Abstract
Objective
According to the American Diabetes Association, the implementation of the standards of care for diabetes has been suboptimal in most clinical settings. Diabetes is a disease that had a total estimated cost of $174 billion in 2007 for an estimated diabetes-affected population of 17.5 million in the United States. With the advent of electronic medical records (EMR), tools to analyze data residing in the EMR for healthcare surveillance can help reduce the burdens experienced today. This study was primarily designed to evaluate the efficacy of employing clinical natural language processing to analyze discharge summaries for evidence indicating a presence of diabetes, as well as to assess diabetes protocol compliance and high risk factors.
Methods
Three sets of algorithms were developed to analyze discharge summaries for: (1) identification of diabetes, (2) protocol compliance, and (3) identification of high risk factors. The algorithms utilize a common natural language processing framework that extracts relevant discourse evidence from the medical text. Evidence utilized in one or more of the algorithms include assertion of the disease and associated findings in medical text, as well as numerical clinical measurements and prescribed medications.
Results
The diabetes classifier was successful at classifying reports for the presence and absence of diabetes. Evaluated against 444 discharge summaries, the classifier’s performance included macro and micro F-scores of 0.9698 and 0.9865, respectively. Furthermore, the protocol compliance and high risk factor classifiers showed promising results, with most F-measures exceeding 0.9.
Conclusions
The presented approach accurately identified diabetes in medical discharge summaries and showed promise with regards to assessment of protocol compliance and high risk factors. Utilizing free-text analytic techniques on medical text can complement clinical-public health decision support by identifying cases and high risk factors.
Keywords: Diabetes mellitus, Natural language processing, Public health informatics
In 2010, it was estimated that 18.8 million people in the United States live with the diagnosis of diabetes.1 The total estimated cost of diabetes in 2007 was $174 billion, including $116 billion in excess medical expenditures and $58 billion in reduced national productivity.2 The burden of diabetes is shared by all sectors of the society – higher insurance premiums paid by employees and employers; reduced earnings due to disease and loss of productivity; and most importantly, reduced overall quality of life for people with diabetes. Disease surveillance can be significantly overcome by the difficulty in identifying the target population.3 Even though manual chart review remains the gold standard for the identification of individuals with diabetes, it can be supported by employing natural language processing (NLP) tools to reduce the burden of labor intensive manual processes.4,5 This preliminary screening can be accomplished by analyzing physician’s notes, medication lists, problem lists, and discharge summaries by NLP tools, as patient records are being increasingly stored in digital format.6,7 In addition, it is also feasible to estimate adherence to quality of care protocols such as ABCS (ie, A1c–glycosylated hemoglobin, Blood pressure, Cholesterol, Smoking) indicators for comparative effectiveness and population health measurement purposes.
In diabetes mellitus, higher amounts of glycosylated hemoglobin indicate poor control of blood glucose in the past and have been associated with cardiovascular disease, nephropathy, and retinopathy. Monitoring the hemoglobin A1c (HbA1c) in patients with type-1 diabetes improves treatment and could also provide potential benefit for the population level quality-of-care assessment.
According to the American Diabetes Association (ADA), the implementation of the standards of care for diabetes has been suboptimal in most clinical settings. A recent study maintains that only about 57.1% of adults with diagnosed diabetes achieved an A1c of <7%, only 45.5% had optimum blood pressure measurements, and about 46.5% had a total cholesterol <200 mg/dL. Most importantly, only 12.2% of people with diabetes achieved all treatment goals of ABCS.8,9 “A major contributor to suboptimal care is a delivery system that too often is fragmented, lacks clinical information capabilities, often duplicates services and is poorly designed for the delivery of chronic care.”10
While a simple solution to these problems is not readily available and beyond the scope of this study, having the patient data in digital format (whether structured or unstructured) provides an opportunity to screen medical records for mention of diabetes and diabetes-related medications. With the provision of richer data sources, it may also be possible to screen medical digital records for adherence to protocols related to quality of care such as ABCS. Electronic health records (EHRs) have the potential to (1) advance the quality of healthcare by providing timely access to medical information, (2) ensure that the patients receive guideline-protocol recommended care, and (3) offer decision support for clinical and administrative healthcare providers. Developing tools to analyze data residing in the EHR for healthcare surveillance can help reduce the burdens experienced today. This potential has not been realized, largely because the medical information is currently stored in an unstructured, free-text format and currently existing structured coding is often driven by financial imperatives.
