Abstract
In postmitotic mammalian cells, protein p53R2 substitutes for protein R2 as a subunit of ribonucleotide reductase. In human patients with mutations in RRM2B, the gene for p53R2, mitochondrial (mt) DNA synthesis is defective, and skeletal muscle presents severe mtDNA depletion. Skin fibroblasts isolated from a patient with a lethal homozygous missense mutation of p53R2 grow normally in culture with an unchanged complement of mtDNA. During active growth, the four dNTP pools do not differ in size from normal controls, whereas during quiescence, the dCTP and dGTP pools decrease to 50% of the control. We investigate the ability of these mutated fibroblasts to synthesize mtDNA and repair DNA after exposure to UV irradiation. Ethidium bromide depleted both mutant and normal cells of mtDNA. On withdrawal of the drug, mtDNA recovered equally well in cycling mutant and control cells, whereas during quiescence, the mutant fibroblasts remained deficient. Addition of deoxynucleosides to the medium increased intracellular dNTP pools and normalized mtDNA synthesis. Quiescent mutant fibroblasts were also deficient in the repair of UV-induced DNA damage, as indicated by delayed recovery of dsDNA analyzed by fluorometric analysis of DNA unwinding and the more extensive and prolonged phosphorylation of histone H2AX after irradiation. Supplementation by deoxynucleosides improved DNA repair. Our results show that in nontransformed cells only during quiescence, protein p53R2 is required for maintenance of mtDNA and for optimal DNA repair after UV damage.
Keywords: DNA precursors, dNTP de novo synthesis, cell cycle, mitochondrial disease
DNA replication and repair require the continued synthesis of the four dNTPs. They are synthesized by evolutionary-related ribonucleotide reductases operating with slightly different mechanisms in aerobic and anaerobic organisms (1). Each ribonucleotide reductase provides the required amounts of all four dNTPs. A similar allosteric mechanism, maintained throughout evolution, regulates both the enzyme’s activity and its substrate specificity. Cells contain small dNTP pools of similar sizes, approximately 10-fold larger during DNA replication than during quiescence. Regulation of pool sizes by ribonucleotide reductases is of great importance for correct DNA replication, and changes in the actual sizes or in their balance lead to increased mutation rates (2). For mammalian cells, the induction of mutations by pool imbalances has been described in detail, along with possible mechanisms (3). In yeast, a recent elegant study (4) linked specific amino acid substitutions in the catalytic subunit of ribonucleotide reductase to defined pool imbalances, which result in increased mutation rates.
In mammalian cells, the canonical ribonucleotide reductase is a complex between two proteins: the large catalytic protein R1 that contains the allosteric sites and the smaller protein R2 that contributes a stable tyrosyl free radical during the reaction (1). Both proteins are transcriptionally activated during early S-phase (5) and are present in roughly equal amounts (6, 7) to deliver the dNTPs required for DNA replication. R2 is degraded during late mitosis (8); thus, postmitotic quiescent cells are essentially devoid of R2 but retain some R1. In the year 2000, a second radical-providing small subunit, termed p53R2, was discovered in mammalian cells (9). p53R2 has the same function as the homologous R2, but is not degraded in mitosis. Quiescent cells contain an undiminished amount of p53R2 (6, 7) but little or no R2. After DNA damage, p53R2 is transcriptionally activated by p53 and was reported to translocate into the nucleus (9). It was therefore thought to be primarily involved in DNA repair. However, with affinity-purified antibodies, we found p53R2, as well as R1 and R2, in the cytosol also after DNA damage (10). Moreover the concentrations of the three proteins at different stages of the cell cycle speak against a specific requirement of p53R2 for DNA repair. During S-phase, murine or human fibroblasts contain roughly equimolar amounts of R1 and R2, whereas p53R2 amounts to only 3% of R2 and increases to 13% after DNA damage (6), indicating that the main catalytically active enzyme is an R1/R2 and not an R1/p53R2 complex. In a study of quiescent human fibroblasts, ribonucleotide reduction was catalyzed largely by an R1/p53R2 complex at a rate amounting to only 2–3% of that of cycling cells (7).
In 2007, Bourdon et al. (11) opened a new chapter in the history of p53R2 by reporting that in humans, genetic inactivation of p53R2 causes a severe mitochondrial disease characterized by profound depletion of mtDNA in differentiated cells and lethality shortly after birth. They found that skeletal muscle of patients with mutations in the RRM2B gene coding for p53R2 completely lacked mtDNA (11). Similar patients were described in later studies from several other laboratories (12–16). It is now clear that p53R2 activity is required for the stability of the mt genome in differentiated tissues, shifting the attention from DNA repair to mtDNA maintenance.
Less clear are the extent to which p53R2 is required for DNA repair and the extent to which it is required only in quiescent cells. Most experiments concerning p53R2’s function have been carried out with transformed cell lines (9, 17–19), which are not suitable for addressing these questions. To investigate p53R2 in nontransformed cells, we recently examined in vitro the consequences of p53R2 inactivation with fibroblasts from a patient with a lethal homozygous missense mutation in the iron-binding center of p53R2 who had died at aged 3 mo with severe muscular mtDNA depletion (16, 20). Compared with age-matched controls, the mutant fibroblasts grew normally in culture and contained a normal complement of mtDNA (20); however, once they became quiescent, their ability to reduce ribonucleotides was strongly curtailed, resulting in smaller dCTP and dGTP pools. The profound changes in deoxyribonucleotide metabolism did not result in a depletion of mtDNA in vitro, unlike in the patient. We hypothesized that this ostensible paradox might be explained by the low copy number of mtDNA in fibroblasts, only <5% of that in skeletal muscle cells, requiring much less dNTPs for its maintenance.
