Abstract
Nature presents various protein fibers that bridge the nanometer to micrometer regimes. These structures provide inspiration for the de novo design of biomimetic assemblies, both to address difficulties in studying and understanding natural systems, and to provide routes to new biomaterials with potential applications in nanotechnology and medicine. We have designed a self-assembling fiber system, the SAFs, in which two small α-helical peptides are programmed to form a dimeric coiled coil and assemble in a controlled manner. The resulting fibers are tens of nm wide and tens of μm long, and, therefore, comprise millions of peptides to give gigadalton supramolecular structures. Here, we describe the structure of the SAFs determined to approximately 8 Å resolution using cryotransmission electron microscopy. Individual micrographs show clear ultrastructure that allowed direct interpretation of the packing of individual α-helices within the fibers, and the construction of a 3D electron density map. Furthermore, a model was derived using the cryotransmission electron microscopy data and side chains taken from a 2.3 Å X-ray crystal structure of a peptide building block incapable of forming fibers. This was validated using single-particle analysis techniques, and was stable in prolonged molecular-dynamics simulation, confirming its structural viability. The level of self-assembly and self-organization in the SAFs is unprecedented for a designed peptide-based material, particularly for a system of considerably reduced complexity compared with natural proteins. This structural insight is a unique high-resolution description of how α-helical fibrils pack into larger protein fibers, and provides a basis for the design and engineering of future biomaterials.
Keywords: fibrous proteins, protein design, helical reconstruction, X-ray crystallography
Filamentous proteins are ubiquitous in nature. They are found across all kingdoms of life, where they occur within cells, attached to cell surfaces, and extracellularly. They perform a wide variety of cellular functions, including giving shape, resilience, and stability to cells and tissues; acting as highways for intracellular transport; providing the machinery underpinning cell movement; and producing glutinous materials for cell attachment. Eukaryotic intracellular fibrous components—which include actin polymers, intermediate filaments, and microtubules—are amongst the best-studied examples at the structural level. In these cases, the monomeric building blocks, typically of nm dimensions and tens of kDa in mass, assemble through spontaneous or assisted processes to form fibers large enough to span cells tens of micrometers across. Some of these proteins and assemblies have yielded to structural definition at high resolution. There are X-ray crystal or electron-diffraction structures for the globular forms of actin (1) and the tubulins (2), and molecular models for their fibrous states based on lower-resolution electron density maps derived from cryotransmission electron microscopy (cryo-TEM) images (3, 4). Likewise, there are experimental structures for the bacterial flagella hook and filament (5, 6). However, others have proved intractable to detailed structural studies. For example, the intermediate filaments, which are assemblies of α-helical coiled coils, remain relatively poorly described at the assembly, molecular, and atomic levels (7–9).
From a more-applied perspective, interest in peptide- and protein-based fibrous biomaterials has increased recently because these materials have potential applications in biotechnology and synthetic biology—for example, as scaffolds for 3D cell culture, tissue engineering, and templating the assembly of functional inorganic materials (10–13). Although natural proteins can and are being used in these areas, much simpler or stripped-down systems are preferable because they reduce complexity, and potentially allow better understanding and control over the folding and assembly processes leading to fiber formation. Indeed, the past decade has witnessed a plethora of successful peptide-based designs of fibrous biomaterials encompassing dipeptides, β-hairpins, protein fragments, peptide-organic hybrids, and peptide amphiphiles, which are based mainly on β-sheet structures (12, 14). However, no design process is complete until the structures of the components and their assemblies have been determined to high resolution to afford molecular and, ideally, atomistic descriptions. Although in silico models for several of the above systems have been presented (11, 15, 16) and structures for amyloid-like assemblies formed from small peptides are being resolved (17, 18), there is currently no high-resolution experimental structure for a fibrous biomaterial of de novo design.
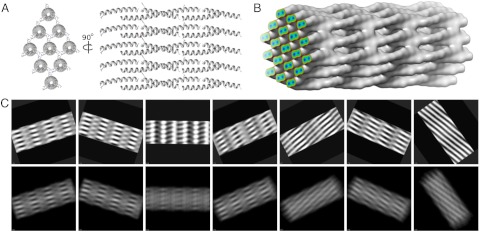
We have used an alternative protein-folding domain, the α-helical coiled coil, to design fibrous biomaterials (19). As depicted in Fig. 1A, coiled coils comprise two or more α-helical chains that wrap, or supercoil, around one another to form extended rope-like structures. Although many of these remain as discrete structures (20), others assemble further into supramolecular structures, as seen for example in intermediate filaments (21, 22). Well-defined sequence-to-structure relationships for coiled coils are available, which provide a solid basis for rational protein design (23).
Fig. 1.
