Abstract
Known viruses build their particles using a restricted number of redundant structural solutions. Here, we describe the Aeropyrum coil-shaped virus (ACV), of the hyperthermophilic archaeon Aeropyrum pernix, with a virion architecture not previously observed in the viral world. The nonenveloped, hollow, cylindrical virion is formed from a coiling fiber, which consists of two intertwining halves of a single circular nucleoprotein. The virus ACV is also exceptional for its genomic properties. It is the only virus with a single-stranded (ss) DNA genome among the known hyperthermophilic archaeal viruses. Moreover, the size of its circular genome, 24,893 nt, is double that of the largest known ssDNA genome, suggesting an efficient solution for keeping ssDNA intact at 90–95 °C, the optimal temperature range of A. pernix growth. The genome content of ACV is in line with its unique morphology and confirms that ACV is not closely related to any known virus.
Keywords: Archaea, hyperthermophile, virion structure
Cells from each of the three domains of life, Archaea, Bacteria, and Eukarya, are infected by a plethora of diverse viruses (1, 2). These viruses are now increasingly recognized to have played a major role in the history of life on our planet, introducing new functions in cellular genomes, promoting gene transfer, and controlling microbial populations (3–5). However, our knowledge of viral biodiversity is still in its infancy. Most of the currently studied viruses infect a limited number of model organisms—certain groups of animals, plants, and bacteria. Furthermore, whereas viruses infecting Eukarya and Bacteria have been studied for a century, the study of viruses infecting Archaea was initiated only 30 y ago and remains limited to a small number of laboratories (2, 6). As a consequence, the number of described species of archeoviruses represents less than 1% of the known viruses from the other two domains of life (1). Despite this limited number, the morphological diversity of archaeal viruses is astounding. Archaeal viruses include all morphotypes of bacterial double-stranded (ds) DNA viruses, with head-tailed, icosahedral, and pleomorphic virions and, additionally, diverse Archaea-specific morphotypes never observed among viruses infecting the Bacteria or Eukarya domains (6, 7). It is noteworthy that exceptional viral morphotypes are associated mainly with the hyperthermophilic Archaea and include droplet-shaped, bottle-shaped, fusiform, and other unexpected morphologies. Viruses similar to the bacterial head-tailed viruses (order Caudovirales) have never been isolated from hyperthermophiles; viruses of this type infect mesophilic Archaea—the extreme halophiles and methanogens (8, 9). Not only the morphotypes but also the genomes of hyperthermophilic archaeal viruses are exceptional: more than 90% of their putative genes have no recognizable functions or detectable homologs in other viruses or cellular life forms (6, 10, 11). Moreover, the ways in which some of these viruses interact with their hosts are also particular, as recently demonstrated by the description of a unique virion release mechanism (12–14).
The distinctive morphological and genomic characteristics of hyperthermophilic archaeal viruses could reflect the features of the ancient virosphere, and their description may contribute to understanding the evolutionary history of viruses and their interactions with hosts (2, 6, 15). It is noteworthy that known hyperthermophilic archaeal viruses carry dsDNA genomes (6). Although isolation procedures are not biased toward dsDNA viruses and a haloarchaeal virus with a single-stranded (ss) DNA genome has been described (16), no virus with an ssDNA or an RNA genome has been isolated from a hyperthermic environment (17). As such, the very possibility of their existence in the harsh conditions of hyperthermic habitats was questionable.
Here, we demonstrate that ssDNA viruses do exist in extreme geothermal environments and describe the unique virion structure and genomic properties of an Aeropyrum coil-shaped virus (ACV), a hyperthermophilic archaeal virus with an ssDNA genome.
Results
Virus Isolation.
The virus ACV was discovered and isolated from a culture of Aeropyrum pernix established from an environmental sample collected at the coastal Yamagawa Hot Spring in Japan, where the temperature reaches 104 °C (18), as described in SI Materials and Methods. The virus could not be replicated in any of the 60 available strains of A. pernix, nor in Aeropyrum camini (SI Materials and Methods). Consequently, ACV was propagated in the original A. pernix culture, precluding formal demonstration of infectivity.
Virion Structure.
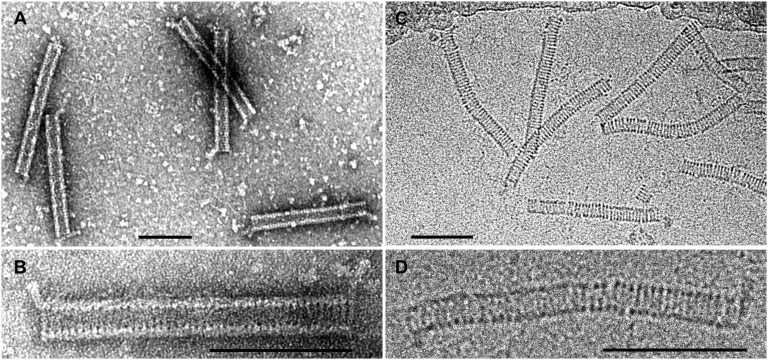
Negatively stained virions of ACV analyzed by transmission electron microscopy (TEM) appeared as rigid cylindrical particles of 220 ± 10 × 28 ± 2 nm with appendages of 20 ± 2 nm protruding from both termini at 45° angles to the axis of the cylinder (Fig. 1A). The appendages in ∼80% of the virions protruded from the same face of the virion, whereas ∼20% did so from the opposite faces (Fig. S1). The virion surface demonstrated a clear periodic pattern, which could represent either stacked discs or a helix with a shallow rise. Approximately 40 discs or 40 turns of the putative helix were readily distinguishable (Fig. 1B).