In the present work, we tested this hypothesis with cycling and quiescent fibroblasts. We induced mtDNA depletion by treating the cells with ethidium bromide (EtBr) (21), and followed the recovery of mtDNA after removal of the drug. In addition, we investigated the involvement of p53R2 in DNA repair by analyzing the ability of the mutated fibroblasts to repair DNA after UV damage. Our data demonstrate the importance of p53R2 for both mtDNA replication and DNA repair in quiescent cells that contain insufficient R2 for dNTP synthesis. The data indicate that correct pool balances are required not only for the fidelity of nuclear DNA replication, but also for optimal mtDNA synthesis and DNA repair after UV damage.
Results
Recovery of mtDNA After Depletion with EtBr.
Cells growing in the presence of EtBr lose their mtDNA (21). In preliminary experiments, p53R2 mutant and control fibroblasts cultured for 7 d in medium containing 10% FCS with either 20 or 50 ng EtBr/mL were rapidly depleted of mtDNA. When growth continued in the absence of EtBr, all cultures rapidly recovered mtDNA (Fig. S1A), with no clear difference between mutant and control cells. We repeated the experiment with quiescent cells. As before, we grew the fibroblasts with EtBr in 10% FCS for 7 d, but when the cultures became confluent we shifted them to 0.1% FCS + EtBr for 7 d longer. The cells reached quiescence, lost protein R2, and became dependent on p53R2. We then removed EtBr and followed the reappearance of mtDNA in low serum. Now only the control cells fully recovered their mtDNA (Fig S1B), suggesting that the cells with mutated p53R2 did not produce sufficient dNTPs for sustained mtDNA synthesis.
Previous studies have shown that quiescent mutant fibroblasts contain smaller dCTP and dGTP pools than controls, whereas dATP is unaffected and dTTP is slightly increased (20). If the inability of the mutant fibroblasts to reconstitute their mtDNA indeed depended on insufficient production of dNTPs, then it might be possible to compensate the deficiency by exploiting the salvage pathway of dNTP synthesis (22). Low micromolar amounts of deoxycytidine (CdR) and deoxyguanosine (GdR) added to the media of quiescent cultures increased the corresponding dNTP pools in both normal and mutant fibroblasts (Table 1). GdR alone diminished the dCTP pool of mutant cells in a concentration-dependent manner with a decrease to 50% after an 18-h incubation with 5 μM GdR (Table S1). The combination of the two deoxynucleosides increased both the dCTP and dGTP pools of mutant cells slightly above the pool sizes of the controls.
Table 1.
Effect of deoxynucleosides in the medium on dNTP pool sizes in quiescent control and p53R2 mutant fibroblasts
| dNTP, pmoles/106 cells |
||||||||
| Control |
Mutant |
|||||||
| Treatment | dCTP | dGTP | dTTP | dATP | dCTP | dGTP | dTTP | dATP |
| None | 2.9 ± 0.63 | 0.5 ± 0.11 | 2.6 ± 0.89 | 2.4 ± 0.56 | 1.2 ± 0.25 | 0.2 ± 0.05 | 2.7 ± 0.79 | 1.4 ± 0.41 |
| 5 μM CdR + 5 μM GdR | 4.5 ± 1.17 | 1.2 ± 0.28 | 2.9 ± 0.32 | 2.0 ± 0.67 | 3.8 ± 0.49 | 0.8 ± 0.16 | 3.4 ± 1.31 | 1.4 ± 0.76 |
Summary of dNTP pools from between five and seven separate experiments with quiescent mutant and control cells. Pools were measured after an 18-h incubation with deoxynucleosides. Values are mean ± SD.
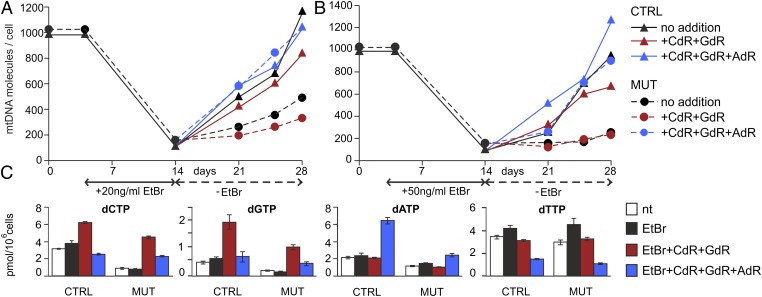
We next tested whether changes in intracellular dNTPs arising from the addition of deoxynucleosides to the medium affected the recovery of mtDNA in the quiescent mutant cells after depletion by EtBr. We found no clear increase in mtDNA after the addition of CdR and GdR either alone or in combination. However, when we also included deoxysadenosine (AdR), the restoration of EtBr-depleted mtDNA was almost complete at 7 d after removal of the drug (data not shown). Thus, in two independent time curves in quiescent fibroblasts, we compared the effects of AdR + CdR + GdR with those of CdR + GdR on the recovery of mtDNA depleted by 20 ng/mL EtBr (Fig. 1A) and 50 ng/mL EtBr (Fig. 1B). At both concentrations, control cells recovered a full complement of mtDNA independent of the presence of deoxynucleosides. In the mutant cells, in agreement with the preliminary results, mtDNA fully recovered only in the presence of all three deoxynucleosides, confirming the importance of AdR. Fig. 1C also shows the changes in dNTP concentrations induced by the two combinations of deoxynucleosides after removal of EtBr. We found no systematic differences in the concentrations at the various times of recovery, and thus report their average values here.