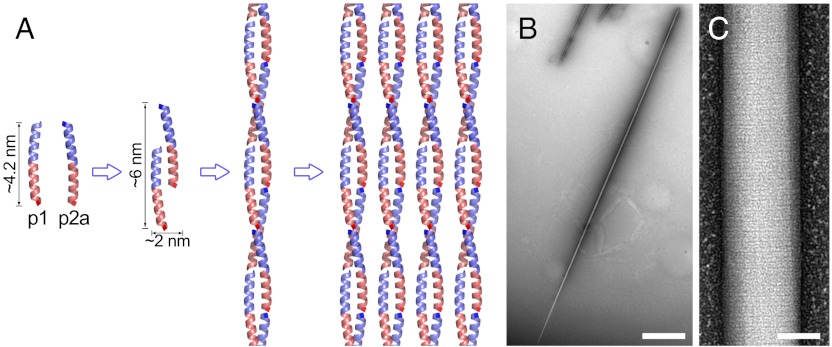
Structure and hierarchy of α-helical SAFs. (A) Two heterodimer-directing peptides (SAF-p1, SAF-p2a) assemble to form a sticky-ended unit that associates further to form coiled-coil fibrils, which are hexagonally packed within the SAFs. Each peptide has two differently charged regions: In both cases, the N and C terminal halves are predominantly basic and acidic, respectively, and are colored blue and pink accordingly. (B and C) Fibrils associate laterally to form fibers (B) that display striations (C) after staining with heavy-metal salts and visualization by TEM. Scale bars, 2 μm (B) and 50 nm (C).
We have used the understanding of coiled coils to construct a de novo designed self-assembling fiber system (SAF) (24). In these so-called SAFs, two complementary 28-residue polypeptides combine to form a “sticky ended,” parallel, dimeric coiled-coil building block of dimensions of approximately 2 by 6 nm (Fig. 1A). These assemble to form stiff fibers of approximately 40 μm in length and, intriguingly, thicken to approximately 80 nm in diameter (Fig. 1B). We have presented several designs on this theme (24–26) and explored the detailed mechanism of assembly (27). Thus far, we have probed the structure of the fibers to low resolution (approximately 2 nm) by combined negative-stain TEM and X-ray fiber diffraction (26).
These studies give the following working model for how the SAFs are structurally organized (Fig. 1A): The sticky-ended blocks assemble end-to-end into fibrils. These fibrils are most likely extended parallel, dimeric, α-helical coiled coils, albeit interrupted every 28 residues along each chain by the end of one peptide and the start of the next. The fibrils pack hexagonally into bundles in the matured fibers. There is considerable order in both dimensions of the fibers (Fig. 1C), which we have suggested is imparted by periodic interfibril side-chain interactions both laterally and longitudinally through the fibers (24–26). Although this model is helpful, our understanding remains incomplete, and any further design and engineering studies of the SAFs are limited by this low-resolution model. Here, we present an 8 Å structure of the SAFs from cryo-TEM data. This was used to generate a 3D electron density map, which, together with side-chain geometry from a 2.3 Å structure for a variant of the peptide building blocks from X-ray crystallography, yielded a molecular model of the complete fiber system. This was validated using techniques developed for single-particle analysis and also molecular-dynamics simulations. Combined, these provide the only high-resolution experimentally derived structural model of a de novo designed protein-based biomaterial.
Results
Cryo-TEM Imaging.
We turned to cryo-TEM to advance our structural studies of the SAFs. Cryo-TEM allows the visualization of proteins in near-native, frozen-hydrated states. First, cryoelectron tomography was performed on frozen-hydrated SAFs (Fig. S1). This showed that the SAFs had approximately circular cross-sections with uniform internal electron density (i.e., they are solid cylinders). Next, imaging the SAFs by cryo-TEM and without stain revealed considerable order in both fiber dimensions (width and length) (Fig. 2A). This is astonishing given that they are supramolecular structures containing millions of peptides held together by noncovalent forces. We estimate that the segment of fiber shown in Fig. 2A has approximately 220,000 peptides, and that intact fibers comprise of the order 30·106 peptides. Note that these calculations assume that coiled-coil fibrils are hexagonally packed solid cylinders, and that there are approximately 58 such fibrils across the fiber width, and approximately 60 coiled-coil peptides along the length of the segment in Fig. 2A.
Fig. 2.