Fig. 1.
Electron micrographs of ACV virions. (A and B) Negatively stained with 2% (wt/vol) uranyl acetate. (C and D) Sample embedded in vitreous ice. (Scale bars, 100 nm.)
ACV virions embedded in vitreous ice and observed with cryo-EM appeared to be more flexible than the negatively stained virions (Fig. 1 C and D). The ice-embedded virions measured 230 ± 10 × 19 ± 1 nm. The closer examination of cryo-EM images revealed periodic, higher density regions along the sides of the cylindrical particles appearing as darker dots in Fig. 1D. The staggered position of these dots on the opposite sides of the particle (Fig. S2) was more consistent with the helical organization of the virion rather than the alternative possibility, i.e., stacking of discs. The helical organization of the ACV virion was even more evident from the analysis of partially disassembled particles (see below). From the examination of TEM and cryo-EM images, we also conclude that the virion of ACV is nonenveloped.
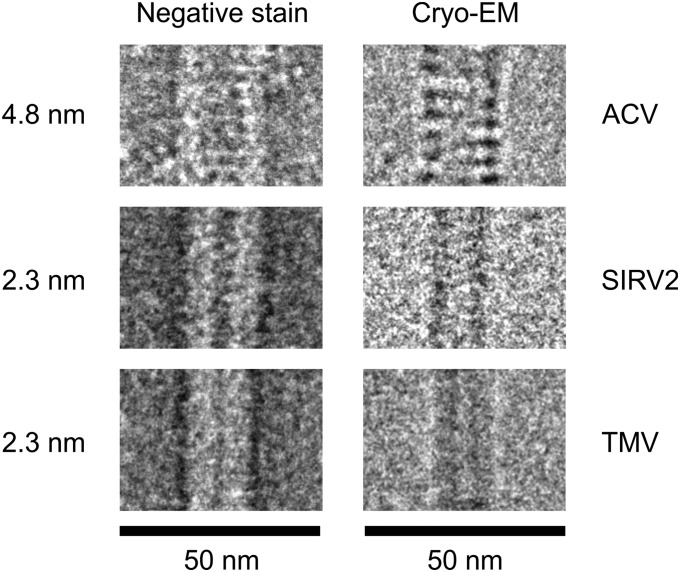
We attempted to measure the pitch of the virion helix using the Fourier transformation of both TEM and cryo-EM images. However, only images of negatively stained ACV virions, but not those analyzed by cryo-EM, could be used for such measurements due to the rigid cylindrical shape of the former. For comparison, virions of other helical viruses, the tobacco mosaic virus (TMV) and the archaeal rudivirus SIRV2 (19), were added to the analyzed sample (Fig. 2 and Fig. S3). The helix pitch of TMV and SIRV2 was estimated to be ∼2.3 nm, whereas the pitch of the ACV helix was nearly twice as wide at ∼4.8 nm. These results suggest an unusual nucleoprotein arrangement in the ACV virion.
Fig. 2.
Comparison of ACV to other helical viruses. Portions of negatively stained virions of ACV, tobacco mosaic virus (TMV), and Sulfolobus islandicus rod-shaped virus 2 (SIRV2) are in the left column, and their cryo-EM images are in the right column. Original micrographs are shown in Fig. S3. The pitch of the virion helix, determined by Fourier transformation of the negatively stained images, is indicated.
Considering the length of the virion and the measured pitch of the helix, we were able to calculate the number of helix turns as 220/4.8 = 45, which is close to the number 40 estimated by manually counting the observable helix turns in negatively stained virions (Fig. 1B). Using these parameters and assuming the diameter to be equal to 28 nm, the helix-forming filament would be ∼4,000 nm in length (http://deepfriedneon.com/tesla_f_calchelix.html).
