway epithelia.
It is possible that changing the expression of only a limited number of genes coregulated by miR-138 and SIN3A would be necessary and sufficient to restore CFTR-ΔF508 function. For example, Sun et al. (48) reported that Derlin-1 (DERL1) interacts with both WT and ΔF508 CFTR, reducing their expression. Knockdown of Derlin-1 with siRNA markedly increased the abundance of immature CFTR-ΔF508 protein (48). In addition, siRNA inhibition of DNAJA1, STIP1, and HSPA8 is associated with enhanced plasma membrane stability for CFTR-ΔF508 (18). These four genes (DERL1, DNAJA1, STIP1, and HSPA8) all showed reduced abundance in response to treatment of epithelia with the miR-138 mimic or SIN3A DsiRNA. Additional selected siRNA and gene addition studies of candidates in the CFTR-associated gene network (Fig. 3C and SI Appendix, Tables S3–S5) may help identify the subset of genes most influential in CFTR-ΔF508 rescue.
Further steps are needed to carry the findings of the present study toward a therapeutic application. Although transient, partial, and airway-specific delivery of an miR-138 mimic or anti-SIN3A siRNA might be therapeutic, the efficient delivery of siRNA or miR mimics to airway epithelia is inefficient with current technology (49, 50). Further advancements in this field will likely lead to new clinical applications. In addition, systemic inhibition of SIN3A function may be undesirable, because the protein is a highly conserved transcriptional repressor that regulates the expression of many genes in a cell- and tissue-specific fashion. An alterative strategy is to focus on the downstream targets of SIN3A that directly mediate the observed CFTR-Δ508 rescue. Drug screening concentrated on identifying small molecules or pharmaceuticals that inhibit SIN3A, or a core subset of gene products responsible for CFTR-ΔF508 rescue, represents another therapeutic approach (51).
Our surprising findings reveal an elegant miRNA-regulated gene network that influences multiple steps in protein biosynthesis. This discovery provides insight into how CFTR expression is regulated, and suggests therapeutic targets for rescuing the function of the mutant CFTR protein most commonly associated with CF. These findings also raise the possibility that manipulating miR-138/SIN3A and their targets might restore function of misprocessed proteins resulting from other genetic diseases.
Materials and Methods
Primary Human Airway Epithelia.
Airway epithelia from human trachea and primary bronchus removed from organs donated for research were cultured at the air–liquid interface as described previously (52).
RNA Isolation.
Total RNA from human primary airway epithelial cultures and cell lines (Calu-3, HEK293T, HeLa, and CFBE) were isolated using the mirVana miRNA Isolation Kit (Ambion) (53). Total RNA was tested on an Agilent 2100 Bioanalyzer. Only samples with an RNA integrity number >7.0 were selected for downstream processing.
Detailed descriptions of all methods are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Beverly Davidson, John Engelhardt, Michael Hildebrand, Jeffrey Murray, Val Sheffield, Patrick Sinn, David Stoltz, and Joseph Zabner for critically reviewing the manuscript. This work was supported by National Institutes of Health Grants R21 HL-91808 (to P.B.M. and Y.X.), P01 HL-51670 (to P.B.M.), P30 DK-054759 (to Y.X.), and P01 HL-091842 (to M.J.W.); the Roy J. Carver Charitable Trust; and the Cystic Fibrosis Foundation Research Development Program (M.J.W.). We also acknowledge the support of the In Vitro Models and Cell Culture Core, Gene Transfer Vector Core, and Cell Morphology Core, which are partially supported by the Center for Gene Therapy for Cystic Fibrosis (National Institutes of Health Grant P30 DK-54759) and the Cystic Fibrosis Foundation. M.J.W. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38956).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210906109/-/DCSupplemental.
References
- 1.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200–dependent pathway in mice. J Clin Invest. 2011;121:1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeri M, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcet B, et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol. 2011;13:693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 7.Lizé M, Herr C, Klimke A, Bals R, Dobbelstein M. MicroRNA-449a levels increase by several orders of magnitude during mucociliary differentiation of airway epithelia. Cell Cycle. 2010;9:4579–4583. doi: 10.4161/cc.9.22.13870. [DOI] [PubMed] [Google Scholar]
- 8.Schembri F, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 10.Riordan JR, et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 11.Anderson MP, et al. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–205. doi: 10.1126/science.1712984. [DOI] [PubMed] [Google Scholar]
- 12.Anderson MP, Rich DP, Gregory RJ, Smith AE, Welsh MJ. Generation of cAMP-activated chloride currents by expression of CFTR. Science. 1991;251:679–682. doi: 10.1126/science.1704151. [DOI] [PubMed] [Google Scholar]
- 13.Kerem B-S, et al. Identification of the cystic fibrosis gene: Genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 14.Welsh MJ, Ramsey BW, Accurso F, Cutting GR. Cystic fibrosis. In: Scriver CR, et al., editors. The Metabolic and Molecular Basis of Inherited Disease. New York: McGraw-Hill; 2001. 8th Ed, Vol 3, pp 5121–5189. [Google Scholar]
- 15.Cheng SH, et al. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 16.Denning GM, et al. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 17.Brown CR, Hong-Brown LQ, Biwersi J, Verkman AS, Welch WJ. Chemical chaperones correct the mutant phenotype of the delta F508 cystic fibrosis transmembrane conductance regulator protein. Cell Stress Chaperones. 1996;1:117–125. doi: 10.1379/1466-1268(1996)001<0117:ccctmp>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okiyoneda T, et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott CJ, et al. Nucleosome occupancy reveals regulatory elements of the CFTR promoter. Nucleic Acids Res. 2012;40:625–637. doi: 10.1093/nar/gkr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell BC, et al. Expression of the cystic fibrosis transmembrane conductance regulator gene in the respiratory tract of normal individuals and individuals with cystic fibrosis. Proc Natl Acad Sci USA. 1991;88:6565–6569. doi: 10.1073/pnas.88.15.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung J, et al. Identification of the human cortactin-binding protein-2 gene from the autism candidate region at 7q31. Genomics. 2001;78:7–11. doi: 10.1006/geno.2001.6651. [DOI] [PubMed] [Google Scholar]