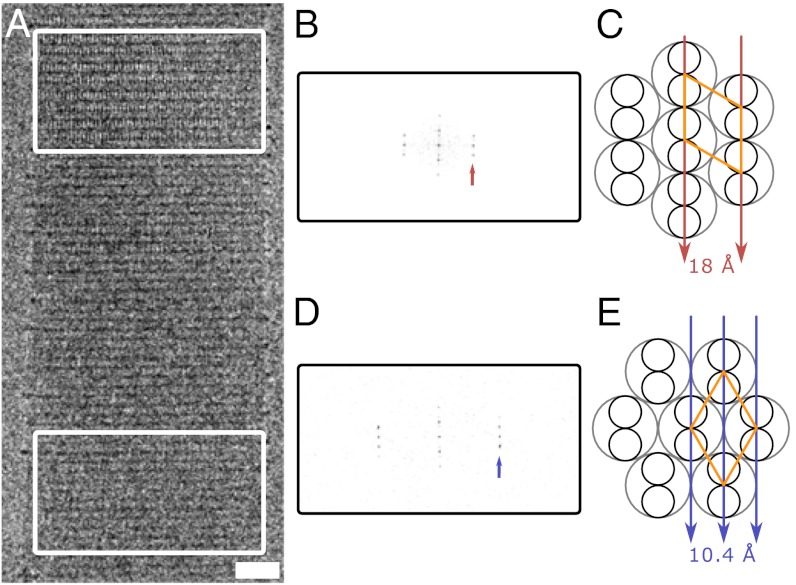
Direct observation of ultrastructure in frozen-hydrated SAFs. (A) Representative cryo-TEM of a single frozen-hydrated SAF, revealing the lateral striations across the width and along the length of the entire fiber (fiber axis is vertical), and two different sets of longitudinal striations (boxed regions). Scale bar, 20 nm. (B) Fourier transform of A, Upper, with the first row line at 18 Å (arrow). (C) Projecting along the [100] axis (arrows) causes alignment of the 18 Å [100] planes of the unit cell. Small circles represent α-helices within the coiled coil (large gray circles); the hexagonal unit cell with sides of 20.8 Å is shown in orange. (D) Fourier transform of A, Lower, with the row line at 10.4 Å (arrow). In both B and D, the first meridional reflection is at 41.8 Å. (E) Projecting along the [110] axis aligns the 10.4 Å [110] planes.
More specifically, in unstained cryo-TEM there were lateral striations with a similar repeat distance to that seen by negative-stain TEM (Figs. 2A and 1C, respectively). The microscope was calibrated at high magnification using a catalase crystal and a sample of graphitized carbon. This allowed the striations in the cryo-TEM micrographs to be determined accurately as a 41.8 Å repeat; this was confirmed using T4 bacteriophage as an in situ standard, which has a striation spacing of 40.6 Å. As we have noted before (26), this corresponds almost precisely to the length of each SAF peptide when configured as a coiled coil—i.e., 41.44 Å (= 28·1.48 Å, the rise per residue in an α-helical coiled coil). Thus, coiled-coil peptides are aligned along the long axis of the fibers as per our original design (24) and subsequent structural analyses of stained and dried fibers (26).
Moreover, order in the other fiber dimension was also visible in the cryo-TEM images. Amongst these, there were striations with a periodicity of 18 Å parallel to the fiber long axis (Fig. 2A, Upper). These had not been seen before, but relate to the hexagonal packing of the coiled coils as observed by wide-angle X-ray fiber diffraction (26). It is important to note here that the images are formed from the projected electron density of the protein only, so these striations are a direct visualization of the supramolecular structure of the SAFs.
Analysis of these new longitudinal striations immediately improved our model of the organization in the SAFs: Not only are the coiled-coil fibrils hexagonally packed, but they are highly aligned. In such a system, particular lattice planes are expected to become visible at specific projection angles through the fibers (i.e., at specific positions along the aligned coiled-coil supercoils) (Table S1). For example, the 18 Å longitudinal striations correspond to projecting through the fibrils along the [100] axis, which allowed the distance between adjacent fibrils to be determined as 20.8 Å (Fig. 2 A–C). At different points along the fibers, other lattice planes from Table S1 are expected to be evident, provided sufficient resolution. Indeed, the resolution of our EM images was sufficient to see the [110] planes as longitudinal striations with a periodicity of 10.4 Å (Fig. 2 A, D, and E, Lower, and the other planes in Table S1).
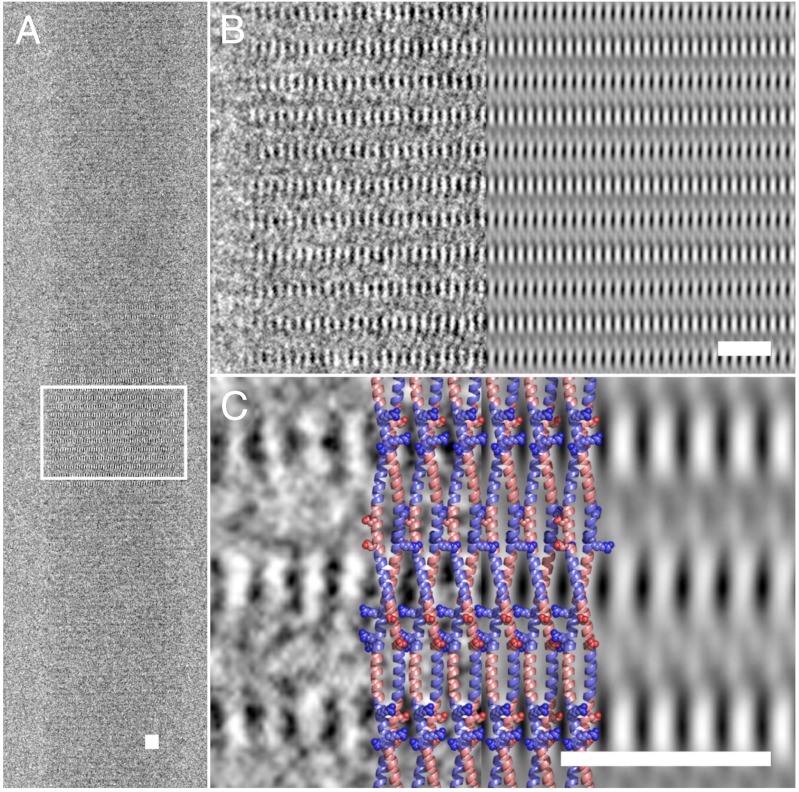
Although the SAFs are 3D objects, with the above organization and alignment we reasoned that methods designed for the analysis of 2D crystals should be applicable to solve the structure of the SAFs. Initially, we focused on the [100] projection of an ultrahigh-resolution image of a SAF fiber (Fig. 3), leaving the other regions to verify our models at a later stage. The Fourier transform of the image shown in Fig. 3A was consistent with a single lattice with reflections extending to 6 Å along the equator, and past 20 Å along the meridian (Fig. S2). The equatorial reflections relate to the aforementioned hexagonal packing of the coiled-coil fibrils (Fig. 2). The meridional reflections report on the coiled-coil helical and superhelical structures along the long axis of the fibers.