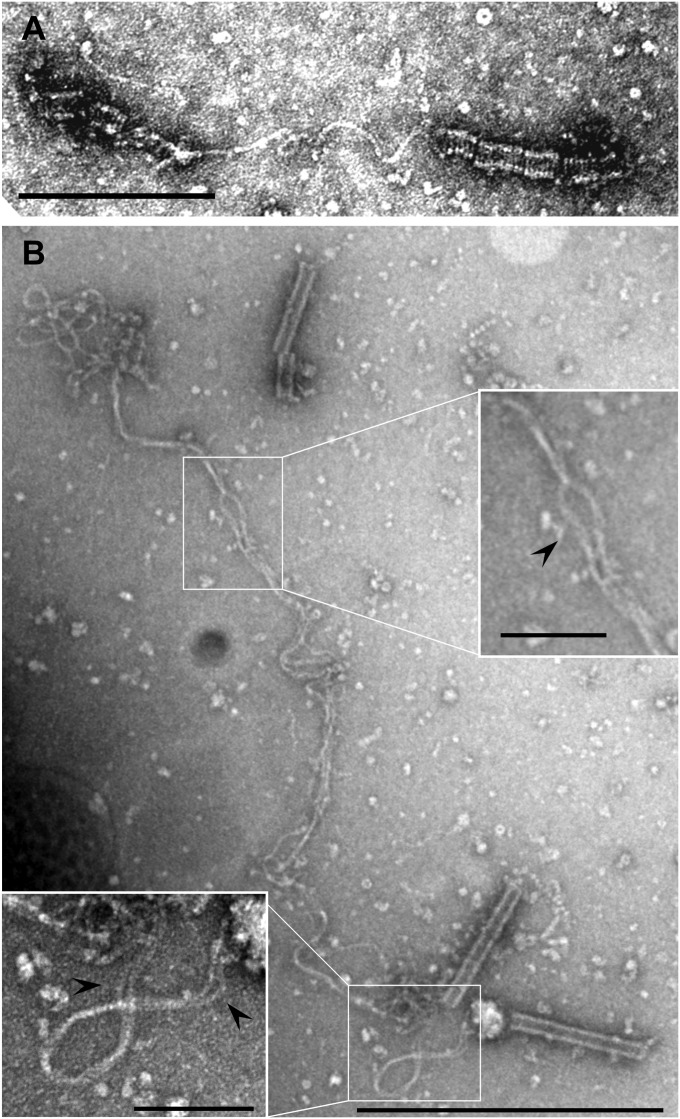
Due to the inherent fragility of the virions, their purification resulted in a fraction of partially or completely disassembled viral particles. The broken particles could be observed in top view, down the long axis of the cylindrical virion; the images were consistent with the absence of an envelope and revealed the existence of a central cavity that apparently extended throughout the full length of the virion (Fig. S4). The helix of some damaged virions was partially unwound, revealing the constituent filament and suggesting that the helix may be right-handed (Fig. 3A). The observation of completely unwound virions (Fig. 3B) allowed for a rough estimation of the length of the helix-forming nucleoprotein filament in the range of 4,000 nm, as is also suggested by the above-described calculations. Moreover, an image analysis revealed that this filament represents a rope-like helical fiber composed of two intertwined strands, which are clearly visible in the Insets in Fig. 3B.
Fig. 3.
Transmission electron micrographs of disrupted ACV virions. (A) Fragments of partially disassembled virions connected by a twisted filament. (B) A completely unwound helix-forming filament; the regions where two constituent strands of the helix-forming filament are clearly distinguishable are shown in Insets and are indicated by arrowheads. [Scale bars: (A) 200 nm; (B) 500 nm; (Insets) 100 nm.]
The virions purified by isopycnic centrifugation in caesium chloride (CsCl) gradient yielded in SDS/PAGE two major bands of approximately equal intensity, corresponding to proteins with molecular mases of ∼23 and 18.5 kDa and a few minor bands of proteins with molecular masses in the range of ∼5–13 kDa (Fig. S5). The identity of the major capsid proteins could not be determined by MALDI-TOF mass spectrometry analysis, possibly due to extensive posttranslational modification of these proteins (see below).
Virus Genome.
The nucleic acid extracted from the ACV virions was completely degraded by DNase I but not by RNase and thus represented DNA. As the first step in the determination of its nucleotide sequence, segments of the viral genome were PCR-amplified using a sequence-independent, single-primer amplification (SISPA) method (20), as described in Materials and Methods. The nucleotide sequence of the largest produced DNA fragment (1.5 kb) was used to design primers for the amplification of the remainder of the presumably circular viral genome (Materials and Methods). The formation of a large PCR product, ∼20 kb in size, confirmed the circular nature of the viral genome. Its nucleotide sequence was determined and assembled using the sequence of the 1.5-kb DNA fragment to produce the complete sequence of the viral genome. The sequences of the smaller PCR products amplified from DNA extracted from the ACV virions also matched the assembled viral genome.
Although ACV DNA was sensitive to DNase I, it was not sensitive to type II restriction endonucleases, which had recognition sequences in the viral genome and were insensitive to DNA methylation (e.g., NdeI). To determine whether the packaged ACV genome consisted of ssRNA or dsDNA, we conducted primer extension experiments. Using the Klenow enzyme and a mixture of random hexamers as primers, it was possible to copy ACV DNA without a template denaturation step (Fig. S6). The results strongly suggested that the ACV DNA was single-stranded.