Fig. 1.
Stimulation of mtDNA recovery in mutant cells by deoxynucleosides present in the medium. We depleted the mtDNA of quiescent mutant or control fibroblasts in 0.1% FCS with EtBr 20 ng/mL (A) or 50 ng/mL (B). After removal of the drug, the cultures were divided into three groups during a 2-wk recovery period. Group 1 (blue) was contained in a medium composed of 5 μM AdR, CdR, and GdR; group 2 (red) was in a medium composed of only CdR and GdR; and group 3 (black) served as control without deoxynucleosides. At the indicated time intervals, in cells from each group we determined the copy number of mtDNA by real-time PCR (A and B) and measured the sizes of the four dNTP pools (C). Pool sizes are mean ± SEM of the values measured at the three time points after removal of EtBr. nt (white), pool sizes in parallel cultures not treated with EtBr.
DNA Repair After UV Damage: dNTP Pools.
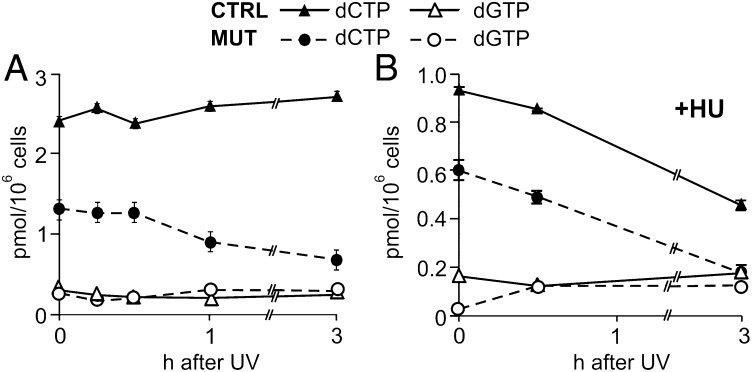
We first investigated whether and how UV irradiation affects the size of the four dNTP pools in quiescent mutant and control cells during a 3-h period after irradiation. In the control fibroblasts, the pools did not change, but in the mutant cells, the dCTP pool (originally 50% of the control pool) decreased further to 25% after irradiation. UV irradiation had no clear effect on the very small dGTP pool (Fig. 2A). Neither dATP nor dTTP was affected (data not shown).
Fig. 2.
Effects of UV irradiation and hydroxyurea on dCTP and GTP pools. (A) Pool changes after irradiation. We irradiated quiescent mutant or control fibroblasts with UV (24 J/m2) and measured the size of the dCTP and dGTP pools during a 3-h repair period. (B) Effect of hydroxyurea. The same experiment as in A was run but with 2 mM hydroxyurea (HU) in the medium starting at 30 min before irradiation and during repair. Bars indicate SEM. In most cases, the values were too low to be visible in the figure.
Does a remaining activity of ribonucleotide reductase support the dNTP pools in the mutant cells? The addition of 2 mM hydroxyurea to the incubation medium at 30 min before irradiation and during the subsequent 3 h decreased the size of the dCTP pool in both mutant and control cells (Fig. 2B). Hydroxyurea inactivates ribonucleotide reductase by sequestering the free tyrosyl radical of R2 (23). Its inhibitory effect on the dCTP pool of both types of cells indicates that in the mutated fibroblasts, ribonucleotide reduction was responsible for maintenance of the dCTP pool, implying either that p53R2 was not completely inactivated by the mutation or that the quiescent cells retained some R2 activity.
DNA Repair After UV Damage: Disappearance of Primary Damage.
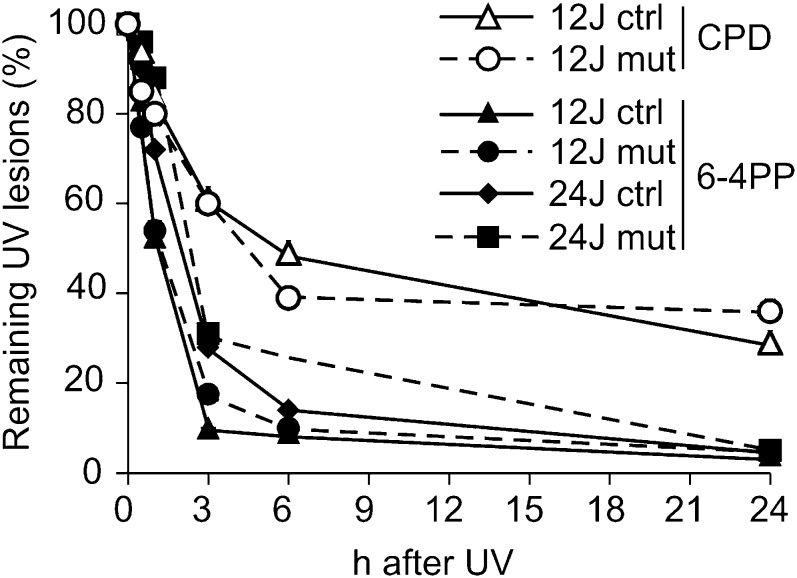
The primary UV-induced damage involves mainly two types of cross-links in the DNA structure (24): cyclobutane pyrimidine dimers (CPDs) and a smaller number of 6-4 pyrimidone photoproducts (6-4PPs). Early during nucleotide excision repair (NER), specific proteins recognize the DNA damages and remove the cross-links, with 6-4PPs disappearing rapidly during the first 6 h after irradiation and CPDs possibly persisting longer (24).