Fig. 3.
Direct imaging of dimeric α-helical coiled coils within the SAF ultrastructure. (A) Region showing [100] planes to high resolution used for further analysis (fiber axis is vertical). (B) After processing the images with 2dx (28), the magnified boxed area (Left) more clearly shows the regular lattice of high-contrast striations (Right). (C) Further magnification of the experimental (Left) and processed (Right) images, with a cartoon overlay of the postulated SAF structure (Middle, colored as in Fig. 1). Light areas indicate high electron density. The lateral striations are proposed to be caused by the charged Arg and Asp residues (spheres) forming salt bridges between adjacent coiled coils. Scale bars, 10 nm.
In fiber diffraction patterns, reflections appear along lines (layer lines). The first of these was at 125.4 Å (Fig. S2), corresponding to the superhelical pitch of the coiled coil. However, the first meridional reflection was at 41.8 Å (i.e., one-third of the pitch), indicating that the coiled-coil fibrils comprise three heterodimeric subunits per single coiled-coil pitch, and, as such, are 31 helices. This second distance is the same as the length of the folded peptides (see above). This is not a coincidence, and it explains the hexagonal packing: Viewing a coiled-coil fibril along its long axis reveals that each peptide motif repeats every 120° (Fig. S3). This leads to repeating side chain-mediated interfibril interactions that cement the hexagonal packing, which could not be achieved with a noninteger, or indeed any other superhelix.
Image processing (28) led to a 2D electron density map that showed clear density consistent with paired α-helices in the coiled-coil fibrils (Fig. 3 B and C). This was direct structural evidence that the SAFs assembled via dimeric coiled coils as designed. Similar density modulations from aligned α-helices within stacked coiled coils have been reported in thin sheets of myosin fragments (29). Our map also highlighted the lateral striations (Fig. 3B). Although the resolution of the map was not sufficient to see amino acid side chains, the positions of these striations indicated regions of increased electron density that are likely to correspond to regions of closely packed and ordered side chains. The sequences of the SAF peptides include Asp and Arg residues (Table S2) that had been designed to fall on opposite faces and at the same level in the fully folded coiled-coil fibril. These charged clusters would therefore repeat with the peptides once every 41.8 Å (i.e., one-third of the supercoil pitch). As such, they provide ideal candidates for causing the lateral striations and for mediating hexagonal packing by intercoiled-coil salt-bridge formation, as introduced above.
X-Ray Crystallography.
Because the SAFs are composed of sticky-ended dimers that assemble to form large filaments, they are not amenable to standard X-ray crystallography. Therefore, to obtain a high-resolution structure of the building blocks, we permuted SAF-p1 to form blunt-ended dimers. This new Blunt-p1-I peptide included an iodophenylalanine residue in place of a tyrosine in the parent peptide (Table S2). The inclusion of iodine did not affect fibrillogenesis: A sticky-ended combination of an iodine-containing variant of SAF-p1 (SAF-p1-I; Table S2) and SAF-p2a formed fibers, which were striated in precisely the same manner and with the same dimensions as the parent fibers (Fig. S4). Although additional electron density from the iodine atoms might be expected in the EM images, this proved not to be obvious. This is possibly because these atoms are proximal to the Asp and Arg residues (Table S2), already a region of high electron density. These data for the iodinated fibers were therefore fully consistent with our developing structural model.
The blunt-ended assembly between Blunt-p1-I and SAF-p2a was crystallized and its X-ray structure solved to 2.3 Å using experimental phasing from the anomalous signal of iodine (Table S3). The resulting structure showed the peptide forms a parallel, dimeric, α-helical coiled coil (Fig. S5A), fully consistent with our original design and subsequent experiments on the SAFs (24, 27). The pitch of this blunt-ended coiled coil was 143 Å, a value significantly higher than that returned from the cryo-TEM analysis (Fig. S5B). As discussed below, this is interesting in terms of how the coiled-coil superhelix is remodeled within the fibers. However, because of these differences, the full X-ray crystal structure could not be used directly as a search model for the next stages of generating an electron density map and building an atomistic model for the SAFs. Nonetheless, the side-chain conformations from the X-ray crystal structure were useful in these processes.