Various NLP technologies have been applied to the clinical domain.11 Table 1 lists related works and lists the distinctive characteristics of each work including: (1) disease domain applied, (2) data source that the classification was being performed against, (3) the classification approach employed (ie, statistical vs. rule-based), (4) if it supports ABCS protocol compliance detection, and (5) if it supports high risk factors detection. Some of these have used ConText12,13 and NegEx14 to perform concept extraction of medical terminology and apply a set of medically relevant descriptive qualifiers, such as negation, to the task. These two regular expression-based concept extraction approaches have been applied as an initial processing step to various tasks including identifying the presence or absence of medical findings in support of clinical trials screening,7 obesity screening,15,16 smoking status,17 medication identification,18 and pathology report analysis.19 This work is an extension of the authors’ approach15 employed in the i2b2 Obesity Challenge and extends the methodology to a different dataset and to novel application domains described below.
Table 1.
Review of related works.
| Title and Principal Investigator/Author | Year | Description | Applied To Disease Condition/ Description | Data Source | Classification Of Approach | Detect ABCS Compliance | Detect High Risk Factor |
|---|---|---|---|---|---|---|---|
| Towards automatic diabetes case detection and ABCS protocol compliance assessment Mishra N. (this work) |
2012 | Rule-based classifier incorporating the clinical natural language processing tool, ConText, to analyze discharge summaries for detecting diabetes, ABCS protocol compliance and high risk factors; extends upon published work originally applied to obesity. | Diabetes/ screening | Medical discharge summaries | Rule-based | Yes | Yes |
| Identification of patients with diabetes from the text of physician notes in the electronic medical record Turchin A.4 |
2005 | DITTO-Diabetes Identification Through Textual element Occurrences; program written in Perl language that takes one or more text files containing patient notes as input. | Diabetes/screening | Electronic medical record | Rule-based | No | No |
| Use of an electronic medical record for the identification of research subjects with diabetes mellitus Wilke R.29 |
2007 | Electronic case-finding algorithm that can be used for the identification of research subjects with diabetes mellitus in the Personalize Medicine Research Project (PMRP) DNA biobank; combined medical record: including indices to all events and encounters with access all textual documentation such as office notes, operative reports and discharge summaries. | Diabetes/ screening | Electronic medical record | Rule-based | No | No |
| Computerized extraction of information on the quality of diabetes care from free text in electronic patient records of general practitioners Voorham J.30 |
2007 | Evaluation of a computerized method for extracting numeric clinical measurements related to diabetes care from free text in electronic patient records converted numeric clinical information to structured data research and quality of care assessments for practices lacking structured data entry; 13 numeric clinical measurements considered relevant for evaluating the quality of diabetes care. | Diabetes care | Electronic medical record | Rule-based | No | Yes |
| Automatic detection of acute bacterial pneumonia from chest X-ray reports Fiszman M.31 |
2000 | Four physicians, three lay persons, a natural language processing system, and two keyword searches (designated AAKS and KS) detected the presence or absence of three pneumonia-related concepts and inferred the presence or absence of acute bacterial pneumonia from 292 chest x-ray reports. | Pneumonia | Chest radiograph reports | Statistical | n/a | n/a |
| A simple algorithm for identifying negated findings and diseases in discharge summaries Chapman W.32 |
2002 | NegEx–describes and tests a computationally simple algorithm that could be implemented quickly and easily to determine whether an indexed term is negated | Various diseases | Medical discharge summaries | Rule-based | n/a | n/a |
| Creating a text classifier to detect radiology reports describing mediastinal findings associated with inhalational anthrax and other disorders Chapman W.33 |
2003 | Identify Patient Sets (IPS) system to create a key word classifier for detecting reports describing mediastinal findings consistent with anthrax and compared their performances on a test set of 79,032 chest radiograph reports | Covert anthrax release | Chest radiograph reports | Statistical | n/a | n/a |
| The role of domain knowledge in automating medical text report classification Wilcox A.34 |
2003 | Inductively created classifiers for medical text reports using varying degrees and types of expert knowledge and different inductive learning algorithms. Six clinical conditions: congestive heart failure, chronic obstructive pulmonary disease, acute bacterial pneumonia, neoplasm, pleural effusion, and pneumothorax | Multiple clinical conditions | Chest radiograph reports and discharge summaries | Both | n/a | n/a |
| Classifying free-text triage chief complaints into syndromic categories with natural language processing Chapman W.35 |
2005 | Natural language processing application for classifying chief complaints into syndromic categories for syndromic surveillance | Syndromic surveillance | Chief complaints | Statistical | n/a | n/a |
| Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system Zeng Q.36 |
2006 | Health Information Text Extraction (HITEx) tool used to extract key findings for a research study on airways disease The principal diagnosis, co-morbidity and smoking status extracted by HITEx from a set of 150 discharge summaries were compared to an expert-generated gold standard. | Asthma | Discharge summaries | Rule-based | n/a | n/a |
| Using A natural language processing system to extract and code family history data from admission reports Friedlin J.37 |
2006 | Extracts and code family history data from hospital admission notes | Family history of 12 diseases | Hospital admission notes | Rule-based | n/a | n/a |
| Identifying smokers with a medical extraction system Clark C.38 |
2007 | Rule-based extraction engine with machine learning algorithms to identify and categorize references to patient smoking in clinical reports | Smoking/screening | Electronic medical record | Both | n/a | n/a |
| Mayo Clinic NLP system for patient smoking status identification Savova G.17 |