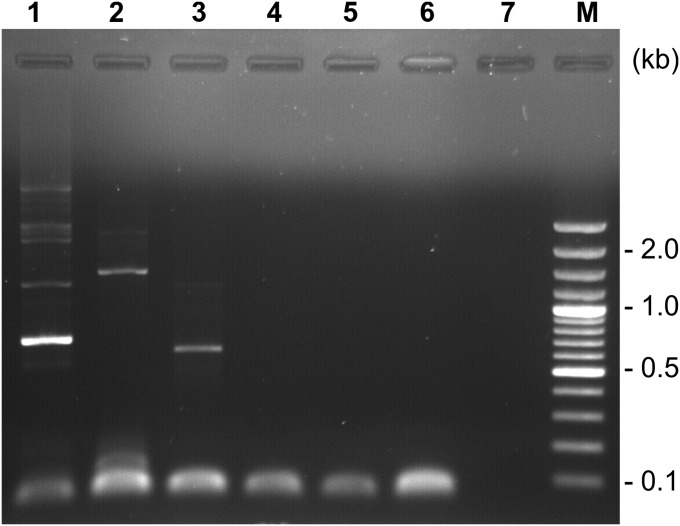
To determine the orientation of the viral ssDNA, we applied the modified SISPA method that we previously developed and verified with the ssDNA genomes of the M13 and phiX174 viruses (21). Briefly, the experiment proceeded as follows: DNA complementary to the segments of ACV DNA was produced by elongation of the semirandom primer FR26RV-N (20). After complete removal of the excessive primer FR26RV-N, this step was followed by PCR amplification of the synthesized molecules using FR20RV (20) and an ACV-specific oligonucleotide as primers (Materials and Methods). Amplification occurred only if the direction of the ACV-specific primer and the ACV genome coincided. The results of the experiment shown in Fig. 4 clearly demonstrated the formation of PCR products with three ACV-specific “forward” primers and the absence of synthesis with three ACV-specific “reverse” primers. The orientation of the fragments was further verified by their sequence. This analysis not only confirmed the single-stranded nature of the genome but also allowed us to determine that ACV is a positive-sense ssDNA virus (see below).
Fig. 4.
Analysis of single-stranded DNA orientation. Agarose gel electrophoresis of PCR products after step 2 of the Strand Orientation–SISPA method. Along with FR20RV, the following primers were used: lane 1, 1F; lane 2, 2F; lane 3, 3F; lane 4, 4R; lane 5, 5R; lane 6, 6R; lane 7, negative control; and lane M, DNA markers.
Genome Sequence.
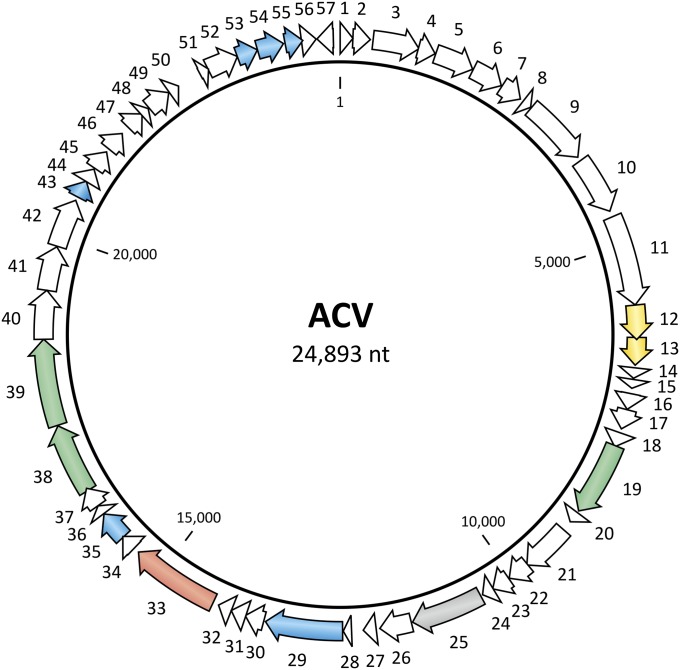
The circular ssDNA genome of ACV consists of 24,893 nt and has a G+C content of 46.7%. The latter value is significantly lower than that of A. pernix K1 (56.3%) (22) and the three A. pernix viruses previously isolated (52.7–56.5%) (18, 23). The genome contains 57 predicted ORFs larger than the 40 codons (Fig. 5) that occupy 93.5% of the genome. All but one of these ORFs are present on the DNA strand that is packaged into the ACV virions, indicating that the genome is positive sense.
Fig. 5.
Circular genome map of ACV. The ORFs are marked with arrows indicating the direction of transcription. The ORFs encoding putative proteins for which functions could be predicted are color-coded as follows: thioredoxins, yellow; carbohydrate metabolism, green; DNA binding, blue; serine protease, gray; and recombinase, red.
Homology searches using the BLASTP program (24) revealed that only four (7%) ACV gene products share significant homology (cutoff of E = 1e-05) with sequences in the nonredundant protein database (Table S1). Three of these proteins were predicted to be involved in carbohydrate metabolism (ORFs 19, 38, and 39), whereas the fourth (ORF25) was significantly similar (E = 3e-39) to trypsin-like serine proteases. To obtain further information on the functions encoded by ACV, we carried out a more sensitive, hidden Markov model-based HHpred analysis (25), which allowed us to identify a set of more remote homologs. As a result, we collectively assigned functions to 14 ACV gene products (25%; Table S1).