Using specific antibodies, we determined the time-dependent disappearance of the two types of DNA damage in quiescent mutant and control cells after exposure to UV irradiation (Fig. 3). The 6-4PPs were gone after 6 h, whereas CPD removal took considerably longer. In both instances, we found no clear difference between mutant and control cells, indicating that the mutant cells were not deficient in the early recognition and removal of the damaged sites.
Fig. 3.
Disappearance of primary DNA damage in mutant and control fibroblasts. We treated quiescent mutant or control cells with UV (12 or 24 J/m2) and used specific monoclonal antibodies to identify the disappearance of the two main types of DNA damage products (CPDs and 6-4PPs) over the subsequent 24 h.
DNA Repair in Mutant Fibroblasts: Fluorometric Analysis of DNA Unwinding.
dNTPs are required during NER to fill the gaps arising from excision of the UV-induced photoproducts. At the end of the process, the nicks are sealed by ligases, restoring the original dsDNA. Before the ligation step, alkali treatment of the nicked DNA produces single-stranded regions at the sites of the initial damage. Thus, the extent of DNA double-strandedness provides a measure of the ongoing but still incomplete repair (25, 26). This can be quantified by fluorometric analysis of DNA unwinding (FADU) (27) from the fluorescence of DNA-bound EtBr. Immediately after the damage occurs, fluorescence of the DNA-bound EtBr shows a rapid drop in alkali. When the nicks are sealed after resynthesis with fresh dNTPs, the ensuing recovery of fluorescence reflects completion of the gap-filling process.
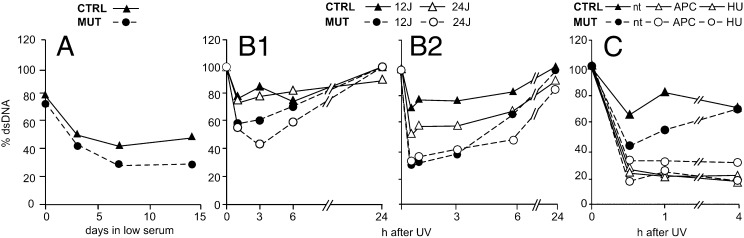
In a series of experiments, we used FADU to evaluate UV-induced DNA repair in cultures of the two cell lines maintained in low serum for various time periods. We irradiated cells with 24 J/m2 UV after 0 d (confluent cultures), 3 d, 7 d, or 14 d in 0.1% FCS, and determined the percentage of dsDNA at 3 h after irradiation (Fig. 4A). At all time points, the mutant contained less dsDNA than the control, suggesting a greater residual damage caused by slower repair. We next investigated the time course of DNA repair after UV irradiation with 12 or 24 J/m2 immediately after serum change (Fig. 4B1) or after 7 d in low serum (Fig. 4B2). In both instances, the fluorescence dropped to low values immediately after irradiation, indicating the rapid loss of dsDNA, followed over the next 6 h by a slow recovery that was almost complete after 24 h. There were some quantitatively distinct features, however; the drop in fluorescence was larger in cells irradiated with the higher UV dose, after incubation in low serum, and in mutant cells. These data suggest that the timing of fluorescence recovery is related to the efficiency of DNA repair. Such a relationship is also supported by the effects of hydroxyurea (23) and aphidicolin (28), two inhibitors of DNA synthesis (Fig. 4C). Neither drug affected the immediate loss of fluorescence after UV irradiation, but both drugs completely abolished the recovery phase in both cell lines. In fact, at the first time point after irradiation (i.e., 30 min), fluorescence was already higher in the cultures without inhibitors than in those with the inhibitors. The difference was particularly marked in the controls, reflecting the efficiency of repair synthesis.
Fig. 4.
Repair of UV-damaged DNA analyzed by FADU, which measures the fraction of dsDNA from the fluorescence of EtBr bound to alkali-treated DNA. (A) Mutant and control fibroblasts were maintained for up to 14 d in low-serum medium, as indicated on the abscissa, before irradiation with UV (24 J/m2). The percentage dsDNA was determined after 3 h of repair. (B) Time course of DNA repair in mutant and control cells after irradiation with UV (12 or 24 J/m2) after 0 (B1) or 7 (B2) days in low serum. (C) Inhibition of DNA repair by 2 mM hydroxyurea (HU) or 2 μM aphidicolin (APC) in quiescent mutant or control cells maintained for 7 d in low serum before UV irradiation (24 J/m2). The drugs were present 30 min before and for 4 h after irradiation. nt, not drug-treated.