2008 | Identifying the smoking status of patients; system of reusable text analysis components built on the Unstructured Information Management Architecture and Weka; system has three layers of sentence classification | Smoking status | Electronic medical record | Rule-based | n/a | n/a |
| Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study Li L.7 |
2008 | Strengths and weaknesses of using structured or textual clinical data for clinical trials eligibility pre-screening. Medical Language Extraction and Encoding System (MedLEE)6 - processes narrative discharge summaries, MedLEE generates structured, computer-recognizable, XML-coded concepts using semantic classes for problem, finding, body location, certainty, degree, region, etc. | Screening for clinical trials | Clinical Data Warehouse (CDW) | Rule-based | n/a | n/a |
| A Rule-based approach for identifying obesity and its comorbidities in medical discharge summaries Mishra N.15 |
2009 | Free-text classifier based on keyword identification, negation detection, and simple scoring rules. | Obesity and comorbidity screening | Medical discharge summaries | Rule-based | n/a | n/a |
| A system for classifying disease comorbidity status from medical discharge summaries using automated hotspot and negated concept detection Ambert K.16 |
2009 | A text mining system for classifying patient comorbidity status, based on the information contained in clinical reports. No aspect of their system requires a priori knowledge of a disease, expert medical knowledge, or manual examination of the patient records beyond the training labels themselves. | Obesity and comorbidity screening | Medical discharge summaries | Statistical | n/a | n/a |
| ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports Harkema H.13 |
2009 | Algorithm for determining whether clinical conditions mentioned in clinical reports are negated, hypothetical, historical, or experienced by someone other than the patient. | Various diseases | Electronic medical record-6 report types- | Rule-based | n/a | n/a |
| Description of a rule- based system for the i2b2 Challenge in natural language processing for clinical data Childs L.39 |
2009 | Lockheed Martin’s rule-based natural language processing capability, ClinREAD; “read” a patient’s clinical discharge summary and replicate the judgments of physicians in evaluating presence or absence of obesity and 15 comorbidities. | Obesity and 15 comorbidities | Clinical discharge summary | Rule-based | n/a | n/a |
| Semi-automated construction of decision rules to predict morbidities from clinical texts Farkas R.40 |
2009 | Development of automatic systems that analyzed clinical discharge summary texts and addressed the following question: “Who’s obese and what co-morbidities do they (definitely/most likely) have?” | Obesity and 15 comorbidities | Clinical discharge summary | Both | n/a | n/a |
| A Text Mining Approach to the Prediction of Disease Status from Clinical Discharge Summaries Yang H.41 |
2009 | Assembled a set of resources to lexically and semantically profile the diseases and their associated symptoms, treatments, etc. These features were explored in a hybrid text mining approach, which combined dictionary look-up, rule-based, and machine-learning methods. | Obesity and 15 comorbidities | Clinical discharge summary | Both | n/a | n/a |
| Semantic Classification of Diseases in Discharge Summaries Using a Context-aware Rule-based Classifier Solt I.28 |
2009 | Context-aware rule-based semantic classification technique for use on clinical discharge summaries | Obesity and 15 comorbidities | Clinical discharge summary | Rule-based | n/a | n/a |
| Natural Language Processing Framework to Assess Clinical Conditions Ware H.42 |
2009 | Natural language processing framework that could be used to extract clinical findings and diagnoses from dictated physician documentation; authors used a combination of concept detection, context validation, and the application of a variety of rules to conclude patient diagnoses. | Obesity and 15 comorbidities | Clinical discharge summary | Both | n/a | n/a |
| Symbolic rule-based classification of lung cancer stages from free-text pathology reports Nguyen A.19 |
2010 | Classify automatically lung tumor-node-metastases (TNM) cancer stages from free-text pathology reports using symbolic rule-based classification. | Lung cancer/ classification of stages | Free-text pathology reports | Rule-based | n/a | n/a |
| Automated Identification of Postoperative Complications Within an Electronic Medical Record Using Natural Language Processing Murff H.43 |
2012 | Classifier focused on the 6 postoperative complications (acute renal failure requiring dialysis, sepsis, deep vein thrombosis, pulmonary embolism, myocardial infarction, and pneumonia; used Multi-threaded Clinical Vocabulary Server natural language processor system. | Postoperative surgical complications | Electronic medical record-surgical inpatient admissions | Rule-based | n/a | n/a |
n/a, data not available
Reviewing the related i2b2 Obesity Challenge approaches, of the top five teams in the textual judgment task in the 2008 i2b2 Obesity Challenge, all used rule-based approaches. At a high level, all of these approaches examined discharge summaries for the occurrence of clinical terms associated with each morbidity and tried to establish the context in which each term was mentioned. The context could be temporal (eg, “history of _”), an indicator of the experiencer (eg, the patient or a family member), or an indicator that the person did or did not have the relevant morbidity, or if it was questionable that the patient had the morbidity. Once the term mentions and their contexts had been identified, rules were applied to integrate the results and establish a judgment for each document. Some of the major differences in the approaches would be due to text pre-processing, content of the dictionaries, the variation in the algorithms to identify positive, negative, and uncertain term mentions, the approach to ranking or ignoring certain sections of the discharge summaries, and the rules applied to integrate the results and establish a judgment for each document
The focus of the current work is to build on our obesity challenge work described above and extend these methodologies to the new domains of compliance assessment and risk factor identification, in addition to the classification task required by the i2b2 task. This work seeks to facilitate prescreening through analysis of free-text discharge summaries by applying ConText to check for linguistic evidence indicating a presence of diabetes, ABCS protocol assessment, and high risk indicators. This work also explores the potential benefits of medication discourse to support the classification process.