Six of the ACV ORFs were predicted to encode DNA-binding proteins containing ribbon-helix-helix (RHH) (ORFs 29, 35, and 51) or winged helix-turn-helix (wHTH) (ORFs 43, 52 and 55) motifs that may be responsible for controlling the expression of viral and/or cellular genes. In addition, ORF33 encodes a protein with an N-terminal DNA-binding HTH motif (Fig. S7A) followed by a domain related to the catalytic domains of diverse tyrosine recombinases, such as siphovirus λ integrase, P1 Cre recombinase, yeast Flp recombinase, bacterial XerD/C recombinases, and eukaryotic type IB topoisomerase. A search against the conserved domain database (26) revealed that all active-site residues typical for this family of integrases/recombinases were conserved in ACV ORF33 (Fig. S7B). Notably, as in the case of the Cre and Flp recombinases, ACV ORF33 possesses an active-site tryptophan instead of the histidine residue found in the active sites of the vast majority of tyrosine recombinases (27, 28). The closest homolog of ORF33 (with the same domain organization) is encoded in the genome of the ammonia-oxidizing soil thaumarchaeon Candidatus Nitrosopumilus koreensis, whereas weak hits to proteins from other organisms are confined to the catalytic domain. Consequently, the product of ORF33 is a highly divergent member of the tyrosine recombinase family, but its exact function during the viral replication cycle is unknown.
ORFs 12 and 13 encode thioredoxin-like proteins with conserved CXXC motifs (cysteines separated by two amino acids). Whereas ORF13 appears to encode a soluble protein, the product of ORF12 possesses a predicted N-terminal transmembrane domain (Table S1) and is therefore likely to be tethered to the cellular membrane.
Finally, ORF16 encodes a putative zinc ribbon protein with a characteristic CXXC...CXXC motif (Fig. S8). Searches against the Protein Data Bank (PDB) database using HHpred revealed a number of potential homologs of ORF16 (prediction probability greater than 95%), including the RNA polymerase subunit P, RpoP (PDB ID: 2WAQ), and the N-terminal domain of the transcription factor II B (PDB ID: 1PFT). Notably, the size of the ORF16 product (72 aa) falls in the typical range of archaeal RpoP proteins (43–80 aa; SI Text).
An all-against-all comparison of the ACV gene products revealed the expansion of three different genes (ORFs 5, 14, 40), all coding for hypothetical proteins (Table S1). Three ORF14-like paralogs exist in the ACV genome, whereas ORF5- and ORF40-like genes are present in two copies each. ORF5 encodes a protein with an N-terminal transmembrane domain. Peculiarly, this domain is not present in the paralogous ORF6, suggesting that gene duplication was followed by functional diversification.
Discussion
The structural and genomic features of the virus ACV are exceptional and clearly distinguish it from all other viruses described to date. The ACV virions differed in their appearance depending on the electron microscopy technique used for their observation. Negatively stained virions observed with TEM appeared as rigid circular cylinders with short appendages at both ends (Fig. 1). The analysis of partially disassembled virions (Fig. 3A) allowed us to suggest that the virion cylinder is hollow and formed from a coiling nucleoprotein filament. The helical structure of the ACV virion was even more apparent in the cryo-EM analysis (Fig. 1 C and D). Notably, virions embedded in vitreous ice appeared more flexible than the negatively stained virions (Fig. 1 A and B). Because cryo-EM allows for the observation of samples in their natural conformation, avoiding possible artifacts caused by dehydration due to negative staining, we consider the images of particles embedded in ice to be closer to the authentic structure of the ACV virion. The flexible coil of the native virion could indeed be prone to contraction and stiffening upon dehydration due to uranyl acetate staining. This process would explain the observed differences in virion appearance under the two conditions. The short appendages observed at the virion termini could result from a modification of the ends of the coil-forming filament.
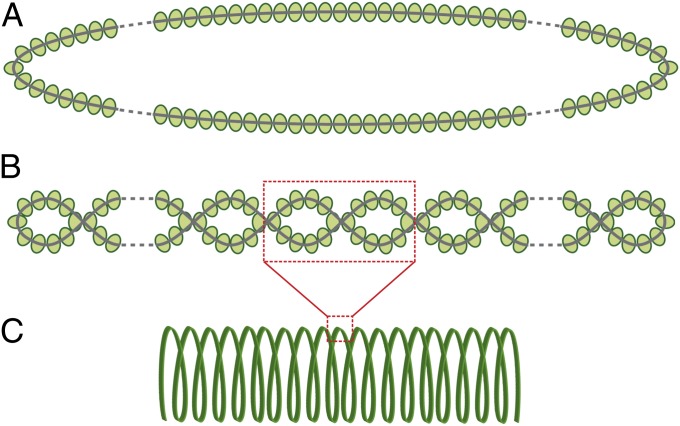
The TEM observations of disassembled virions (see, e.g., Fig. 3) highlighted that at least two levels of helical organization must exist in the virion and that the filament, which forms the virion helix, can itself have a helical structure, being composed of two intertwining strands. Each of these strands is most likely a nucleoprotein containing a strand of ssDNA. Because the viral ssDNA is circular, the two strands likely represent the two halves of the same circular nucleoprotein. On the basis of these considerations, a tentative model for the structural organization of the ACV virion can be proposed (Fig. 6).