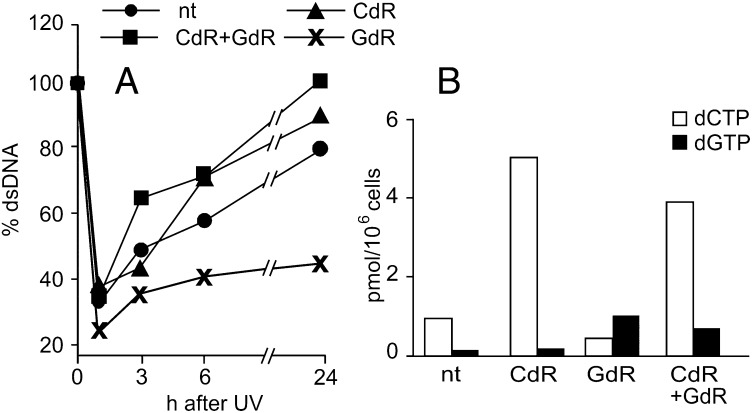
CdR and GdR were converted to their triphosphates, and in the mutant fibroblasts, their combination “cured” the dCTP and dGTP pool deficiencies (Table 1). In a FADU experiment with mutant cells, the two deoxynucleosides did not affect the initial decay of DNA fluorescence (Fig. 5A), suggesting that they do not affect the detection and removal of DNA cross-links, that is, the early steps of NER that do not require dNTPs. However, after 3 h, DNA fluorescence was already higher in the presence of deoxynucleosides than in the absence of deoxynucleosides and continued to increase until the end of the experiment, indicating faster repair. GdR alone had the opposite effect (Fig. 5A), reducing the recovery of fluorescence. This suggests that a deficiency of dCTP delays repair (Fig. 5B and Table S1).
Fig. 5.
Effects of CdR and/or GdR on UV-induced DNA repair in quiescent mutant fibroblasts. After 7 d in low serum, we irradiated mutant cells with UV (12 J/m2). The indicated deoxynucleosides were added at final concentrations of 5 μM 18 h before irradiation and during DNA repair. (A) Fraction of dsDNA determined by FADU at various time points after irradiation. (B) dCTP and dGTP pool sizes at the time of irradiation. nt, cells not treated with deoxynucleosides.
We conducted three additional FADU experiments, comparing results from mutant and control cells kept in low serum for 4, 7, or 11 d before irradiation, to substantiate a connection between dNTP pools and DNA repair (Fig. S2). In all cases, mutant cells showed delayed DNA repair, with progressively more marked effects with increasing quiescence time. The addition of CdR + GdR counteracted the delay. The combined FADU data strongly suggest that the p53R2 mutation reduces the cells’ ability to repair UV-induced DNA damage because of limitations in the supply of dNTPs.
DNA Repair in Mutant Fibroblasts: Histone H2AX Phosphorylation.
The phosphorylation of histone H2AX on Ser-139 (γH2AX) is considered a marker of repair of double-strand breaks induced by ionizing radiation (29, 30). DNA damage by UV also induces γH2AX in both proliferating and quiescent cells (31, 32), but its relevance to this connection is unclear. Thus, it was proposed that γH2AX might act as a biomarker of UV damage rather than as a participant in repair (33). During NER, the presence of γH2AX signals a persistence of DNA gaps created by excision of the UV-induced damage, and its disappearance signals filling of the gaps by dNTP polymerization. A comparison of γH2AX kinetics in mutant and control fibroblasts after UV irradiation further illuminates the importance of dNTPs and the p53R2 mutation for DNA repair.
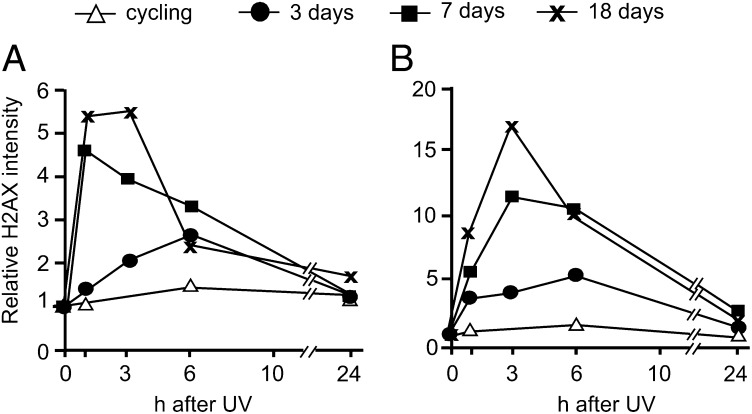
We analyzed the γH2AX content in quiescent mutant and control cells at different times after UV irradiation by flow cytometry and confocal fluorescence microscopy. In cells maintained in low-serum medium, flow cytometry showed that G1/G0 cells accounted for 95% or more of the total cell population; thus, we limited our analysis to these phases of the cell cycle. In cycling cultures, the intensity of the γH2AX signal remained almost unchanged after irradiation, with no differences between mutant and control fibroblasts (Fig. 6). In contrast, in quiescent cultures, the γH2AX signal increased in the irradiated cells and reached higher values in the mutant fibroblasts compared with the control fibroblasts. This difference increased with the time of quiescence (Fig. 6).
Fig. 6.
Histone H2AX phosphorylation after UV-induced DNA damage in cycling and quiescent mutant and control fibroblasts. Control (A) and mutant (B) cells were irradiated with UV (12 J/m2) during proliferation in 10% serum (cycling) or after 3, 7, or 18 d in 0.1% FCS medium. γH2AX was measured during a 24-h repair period by flow cytometry after staining with a specific monoclonal antibody. Note the difference in ordinates in the two panels.
We then examined the effects of deoxynucleosides on H2AX phosphorylation. Mutant and control fibroblasts were incubated with or without CdR + GdR for 7 d, from the shift to low serum to the time of UV irradiation, and during the 24 h after the irradiation. Parallel sets of cultures were maintained without added deoxynucleosides. Direct microscopic analysis of the accumulation and decay of γH2AX in irradiated mutant and control fibroblasts showed that addition of deoxynucleosides reduced the intensity and the persistence of the signal in the mutant cells, whereas it had no effect in the control cells (Fig S3).