Electron-Density Maps.
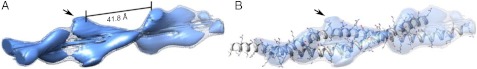
Cryoelectron tomography can be used to generate 3D electron density maps of protein complexes. However, in our microscope, the cumulative electron dose required for a tomographic series showing the striations at sufficient signal-to-noise, even in low-dose conditions, destroyed the fibers. Therefore, we used the processed image shown in Fig. 3 in helical reconstruction (30) (Fig. S6 and Table S4). The map revealed density consistent with the α-helices of a dimeric coiled coil, into which an all-atom model could readily be placed (Fig. 4). A search model for this was generated using MAKECCSC (31), a program that builds a sequence of amino acids as a parallel coiled coil of given geometry. Because the coiled coils in the SAFs are 31 helices, there were 84 residues in each α-helix in one coiled-coil pitch, and, therefore, 23 turns of each α-helix. Combined with the supercoil pitch and radius, all parameters that govern coiled-coil geometry can be derived except the relative orientation of α-helices, which was set as 210° as defined as typical for natural coiled coils (31). Coordinates for the side chains were taken from the X-ray crystal structure of the Blunt-p1-I:SAF-p2a complex, but with the iodophenylalanine replaced by the parent tyrosine. Breaks in the backbone were then added to form the correct N and C termini for the SAF-p1 and SAF-p2a peptides.
Fig. 4.
Electron density map of composite coiled-coil fibrils comprising multiple SAF peptides. (A) Electron density map (EMDB entry number 1995) generated using the Burnham Brandeis Helical Package (30), showing the lobes of density spaced 41.8 Å apart. Solid surface and net contain 60% and 100% of the mass of the protein, respectively. (B) The arrows indicate the lobe of electron density into which the Asp and Arg salt bridges fit, as well as the tyrosine residue, after docking the model using Chimera (University of California, San Francisco) (32).
Note that we considered the possibility of fibrils comprising antiparallel coiled coils, but excluded it on the basis that: (i) This is not consistent with the above X-ray crystal structure; (ii) the pairing of asparagine residues in the hydrophobic core would be broken, leaving two orphaned and therefore destabilizing asparagines every peptide-peptide interface (24); and (iii) all of the designed g:e salt-bridge interactions would be broken and replaced by repulsive, like-charge interactions (g:g and e:e) (24).
This parallel coiled-coil model for the fibrils was docked as a rigid structure into the map (32). A threshold value for the surface representation was set to enclose the volume corresponding to either 100% (net; Fig. 4A) or 60% (solid surface; Fig. 4A) of the mass of the protein model. The resulting fit, shown in Fig. 4B, was obtained regardless of the starting position of the search model and showed that the Arg, Asp, and proximal tyrosine residues best filled the large and prominent lobes of electron density regularly spaced on the surface of the fibrils (Fig. 4). Again, this is consistent with these being the cause of the lateral striations and the resultant hexagonal packing. Although the direction of the coiled-coil fibrils within the map could not be unambiguously assigned through this modeling, this does not affect the positioning of this constellation of charged and aromatic residues within the envelope of electron density.
All-Atom Model.
To test further the hypothesis that outwardly and oppositely facing Asp and Arg residues mediate hexagonal packing of the coiled-coil fibrils, an all-atom 3D model of the SAFs was generated (Fig. 5A). A hexagonal array of fibrils from the above helical reconstruction was generated based on the dimensions of the lattice from cryo-TEM images (Fig. 2 and Table S1), and with the fibrils in an all-parallel arrangement. The modeled interfibril interactions centered on the aforementioned Asp:Arg salt bridges (Fig. 5A). After an equilibration period of 20 ns, the rmsd over all 25,569 backbone and side-chain atoms to the initial energy-minimized structure remained at 3 Å for the remaining 100 ns of the simulation (Fig. S7), indicating a viable configuration for the coiled coils within assembled fibers.
Fig. 5.
Model of the SAF superstructure used to simulate cryo-TEM images. (A) The hexagonal array of coiled coils used for the molecular-dynamics simulations, shown as a sticky-ended unit cell, held together with Asp:Arg salt bridges. (B) Model filtered to 10 Å used to simulate projection images, which is identical to the molecular-dynamics simulation but contains more coiled coils. The surface threshold is set to contain 100% of the mass of the protein array. Protein occupies 50% of the volume of the array and contains pores of approximately 100 Å2. The colored plane shows the density gradient and clearly shows the two α-helices comprising the coiled coils. (C) Projection images of the model generated by EMAN2 (16) (Bottom) and matching 2dx-processed images (28) (Top).
Because of the unusual structure of the SAFs, neither classical electron crystallography nor single-particle analysis could be used: Electron crystallography requires thin 2D crystals and single-particle analysis requires a homogenous population of large “single” particles, whereas the SAFs are thick, 3D, heterogeneous fibrous assemblies of 3 kDa peptides.