Fig. 6.
Schematic representation of different levels of organization of the circular ACV nucleoprotein. As described in the text, the two halves of the circular nucleoprotein (A) intertwine with each other and form a nucleoprotein filament (B), which is condensed into the helical coil of the virion (C).
To our knowledge, the virion architecture suggested for ACV has not been reported for any known virus with either a DNA or an RNA genome. The only known filamentous viruses with ssDNA genomes, members of the Inoviridae family, have virions that differ significantly in structure from the ACV virion. The former have cylindrical protein shells encasing two antiparallel strands of circular ssDNA that are partly base-paired and adopt a conformation similar to classical A- and B-form dsDNA (29). By contrast, the protein shell of the ACV virion apparently encases a single strand of DNA, precluding pseudo double-helix formation. Although the inoviral nucleoprotein filament has no further level of organization, the circular ACV nucleoprotein adopts two more levels of organization by intertwining the two halves of the circular molecule and arranging the resulting helix into a superhelix of higher order to produce the cylindrical helix of the virion (Fig. 6). The differences in structural design appear to be the major reason for the size differences between the virion types. The inovirus virions are ∼7 nm in width, compared with ∼28 nm for ACV virions. Additionally, the ∼3,700 nm-long virion of the inovirus Pf4 is nearly 17 times longer than the ACV virion, even though the ACV genome is double the size of the Pf4 genome (30). It is noteworthy that, in inoviruses and all other known nonenveloped filamentous viruses (Rudiviridae, Alfaflexiviridae, Betaflexiviridae, Gammaflexiviridae, Closteroviridae, Potyviridae, Virgaviridae), the nucleoprotein helix does not adopt any additional levels of organization (1), thereby distinguishing ACV from these viruses. Organization of a nucleoprotein filament into a coil-shaped helical structure has been reported only for enveloped viruses, e.g., members of the order Mononegavirales, such as Ebola and Marburg viruses (31, 32). However, the helical nucleocapsids of these enveloped visuses contain a single strand of an RNA genome and thus are radically different from the coil-forming filament of ACV, composed of two intertwining nucleoprotein strands (Fig. 6B).
The unique architectural solution used by ACV to pack its circular ssDNA does not impose a strict constraint on genome size. Indeed, ACV has the largest genome among the known ssDNA viruses at twice the size (24.8 versus 12.4 kb) of the previous record holder, the above-mentioned inovirus Pf4 (30) (Table S2). Similar to other prokaryotic viruses, including the filamentous inoviruses (10), genome duplications and the occasional acquisition of new genes appear to have contributed to the evolution of ACV. However, the functional complexity of the ACV genome is far greater than that observed for other known ssDNA—and even certain dsDNA—viruses. Indeed, there are examples of bacterial (33), archaeal (18), and eukaryotic (34) viruses with dsDNA genomes that are considerably smaller than that of ACV. The ability to maintain a relatively large genome allows ACV to encode auxiliary functions that, although not typical for ssDNA viruses, are rather frequent in complex dsDNA viruses (SI Text). Such additional functions may modulate virus–host interactions and increase ACV’s chances in the arms race with its host. For example, the three carbohydrate metabolism enzymes encoded by ACV, as in the case of algae-infecting phycodnaviruses (35), might be involved in the glycosylation of viral and/or cellular proteins. Such modification would explain the inability to determine the identity of the major capsid proteins by MALDI-TOF analysis. The serine protease encoded by ORF25 may play a role in coordinating the assembly of ACV virions. The two thioredoxin-like proteins may be responsible for the oxidation of thiols in ACV proteins, as has been shown for vaccinia virus thioredoxin (36). Notably, the assembly of filamentous inoviruses strictly depends on the Escherichia coli thioredoxin (37), suggesting that the ACV-encoded proteins might also participate in this process.
All known ssDNA viruses replicate—or are believed to replicate—their genomes via a rolling-circle (RCR) or rolling-hairpin mechanism (1) catalyzed by homologous proteins (38). Surprisingly, we were unable to identify a potential ACV Rep candidate that shared significant sequence homology or conserved motifs with known RCR Rep proteins, suggesting that ACV might use a unique mechanism of genome replication. Interestingly, the divergent member of the tyrosine recombinase family encoded by ACV ORF33 might be directly involved in this process (SI Text).
In conclusion, the results of the genome analysis are in line with the unique morphology of the ACV virus and confirm that ACV is not closely related to any other characterized archaeal, bacterial, or eukaryal virus. Notably, not only is ACV the first hyperthermophilic virus with a ssDNA genome, but also its genome is the largest among the known ssDNA viruses (Table S2)—a rather unexpected discovery considering the harshness of the extreme geothermal environment in which ACV thrives.