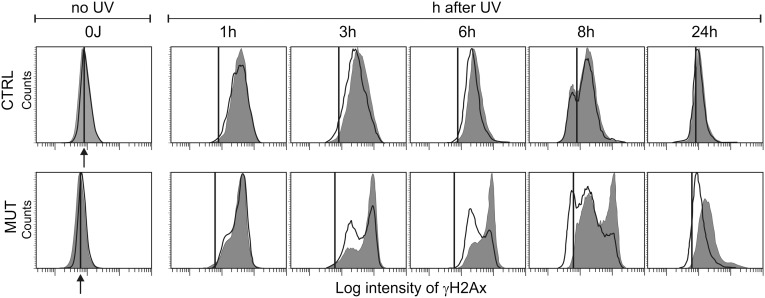
In agreement with earlier observations (31, 33) immunofluorescence revealed that the cell distribution of γH2AX at each time point was not homogeneous (Fig S3). In a parallel experiment, flow cytometry demonstrated greater heterogeneity of the fluorescent signal in mutant fibroblasts than in control fibroblasts during the 24-h repair period (Fig. 7). In the control fibroblasts, the distribution of the γH2AX fluorescence was unimodal at all times after UV irradiation exception the 8-h time point. Peak fluorescence was already reached by 3 h and then shifted back to the original preirradiation value by 24 h. In the mutant fibroblasts, a bimodal pattern already began to appear by 1 h, with a high fluorescence peak that increased in intensity for up to 8 h postirradiation. The subpopulation with lower fluorescence expanded progressively, shifting toward lower signal intensities. The addition of deoxynucleosides enhanced expansion of the lower fluorescence subpopulation, suggesting that the distinctive heterogeneity of the γH2AX signal in the mutant fibroblasts is related to a limitation of DNA precursors during repair synthesis. Here again, deoxynucleosides had no effect on the behavior of the p53R2-proficient cells.
Fig. 7.
Heterogeneity of the γH2AX signal after UV irradiaton of quiescent cells; effect of deoxynucleosides. Control and mutant cells were irradiated with UV (12 J/m2), and the distribution of γH2AX in the nuclei of G1/G0 cells during the subsequent 24 h was detected by flow cytometry after immunostaining. The gray histograms refer to cultures incubated without deoxynucleoside addition; the white histograms, to cultures incubated with 5 μM CdR + GdR for 7 d before and 24 h after irradiation. The vertical line indicates the median value of γH2AX fluorescence in nonirradiated cells, denoted by an arrow.
Discussion
Several levels of control regulate the intracellular concentrations of the four dNTPs required for DNA synthesis. Pool imbalances lead to mutations (2–4), and thus the supply of dNTPs is tightly controlled. The de novo synthesis of dNTPs is regulated by the sophisticated allosteric regulation of ribonucleotide reduction (1). In mammalian cells, their degradation to and resynthesis from deoxynucleosides in substrate cycles (34) provides further fine-tuning of pool sizes. Attesting to the importance of appropriate pool sizes are human genetic diseases caused by both deficiencies in and overproduction of dNTPs (35).
The p53R2 protein is a recent addition to this picture (9). This protein is a subunit of mammalian ribonucleotide reductase with the same radical-providing function as the canonical R2 protein. In postmitotic resting cells in which R2 has disappeared, only p53R2 can provide the free radical required for ribonucleotide reduction, acting as the functioning small subunit of mammalian ribonucleotide reductase (6, 7).
The results of our experiments with the mutant human fibroblasts reported here strongly support this concept. These cells carry a homozygous RR2MB mutation, resulting in the exchange of a highly conserved glycine residue located in the iron center of p53R2 with a valine (16). We found that the loss of p53R2 activity has no consequences for cycling cells containing a large excess of R2 over p53R2; however, in quiescent cells, when R2 function should be taken over by p53R2, mtDNA synthesis and DNA repair are disturbed. In our experiments, the final degradation of R2 occurred during maintenance of the cells in serum-depleted medium after they had reached confluency. At confluency, the cultures still contained a minor fraction of S-phase cells (20), yet the mutant already exhibited delayed repair (Fig. 4B1). After 1 wk in 0.1% FCS, the cells still contained a small amount of R2, along with small dNTP pools (20). Hydroxyurea, a known inhibitor of ribonucleotide reductase, blocked dNTP synthesis (Fig. 2B), demonstrating that the dNTPs are synthesized de novo. Additional evidence for some residual ribonucleotide reductase activity was the decrease in dCTP and dTTP pools after addition of GdR to the medium (Table S1). GdR increased the intracellular dGTP pool, resulting in diminished size of the two pyrimidine dNTPs by allosteric regulation of the substrate specificity of a functioning ribonucleotide reductase, probably containing R2 (1). Despite the small remaining R2 activity in the mutant cells, the absence of p53R2 resulted in deficiencies of dCTP and dGTP, which were further accentuated by UV irradiation (Fig. 2A).
The mutant fibroblasts had been obtained from an individual completely lacking mtDNA in muscle cells. Nevertheless, they grew normally in culture and contained a normal amount of mtDNA even during prolonged quiescence (20). We hypothesized that the low mtDNA content in fibroblasts compared with muscle cells made the fibroblasts less sensitive to a deficiency of dNTPs. Thus, we set up conditions that forced the cells to renew all of their mtDNA during a relatively short period after depletion with EtBr. The R2-containing cycling fibroblasts efficiently resynthesized the mtDNA independent of their p53R2 status, whereas quiescent mutant fibroblasts were deficient (Fig. 1). In response to the increased demand for mtDNA precursors, the supply of dNTPs in the mutant cells was insufficient. This deficiency was cured by adding deoxynucleosides, which expanded the dNTP pools. The combination of CdR, GdR, and AdR was the most effective addition.