Instead, a hybrid approach was designed so that the processed images were used as high-resolution “class-averages,” which were then compared to the projected electron density of the all-atom model (Fig. 5B and C). The model hexagonal array of coiled coils was used to simulate an electron density map filtered to 10 Å. The electron density of the model was projected over angular steps of 5° (Movie S1) and the resulting projection images compared and aligned to the processed images output from 2dx (28). Matching projection images and processed micrographs (shown in Fig. 5C) illustrate the similarity between the two sets. Taking together the facts that our experimental data extend out to 6 Å and that individual α-helices are visible in both projected and processed images, we assign a conservative resolution estimate of 8 Å to the data presented herein.
Interestingly, some of the experimental images of fibers did not have lateral striations (Fig. S8). Using the model, however, these were shown to represent fibers that are oriented out of the imaging plane. Hence, although lateral striations are visible over the full angular range around the fiber axis, they rapidly disappear as the fiber tilts away from the horizontal. Conversely, the longitudinal striations are visible over only a narrow in-plane rotation, but are visible over a larger out-of-plane tilt because of the alignment of α-helices (Movie S1).
Finally, we made a model array with an antiparallel arrangement of coiled-coil fibrils. In contrast to the all-parallel model, projections from this model displayed similar longitudinal striations to the experimental micrographs, but none exhibited lateral striations (Fig. S9). This indicates further that SAFs comprise parallel arrays of coiled-coil fibrils (25).
Discussion
Using a combination of cryo-TEM, 2D-crystal image analysis, and helical reconstruction, we have generated a unique high-resolution structure of a de novo designed self-assembling peptide fiber. Cryoelectron tomography showed that the SAFs are large, solid cylinders, and not sheets or thick 2D crystals. As a consequence of this remarkable structure, neither classical electron crystallographic techniques nor single-particle analysis could be used to analyze micrographs of the SAFs. Instead, we used an approach that combined various image analysis and modeling techniques. Cryo-TEM showed that the peptides assembled to long, parallel, dimeric coiled-coil fibrils. Fourier transforms of raw images indicated that, within these fibrils, peptide heterodimers repeated three times per coiled-coil pitch, measured at 125.4 Å, and that the fibrils were ordered further, being hexagonally packed and separated by 20.8 Å. Because of this high level of order, 2D electron crystallographic techniques could be applied to increase the resolution of the raw images, which, combined with helical reconstruction, allowed the generation of a 3D electron density map.
A coiled-coil model was built using the above data, together with verified aspects of the SAF design (specifically that the peptide termini are very close in space, per ref. 26) as well as standard parameters from known parallel, dimeric coiled coils, including the relative orientation of helices (31) and restraints on the coiled-coil radius from the distance between fibrils (coiled-coil fibrils are 20.8 Å apart, which restricts the radius to approximately 4.4 Å, typical for dimeric coiled coils). Fitting this model into the 3D electron density map confirmed that the lateral striations and hexagonal packing are caused by salt bridges between the Asp and Arg residues, which are on the surface of the coiled-coil fibrils and proximal in an extended array of fibrils. With this information, and an X-ray crystal structure of a related peptide dimer, an all-atom model for the SAFs has been generated and tested for robustness in extended molecular-dynamics simulations. Finally, the structure has been validated using an approach that combines analysis of individual micrographs using 2D crystallography and comparison of the images to projections from a 3D model (i.e., similar to single-particle analysis).
In terms of methods development, we envisage that this approach will be of use to others working on similar problems in the structural biology of fibrous systems. In relation to supramolecular peptide assembly, these unique structural insights and the recently determined assembly pathway (27) mean that the SAFs represent the best-defined, designed, peptide-based, fiber-forming system presented to date. Nonetheless, there remain open questions in this system—most notably, why the thickness of the fibers, which we show here is based on perfect hexagonal packing and would be expected to extend indefinitely, is limited. An answer to this question awaits detailed modeling, which will now be better informed by the kinetic analysis and the structural model presented herein.
In addition, a key finding of our work is that coiled-coil fibrils can be remodeled within these assemblies. Specifically, the helical supercoil can be compressed to allow better packing within the larger supramolecular structure. In this respect, it is worth comparing the values for the coiled-coil pitch derived from the “isolated or free” blunt-ended coiled coil determined by X-ray crystallography and the coiled coils packed within the fibers as determined by cryo-TEM (Fig. S5B): This is larger for the former, at 143 Å, versus 125.4 Å in the latter; the coiled-coil radius was also reduced between the X-ray crystallography and cryo-TEM structures, from 4.9 to 4.4 Å, but given the lower resolution of the latter we do not interpret this any further. To place the pitch values in a broader context, we calculated the superhelical pitches for a larger, manually validated set (33) of X-ray crystal structures of parallel dimeric coiled coils from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB), culled using the coiled-coil database CC+ (34) and parameterized using the program TWISTER (35). These are shown in Fig. S5B, where it is clear that the parameters for the SAF X-ray crystal and cryo-TEM structures fall comfortably within the distribution for PDB set, but that the former falls in the upper half and the latter in the lower half of the values. We suggest that this compression in the SAF system is a direct consequence of the coiled-coil fibrils being constrained within the SAFs by the necessity for forming hexagonal packing contacts, with any strain energy being offset by improved interfibril interactions in the fibers. Indeed, the requirement for repeating contacts every 120° necessitates the peptides to repeat three times per pitch, decreasing the pitch and compressing the fibrils during fiber formation. The structural viability of this compressed assembly was confirmed by the aforementioned molecular-dynamics simulations, which showed motions consistent with simple thermal fluctuations about the model structure (Fig. S7). Thus, the fibrils appear to be remodeled in moving from the X-ray structure, which possibly reflects the situation in a solution of preassembled SAF-peptide heterodimers, through to the assembled, fibrous SAFs.