Due to the unique features of the ACV genome and virion organization, we propose that ACV be considered as a representative of a distinctive viral family, which we tentatively name “Spiraviridae” (from Latin spira, “a coil”).
The structural characterization of viruses belonging to different families and infecting hosts from all three cellular domains has revealed that the number of known architectural solutions allowing the construction of functional virions is rather limited in the virosphere (39, 40). Indeed, new structural folds of the major capsid proteins are being described with decreasing frequency, and reports on entirely novel architectural principles underlying virion formation are perhaps even more infrequent. In contrast to this general trend, the incredible morphological diversity of archaeal viruses continues to surprise us, even after a few decades of their studies (6). The exceptional solution for the packing of a circular viral genome described here suggests that the morphological diversity of viruses is still unexplored and that much remains to be learned from these tiny molecular architects.
Materials and Methods
ACV Isolation.
An enrichment culture for A. pernix (41) was established from a sample isolated from the coastal Yamagawa Hot Spring in Japan, using 3ST medium at 90 °C (18). On the basis of 16S rRNA sequence analysis, only A. pernix strains were determined to be growing in the enrichment culture. The virus ACV was isolated from this culture as described in SI Materials and Methods.
Transmission and Cryoelectron Microscopy.
The samples were prepared and analyzed as described previously (18, 23).
Purification and Analysis of ACV DNA.
DNA was purified from virions using the modified method for λ-phage DNA extraction as described by Sambrook and Russel (42). Before treatment with SDS and proteinase K, the virions were treated with DNase I to remove any contamination with chromosomal or plasmid DNA. DNase I was inactivated by incubation at 90 °C for 10 min. ACV DNA was analyzed in a 0.6% agarose gel at 4 °C.
The sequence-independent SISPA method was applied to obtain partial genome sequences of ACV DNA (20). In brief, the first step was performed using the viral DNA and semirandom primer FR26RV-N (5′-GCC GGA GCT CTG CAG ATA TCN NNN NN-3′) with two PCR cycles using the following conditions: denaturation at 96 °C for 30 s, annealing at 40 °C for 30 s, and elongation at 72 °C for 58 s. The second step was performed using the PCR product from the first step and the primer FR20RV (5′-GCC GGA GCT CTG CAG ATA TC-3′) for 40 cycles under the following conditions: denaturation at 96 °C for 30 s, annealing at 45 °C for 30 s, and elongation at 72 °C for 60 s. Both PCR steps were performed using Ex-Taq polymerase (TaKaRa). The PCR products were purified after agarose gel electrophoresis and cloned into the E. coli cloning vector pCR-II TOPO (Invitrogen). The sequences of the cloned fragments were determined. On the basis of the sequence of the largest fragment, primers were designed for PCR amplification of the rest of the genome (Table S3). PCR was performed using LA-Taq polymerase (TaKaRa) under the following conditions: 40 cycles of denaturation at 96 °C for 30 s and annealing and elongation at 68 °C for 20 min. The PCR product was purified and directly sequenced by GS-FLX Titanium (Beckman Coulter Genomics). Nucleotide sequences were assembled using Mira v.3. The complete genome sequence of ACV has been deposited in the GenBank database under the accession no. HE681887.
ORFs were predicted using GLIMMER ver. 3.02 and GeneMark hmm (ver.2.6r). The validity of each predicted ORF was confirmed manually by searching for a putative ribosome-binding site ∼5 nt upstream of the start codon. The sequence was analyzed as described in SI Materials and Methods.
Primer Extension.
Primer extension was performed to identify the number of strands on the genome using the DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche). Viral DNA (14 ng) was treated in a 20-μL assay containing 2.7 U of Klenow fragment, 0.02 mM dNTP, and 0.12 mM digoxigenin (DIG)-labeled dUTP for 6 h at 37 °C. The product was subjected to electrophoresis in a 0.9% agarose gel and transferred onto a Hybond-N+ membrane (Amersham). The DIG-labeled nucleotides were detected as suggested by the manufacturer. In the same way, the following control samples were treated and analyzed: (i) 14 ng of single-stranded M13; (ii) 14 ng of double-stranded M13; (iii) 25 ng of linearized double-stranded plasmid pBR328 (provided with the kit); and (iv) 25 ng of linearized double-stranded plasmid pBR328, heat-denatured before elongation.
Identification of Genome-Strand Orientation.
Orientation of the single-stranded DNA was identified using the modified SISPA method. In brief, partial copies of the complementary strand were first produced using the semirandom primer FR26RV. After removal of excess primers using a PCR purification kit (Machery-Nagel), a second step was performed with 45 PCR cycles using the purified product from the first step as a template. The used primers were FR20RV and one of the viral sequence-specific primers designed on the basis of the viral genome sequence to cover its different parts (Table S3). The resulting PCR product was visualized by electrophoresis in an agarose gel. The PCR product was formed and observed only if the sequence-specific primer used in the second step coincided in its direction with the direction of the single-stranded template (viral genome).