The mutant cells also exhibited defective repair of DNA damage after UV irradiation. Deoxynucleotides participate in NER only toward the end of the process, after excision of the damaged sites. The early steps involving recognition and removal of the cross-links in the damaged DNA functioned normally in the quiescent mutant cells (Fig. 3); however, the later steps requiring the participation of dNTPs were delayed, and the damage was more extensive and persistent. This delay was evident in the FADU experiments when the irradiated mutant cells contain more single-stranded DNA after alkaline treatment and required more time to reseal the nicked DNA. The addition of deoxynucleosides had a beneficial effect and favored repair. Particularly remarkable are the divergent results seen after the addition of GdR + CdR or GdR alone. The combination of the deoxynucleosides normalized the dCTP and dGTP pools (Table 1) and improved DNA repair. GdR alone increased the dGTP pool but decreased the dCTP pool, resulting in delayed repair. Such a close correlation between the size of the dCTP pool and the cellular ability to repair DNA is strong evidence for a causal relation between the two parameters.
Surprisingly, the mutant cells required a different constellation of deoxynucleosides for mtDNA maintenance (AdR + CdR + GdR) and DNA repair (CdR + GdR). Given that the pool differences between mutant and control fibroblasts concern mainly dCTP and dGTP, we did not expect to see a requirement for AdR. However, considering that CdR is a precursor of both pyrimidine dNTPs via the conversion of dCMP to dTMP, whereas GdR feeds only the dGTP pool, we reasoned that adding AdR to the deoxynucleoside mix would create a condition favoring the synthesis of all four DNA precursors. The main effect of AdR was a twofold to threefold increase in dATP and concomitant similar decreases in the dCTP and dGTP pool expansions caused by the presence of CdR and GdR (Fig. 1C). The dTTP pool was also decreased by the addition of AdR. Thus, AdR provokes a large disturbance of the balance between dNTP pools. An important consideration in relation to the different requirements for deoxynucleosides to support mtDNA reexpansion and nuclear DNA repair is that different DNA polymerases are involved in the two processes.
The mutant cells also differed from the control cells in the level of phosphorylated histone H2AX. Kinetic flow cytometry analyses of UV-irradiated quiescent cells demonstrated larger amounts and a more extended persistence of γH2AX in the mutant fibroblasts than in the control fibroblasts. The distribution of γH2AX was not homogeneous and differed between the two cell populations. Confocal fluorescence microscopy yielded essentially similar results. The most important finding with both types of analysis was that the presence of CdR + GdR in the medium had little affect on the control cells, but shifted the behavior of the mutants toward that of the controls, again indicating that the defective DNA repair exhibited by the mutant fibroblasts in the absence of added deoxynucleosides depends on insufficient de novo synthesis of dNTPs.
Materials and Methods
The fibroblasts from a patient with an inactivating mutation in the gene for p53R2 (16) were identical to those used in a previous study (20). G. Kollberg and E. Holme (Sahlgrenska University Hospital, Goteborg, Sweden) provided the original frozen cells (second passage) and suitable age-matched normal controls. Before use, both cell lines were immortalized with plasmid CMV-hTERT/PGK-Pura, as described previously (36). The cells for the experiments were seeded at 0.35 × 106 cells/10-cm dish and grown in MEM/10% FCS. Quiescent cultures were obtained by growing cells to confluence (usually for 7 d), changing the medium to MEM/0.1% dialyzed FCS, and keeping the cells in this medium until use in the experiments at different times, usually after 7 d. Cultures received fresh medium every third day. Where indicated, the medium was supplemented with 5 μM deoxynucleosides along with a medium change. To prevent degradation of individual deoxynucleosides, 0.5 μM immucillin (37) was added together with GdR and 10 μM erythro-9-2-hydroxy-3-nonyladenine (38) with AdR.
Details of other methodologies used in this study, including sources of materials, analytical methods involving determination of dNTPs, quantification of mtDNA, depletion of mtDNA from cells, UV irradiation of cells, analysis of photoproducts to determine DNA damage, FADU after DNA damage, and determination of histone H2AX phosphorylation by flow cytometry or immunofluorescence microscopy, are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Arianna Calistri for advice and help with the immortalization of the fibroblasts. This work was supported by Italian Telethon Grant GGP09019 (to V.B.), Italian Association for Cancer Research (AIRC) Grant 1091 (to V.B.), the University of Padova, Strategic Project 2008 “Models of Mitochondrial Diseases,” and the Cariparo Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211289109/-/DCSupplemental.