This coiled-coil remodeling mechanism may also be applicable to the structure of intermediate filaments (IFs). Although it is known that IFs are predominantly coiled coil in structure (36), their structure within filaments has proven difficult to solve, partly because of structural heterogeneity and the lengths of the molecules thwarting crystallization. However, it is known that IFs undergo a radial compaction stage during assembly (37). This may reflect the transition from a loose assembly of eight tetramers to a more formal assembly, as the filaments form stronger, more regular contacts between adjacent coiled-coil fibrils. Our results suggest a potential packing regime within the mature fibril, whereby adjacent coiled coils may be held together with salt bridges that repeat with the same frequency as the coiled-coil pitch. Indeed, charge interactions have been proposed to result in staggered coiled-coil alignment in myosin thick filaments when offset by a multiple of a quarter of one pitch (38).
Finally, the structures and models that we present offer a platform for improved engineering of peptide-based fibrous materials, such as the SAFs, and put us a step closer to realizing their application in bionanotechnology through rational materials design.
Materials and Methods
Peptide Synthesis.
Peptides were synthesized by solid-phase peptide synthesis and purified by high-performance liquid chromatography (SI Text).
X-Ray Crystal Structure Determination.
Diffraction-quality crystals for the peptides were obtained using standard high-throughput sparse matrix screens, and structure solution was performed using anomalous phasing methods (SI Text). A summary of the refinement and model-building statistics can be found in Table S3. The refined model and relevant crystallographic data have been deposited in the PDB with accession code 3RA3.
Cryotransmission Electron Microscopy and Tomography.
Peptides were prepared as described previously (26). Samples were frozen and imaged as described in SI Text.
Image Processing.
Micrographs of fibers displaying striations were treated as 2D crystals and analyzed using 2dx (28). Layer and row lines were identified by eye and followed by automatic spot selection and unbending. The contrast transfer function (CTF) modulates the phases of the TEM images based on the amount of defocus and the spherical aberration constant of the objective lens, and was corrected for each image individually. Because the micrographs did not represent identical views of a thin 2D crystal, crystallographic merging of the data was not viable. Therefore, the best images (those with clear reflections and phases out to the highest resolution) were chosen for model refinement. EMAN2 was used to determine the orientation of the processed images and validate the all-atom model (16). Processed images containing 5 by 2 unit cells were padded into 400- by 400-pixel boxes. An all-atom PDB model containing 19 coiled coils was filtered to 10 Å and converted to Medical Research Council format using e2pdb2mrc.py with a box size of 400 by 400 and pixel size of 0.627 Å to match the processed images. Projections of the model sampled C1 space in 5° increments. Using e2refine.py, micrographs were aligned to the best-matching projection image and identified using e2display.py. Tomographic tilt series were collected using FEI software and processed using IMOD (39).
Helical Reconstruction.
Bessel orders were assigned with prior knowledge of coiled-coil geometry (Fig. S6 A and B). The helical lattice (Fig. S6 C and D) and Bessel orders of layer lines (Table S4) were assigned and the Burnham Brandeis Helical Package (30) was used to reconstruct the 3D electron density map from an individual CTF-corrected map at 9.5 Å resolution to avoid Bessel function overlap. The map has been deposited in the Electron Microscopy Data Bank with accession code 1995.
Model Building and Fitting.
Coiled coils were modeled using MAKECCSC and docked into the 3D map as described in SI Text.
Molecular-Dynamics Simulation.
Molecular-dynamics simulations were performed using periodic boundary conditions as an NPT ensemble at 278 K, described further in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
The authors thank Dr. Frank Booy for tutelage on cryo-TEM; Dr. Gail Bartlett for help with programming; and Prof. Helen Saibil, Dr. John Squire, and Dr. Gerald Offer for valuable discussions. We thank the Advanced Computing Research Centre of the University of Bristol for the provision of high-performance computing time. We also thank the Biotechnology and Biological Sciences Research Council for funding (Grant BB/E22359/1). T.H.S. thanks the Engineering and Physical Sciences Research Council for a PhD studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been depoted in the Protein Data Bank, www.pdb.org (PDB ID code 3RA3; EMDB code 1995).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118622109/-/DCSupplemental.