Supplementary Material
Acknowledgments
We thank Hiroshi Nishimura, Satoshi Kawaichi, and Yasuko Yoneda for sample and strain preparations; Elina Roine for comments on the viral DNA analysis; and Soizick Lucas-Staat for assistance with electron microscopy. This work was supported by the Programme Blanc of the Agence Nationale de la Recherche, France (Grant ANR 09-BLN-0288.01). T.M. was supported by Bourse du Gouvernement Français (Dossier 2008661) and Allocations Pasteur-Weizmann. M.K. was supported by the European Molecular Biology Organization (ALTF 347-2010).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the European Molecular Biology Laboratory (EMBL) database, http://www.ebi.ac.uk/embl (accession no. HE681887).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203668109/-/DCSupplemental.
References
- 1.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press; 2011. [Google Scholar]
- 2.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: A unifying view. Nat Rev Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses: Major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol. 2009;11:2771–2774. doi: 10.1111/j.1462-2920.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- 5.Forterre P, Prangishvili D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann N Y Acad Sci. 2009;1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x. [DOI] [PubMed] [Google Scholar]
- 6.Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011;35:1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- 7.Ackermann HW. 5500 phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 8.Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. Global network of specific virus-host interactions in hypersaline environments. Environ Microbiol. 2012;14:426–440. doi: 10.1111/j.1462-2920.2011.02603.x. [DOI] [PubMed] [Google Scholar]
- 9.Pfister P, Wasserfallen A, Stettler R, Leisinger T. Molecular analysis of Methanobacterium phage psiM2. Mol Microbiol. 1998;30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- 10.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. Genomics of bacterial and archaeal viruses: Dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev. 2011;75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prangishvili D, Garrett RA, Koonin EV. Evolutionary genomics of archaeal viruses: Unique viral genomes in the third domain of life. Virus Res. 2006;117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Bize A, et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brumfield SK, et al. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J Virol. 2009;83:5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quax TE, et al. Simple and elegant design of a virion egress structure in Archaea. Proc Natl Acad Sci USA. 2011;108:3354–3359. doi: 10.1073/pnas.1018052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prangishvili D. Evolutionary insights from studies on viruses of hyperthermophilic archaea. Res Microbiol. 2003;154:289–294. doi: 10.1016/S0923-2508(03)00073-1. [DOI] [PubMed] [Google Scholar]
- 16.Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. An ssDNA virus infecting Archaea: A new lineage of viruses with a membrane envelope. Mol Microbiol. 2009;72:307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- 17.Prangishvili D. Hyperthermophilic virus-host systems: Detection and isolation. In: Rainey FA, Oren A, editors. Methods in Microbiology: Extremophiles. Vol 35. London: Elsevier Academic Press; 2006. pp. 331–347. [Google Scholar]
- 18.Mochizuki T, et al. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology. 2010;402:347–354. doi: 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 19.Prangishvili D, et al. A novel virus family, the Rudiviridae: Structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics. 1999;152:1387–1396. doi: 10.1093/genetics/152.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reyes GR, Kim JP. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol Cell Probes. 1991;5:473–481. doi: 10.1016/s0890-8508(05)80020-9. [DOI] [PubMed] [Google Scholar]
- 21.Mochizuki T, Prangishvili D. A simple and sensitive method for determining the strand orientation of single-stranded viral genomes. J Virol Methods. 2012 doi: 10.1016/j.jviromet.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayasi Y, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101, 145–152. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki T, Sako Y, Prangishvili D. Provirus induction in hyperthermophilic Archaea: Characterization of Aeropyrum pernix spindle-shaped virus 1 and Aeropyrum pernix ovoid virus 1. J Bacteriol. 2011;193:5412–5419. doi: 10.1128/JB.05101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 26.Marchler-Bauer A, Bryant SH. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004;32(Web Server issue):W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopaul DN, Guo F, Van Duyne GD. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunes-Düby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day LA. Inoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press; 2011. pp. 375–383. [Google Scholar]
- 30.Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bharat TA, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bharat TAM, et al. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci USA. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krupovic M, et al. Genome characterization of lipid-containing marine bacteriophage PM2 by transposon insertion mutagenesis. J Virol. 2006;80:9270–9278. doi: 10.1128/JVI.00536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fanning E, Zhao K. SV40 DNA replication: from the A gene to a nanomachine. Virology. 2009;384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Etten JL, Gurnon JR, Yanai-Balser GM, Dunigan DD, Graves MV. Chlorella viruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim Biophys Acta. 2010;1800:152–159. doi: 10.1016/j.bbagen.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Senkevich TG, White CL, Koonin EV, Moss B. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc Natl Acad Sci USA. 2002;99:6667–6672. doi: 10.1073/pnas.062163799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russel M. Moving through the membrane with filamentous phages. Trends Microbiol. 1995;3:223–228. doi: 10.1016/s0966-842x(00)88929-5. [DOI] [PubMed] [Google Scholar]
- 38.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from Eubacteria, eucaryotes and Archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krupovič M, Bamford DH. Order to the viral universe. J Virol. 2010;84:12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krupovic M, Bamford DH. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol. 2011;1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Sako Y, et al. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int J Syst Bacteriol. 1996;46:1070–1077. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.