References
- 1.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Niida H, Shimada M, Murakami H, Nakanishi M. Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci. 2010;101:2505–2509. doi: 10.1111/j.1349-7006.2010.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181:305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 4.Kumar D, et al. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39:1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chabes AL, Björklund S, Thelander L. S phase-specific transcription of the mouse ribonucleotide reductase R2 gene requires both a proximal repressive E2F-binding site and an upstream promoter activating region. J Biol Chem. 2004;279:10796–10807. doi: 10.1074/jbc.M312482200. [DOI] [PubMed] [Google Scholar]
- 6.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 7.Pontarin G, et al. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J Biol Chem. 2007;282:16820–16828. doi: 10.1074/jbc.M701310200. [DOI] [PubMed] [Google Scholar]
- 8.Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex Cdh1-mediated proteolysis. Proc Natl Acad Sci USA. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 10.Pontarin G, et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci USA. 2008;105:17801–17806. doi: 10.1073/pnas.0808198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourdon A, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 12.Bornstein B, et al. Mitochondrial DNA depletion syndrome due to mutations in the RRM2B gene. Neuromuscul Disord. 2008;18:453–459. doi: 10.1016/j.nmd.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acham-Roschitz B, et al. A novel mutation of the RRM2B gene in an infant with early fatal encephalomyopathy, central hypomyelination, and tubulopathy. Mol Genet Metab. 2009;98:300–304. doi: 10.1016/j.ymgme.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Tyynismaa H, et al. A heterozygous truncating mutation in RRM2B causes autosomal-dominant progressive external ophthalmoplegia with multiple mtDNA deletions. Am J Hum Genet. 2009;85:290–295. doi: 10.1016/j.ajhg.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaibani A, et al. Mitochondrial neurogastrointestinal encephalopathy due to mutations in RRM2B. Arch Neurol. 2009;66:1028–1032. doi: 10.1001/archneurol.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kollberg G, et al. A novel homozygous RRM2B missense mutation in association with severe mtDNA depletion. Neuromuscul Disord. 2009;19:147–150. doi: 10.1016/j.nmd.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Devlin HL, et al. Impairment of the DNA repair and growth arrest pathways by p53R2 silencing enhances DNA damage-induced apoptosis in a p53-dependent manner in prostate cancer cells. Mol Cancer Res. 2008;6:808–818. doi: 10.1158/1541-7786.MCR-07-2027. [DOI] [PubMed] [Google Scholar]
- 18.Kunos CA, et al. Ribonucleotide reductase inhibition enhances chemoradiosensitivity of human cervical cancers. Radiat Res. 2010;174:574–581. doi: 10.1667/RR2273.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano K, Bálint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 20.Pontarin G, et al. Deoxyribonucleotide metabolism in cycling and resting human fibroblasts with a missense mutation in p53R2, a subunit of ribonucleotide reductase. J Biol Chem. 2011;286:11132–11140. doi: 10.1074/jbc.M110.202283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 22.Arnér ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 23.Krakoff IH, Brown NC, Reichard P. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res. 1968;28:1559–1565. [PubMed] [Google Scholar]
- 24.Friedberg E, et al. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 25.Erixon K, Ahnström G. Single-strand breaks in DNA during repair of UV-induced damage in normal human and xeroderma pigmentosum cells as determined by alkaline DNA unwinding and hydroxylapatite chromatography: Effects of hydroxyurea, 5-fluorodeoxyuridine and 1-beta-D-arabinofuranosylcytosine on the kinetics of repair. Mutat Res. 1979;59:257–271. doi: 10.1016/0027-5107(79)90164-7. [DOI] [PubMed] [Google Scholar]
- 26.Baumstark-Khan C, Hentschel U, Nikandrova Y, Krug J, Horneck G. Fluorometric analysis of DNA unwinding (FADU) as a method for detecting repair-induced DNA strand breaks in UV-irradiated mammalian cells. Photochem Photobiol. 2000;72:477–484. doi: 10.1562/0031-8655(2000)072<0477:faoduf>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 27.Birnboim HC, Jevcak JJ. Fluorometric method for rapid detection of DNA strand breaks in human white blood cells produced by low doses of radiation. Cancer Res. 1981;41:1889–1892. [PubMed] [Google Scholar]
- 28.Huberman JA. New views of the biochemistry of eucaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase alpha. Cell. 1981;23:647–648. doi: 10.1016/0092-8674(81)90426-8. [DOI] [PubMed] [Google Scholar]
- 29.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 30.Paull TT, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 31.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto M, et al. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- 33.Cleaver JE. γH2Ax: Biomarker of damage or functional participant in DNA repair “all that glitters is not gold!”. Photochem Photobiol. 2011;87:1230–1239. doi: 10.1111/j.1751-1097.2011.00995.x. [DOI] [PubMed] [Google Scholar]
- 34.Rampazzo C, et al. Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat Res. 2010;703:2–10. doi: 10.1016/j.mrgentox.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Spinazzola A, et al. Clinical and molecular features of mitochondrial DNA depletion syndromes. J Inherit Metab Dis. 2009;32:143–158. doi: 10.1007/s10545-008-1038-z. [DOI] [PubMed] [Google Scholar]
- 36.Chaouch S, et al. Immortalized skin fibroblasts expressing conditional MyoD as a renewable and reliable source of converted human muscle cells to assess therapeutic strategies for muscular dystrophies: Validation of an exon-skipping approach to restore dystrophin in Duchenne muscular dystrophy cells. Hum Gene Ther. 2009;20:784–790. doi: 10.1089/hum.2008.163. [DOI] [PubMed] [Google Scholar]
- 37.Schramm VL. Development of transition state analogues of purine nucleoside phosphorylase as anti–T-cell agents. Biochim Biophys Acta. 2002;1587:107–117. doi: 10.1016/s0925-4439(02)00073-x. [DOI] [PubMed] [Google Scholar]
- 38.Carson DA, Seegmiller JE. Effect of adenosine deaminase inhibition upon human lymphocyte blastogenesis. J Clin Invest. 1976;57:274–282. doi: 10.1172/JCI108278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.