References
- 1.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 2.Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 Å resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 3.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 4.Sui HX, Downing KH. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure. 2010;18:1022–1031. doi: 10.1016/j.str.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 6.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Kreplak L, Aebi U, Herrmann H. Molecular mechanisms underlying the assembly of intermediate filaments. Exp Cell Res. 2004;301:77–83. doi: 10.1016/j.yexcr.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Goldie KN, et al. Dissecting the 3-D structure of vimentin intermediate filaments by cryo-electron tomography. J Struct Biol. 2007;158:378–385. doi: 10.1016/j.jsb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Nicolet S, Herrmann H, Aebi U, Strelkov SV. Atomic structure of vimentin coil 2. J Struct Biol. 2010;170:369–376. doi: 10.1016/j.jsb.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Schneider JP, et al. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc. 2002;124:15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 11.Zhou M, et al. Self-assembled peptide-based hydrogels as scaffolds for anchorage-dependent cells. Biomaterials. 2009;30:2523–2530. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Woolfson DN, Mahmoud ZN. More than just bare scaffolds: Towards multi-component and decorated fibrous biomaterials. Chem Soc Rev. 2010;39:3464–3479. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]
- 13.Boyle AL, Woolfson DN. De novo designed peptides for biological applications. Chem Soc Rev. 2011;40:4295–4306. doi: 10.1039/c0cs00152j. [DOI] [PubMed] [Google Scholar]
- 14.Ulijn RV, Smith AM. Designing peptide based nanomaterials. Chem Soc Rev. 2008;37:664–675. doi: 10.1039/b609047h. [DOI] [PubMed] [Google Scholar]
- 15.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopal K, Lamm MS, Haines-Butterick LA, Pochan DJ, Schneider JP. Tuning the pH responsiveness of beta-hairpin peptide folding, self-assembly, and hydrogel material formation. Biomacromolecules. 2009;10:2619–2625. doi: 10.1021/bm900544e. [DOI] [PubMed] [Google Scholar]
- 17.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 18.Wiltzius JJW, et al. Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woolfson DN. Building fibrous biomaterials from alpha-helical and collagen-like coiled-coil peptides. Biopolymers. 2010;94:118–127. doi: 10.1002/bip.21345. [DOI] [PubMed] [Google Scholar]
- 20.Moutevelis E, Woolfson DN. A periodic table of coiled-coil protein structures. J Mol Biol. 2009;385:726–732. doi: 10.1016/j.jmb.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: From cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 22.Parry DAD, Strelkov SV, Burkhard P, Aebi U, Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Exp Cell Res. 2007;313:2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 24.Pandya MJ, et al. Sticky-end assembly of a designed peptide fiber provides insight into protein fibrillogenesis. Biochemistry. 2000;39:8728–8734. doi: 10.1021/bi000246g. [DOI] [PubMed] [Google Scholar]
- 25.Smith AM, Banwell EF, Edwards WR, Pandya MJ, Woolfson DN. Engineering increased stability into self-assembled protein fibers. Adv Funct Mater. 2006;16:1022–1030. [Google Scholar]
- 26.Papapostolou D, et al. Engineering nanoscale order into a designed protein fiber. Proc Natl Acad Sci USA. 2007;104:10853–10858. doi: 10.1073/pnas.0700801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bromley EHC, et al. Assembly pathway of a designed alpha-helical protein fiber. Biophys J. 2010;98:1668–1676. doi: 10.1016/j.bpj.2009.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2dx: User-friendly image processing for 2D crystals. J Struct Biol. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 29.Quinlan RA, Stewart M. Crystalline tubes of myosin subfragment-2 showing the coiled-coil and molecular interaction geometry. J Cell Biol. 1987;105:403–415. doi: 10.1083/jcb.105.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owen CH, Morgan DG, DeRosier DJ. Image analysis of helical objects: The Brandeis Helical Package. J Struct Biol. 1996;116:167–175. doi: 10.1006/jsbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 31.Offer G, Sessions R. Computer modeling of the alpha-helical coiled-coil: Packing of side-chains in the inner-core. J Mol Biol. 1995;249:967–987. doi: 10.1006/jmbi.1995.0352. [DOI] [PubMed] [Google Scholar]
- 32.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong CT, Vincent TL, Green PJ, Woolfson DN. SCORER 2.0: An algorithm for distinguishing parallel dimeric and trimeric coiled-coil sequences. Bioinformatics. 2011;27:1908–1914. doi: 10.1093/bioinformatics/btr299. [DOI] [PubMed] [Google Scholar]
- 34.Testa OD, Moutevelis E, Woolfson DN. CC plus: A relational database of coiled-coil structures. Nucleic Acids Res. 2009;37:D315–D322. doi: 10.1093/nar/gkn675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J Struct Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann H, Aebi U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 37.Herrmann H, Haner M, Brettel M, Ku NO, Aebi U. Characterization of distinct early assembly units of different intermediate filament proteins. J Mol Biol. 1999;286:1403–1420. doi: 10.1006/jmbi.1999.2528. [DOI] [PubMed] [Google Scholar]
- 38.McLachlan AD, Karn J. Periodic features in the amino-acid-sequence of nematode myosin rod. J Mol Biol. 1983;164:605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- 39.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.