- 22.Yan W, et al. Identification of Gasz, an evolutionarily conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile-alpha motif, and a basic leucine zipper. Mol Endocrinol. 2002;16:1168–1184. doi: 10.1210/mend.16.6.0864. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Ott CJ, Lewandowsa MA, Leir SH, Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J Cell Mol Med. 2011;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewandowska MA, et al. Multiple mechanisms influence regulation of the cystic fibrosis transmembrane conductance regulator gene promoter. Am J Respir Cell Mol Biol. 2010;43:334–341. doi: 10.1165/rcmb.2009-0149OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinzel T, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 26.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 27.Lutz M, Baniahmad A, Renkawitz R. Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor. Biochem Soc Trans. 2000;28:386–389. [PubMed] [Google Scholar]
- 28.Lutz M, et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackledge NP, et al. CTCF mediates insulator function at the CFTR locus. Biochem J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen BQ, Finkbeiner WE, Wine JJ, Mrsny RJ, Widdicombe JH. Calu-3: A human airway epithelial cell line that shows cAMP-dependent Cl- secretion. Am J Physiol. 1994;266:L493–L501. doi: 10.1152/ajplung.1994.266.5.L493. [DOI] [PubMed] [Google Scholar]
- 31.Varga K, et al. Efficient intracellular processing of the endogenous cystic fibrosis transmembrane conductance regulator in epithelial cell lines. J Biol Chem. 2004;279:22578–22584. doi: 10.1074/jbc.M401522200. [DOI] [PubMed] [Google Scholar]
- 32.Ostedgaard LS, et al. The ΔF508 mutation causes CFTR misprocessing and cystic fibrosis-like disease in pigs. Sci Transl Med. 2011;3:74ra24. doi: 10.1126/scitranslmed.3001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farmen SL, et al. Gene transfer of CFTR to airway epithelia: Low levels of expression are sufficient to correct Cl− transport and overexpression can generate basolateral CFTR. Am J Physiol Lung Cell Mol Physiol. 2005;289:L1123–L1130. doi: 10.1152/ajplung.00049.2005. [DOI] [PubMed] [Google Scholar]
- 34.Ellison-Zelski SJ, Solodin NM, Alarid ET. Repression of ESR1 through actions of estrogen receptor alpha and Sin3A at the proximal promoter. Mol Cell Biol. 2009;29:4949–4958. doi: 10.1128/MCB.00383-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: Master scaffold and transcriptional corepressor. Biochim Biophys Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackledge NP, Ott CJ, Gillen AE, Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–1094. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 38.Pedemonte N, et al. Small-molecule correctors of defective DeltaF508-CFTR cellular processing identified by high-throughput screening. J Clin Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Goor F, et al. Rescue of DeltaF508-CFTR trafficking and gating in human cystic fibrosis airway primary cultures by small molecules. Am J Physiol Lung Cell Mol Physiol. 2006;290:L1117–L1130. doi: 10.1152/ajplung.00169.2005. [DOI] [PubMed] [Google Scholar]
- 40.Granio O, et al. Cellular localization and activity of Ad-delivered GFP-CFTR in airway epithelial and tracheal cells. Am J Respir Cell Mol Biol. 2007;37:631–639. doi: 10.1165/rcmb.2007-0026TE. [DOI] [PubMed] [Google Scholar]
- 41.Ostedgaard LS, et al. Processing and function of CFTR-DeltaF508 are species-dependent. Proc Natl Acad Sci USA. 2007;104:15370–15375. doi: 10.1073/pnas.0706974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rommens JM, et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 43.Yoshimura K, et al. Expression of the cystic fibrosis transmembrane conductance regulator gene in cells of non-epithelial origin. Nucleic Acids Res. 1991;19:5417–5423. doi: 10.1093/nar/19.19.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh J, Sferra TJ, Collins FS. Characterization of the cystic fibrosis transmembrane conductance regulator promoter region: Chromatin context and tissue-specificity. J Biol Chem. 1993;268:15912–15921. [PubMed] [Google Scholar]
- 45.Trapnell BC, et al. Down-regulation of cystic fibrosis gene mRNA transcript levels and induction of the cystic fibrosis chloride secretory phenotype in epithelial cells by phorbol ester. J Biol Chem. 1991;266:10319–10323. [PubMed] [Google Scholar]
- 46.Ott CJ, et al. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci USA. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ott CJ, Blackledge NP, Leir SH, Harris A. Novel regulatory mechanisms for the CFTR gene. Biochem Soc Trans. 2009;37:843–848. doi: 10.1042/BST0370843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun F, et al. Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J Biol Chem. 2006;281:36856–36863. doi: 10.1074/jbc.M607085200. [DOI] [PubMed] [Google Scholar]
- 49.Moschos SA, et al. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol Ther. 2011;19:2163–2168. doi: 10.1038/mt.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 52.Karp PH, et al. An in vitro model of differentiated human airway epithelia: Methods for establishing primary cultures. Methods Mol Biol. 2002;188:115–137. doi: 10.1385/1-59259-185-X:115. [DOI] [PubMed] [Google Scholar]
- 53.Ramachandran S, Clarke LA, Scheetz TE, Amaral MD, McCray PB., Jr Microarray mRNA expression profiling to study cystic fibrosis. Methods Mol Biol. 2011;742:193–212. doi: 10.1007/978-1-61779-120-8_12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.