The primary contributions of this work includes the novel application of clinical NLP to support ABCS protocol assessment and high risk detection, the development of concept dictionaries for ConText and heuristics diabetes classification rules to support the above classifiers in a limited data and resource environment.
Methods
We used a rule-based clinical NLP approach to classify discharge summaries to (1) identify reports indicating diabetes, (2) identify reports demonstrating ABCS protocol compliance, and (3) identify reports with high risk factors such as high blood pressure or cholesterol values (low density lipoprotein [LDL]>100 mg/dL).
Classification Algorithms
Diabetes Classifier
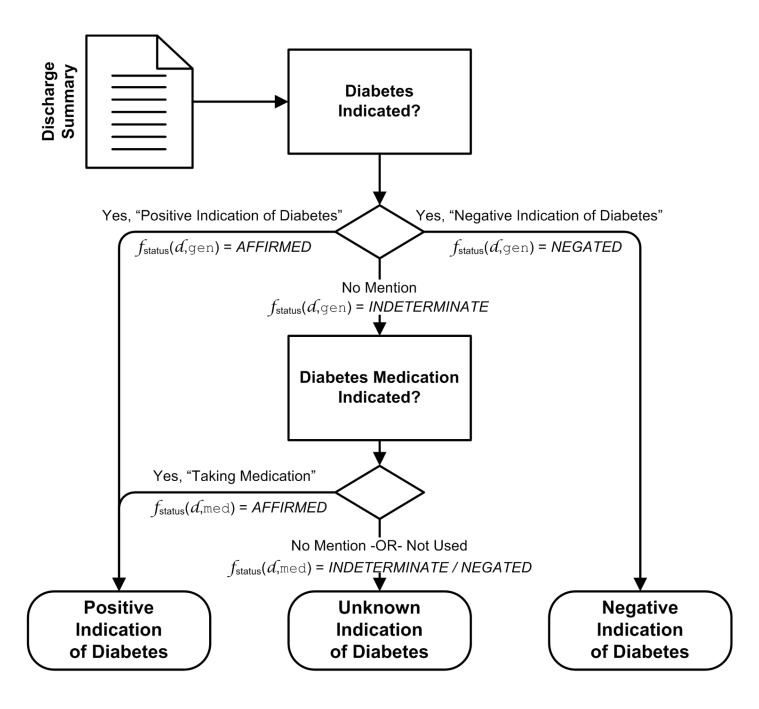
The diabetes classifier, given a discharge summary, utilizes discourse evidence, both direct utterances of diabetes and diabetes medication utterances to identify reports associated with diabetes. In this study, discharge summaries are classified into one of three possible “judgment categories” (also referred to as “indication classes”) with respect to diabetes:
Positive: positive indications of diabetes (eg, diabetes medication listed in discharge report; explicit statement of potential/existing diabetes condition)
Negative: negative indications of diabetes (eg, explicit statements supporting the absence of diabetes)
Unknown: indeterminate indications of diabetes (eg, no mention of diabetes or related medications supporting or refuting the presence of diabetes).
Figure 1 illustrates the rules utilized to perform the classification. As shown in the diagram, explicit discourse by the physician indicating the presence or absence of diabetes takes precedence. Diabetes medication discourse can also be employed to affirm diabetes, but discourse mentioning non-use of such medications cannot exclude diabetes (eg, non-insulin-dependent diabetes mellitus). The fstatus(d, type) notation in figure 1 is discussed later in this paper. Discharge summaries that are deemed “positive” can be further analyzed to assess adherence to the ABCS protocol, as well as identify potential high risk factors.
Figure 1.
Diabetes indication classifier rule.
ABCS Protocol Compliance Classifier
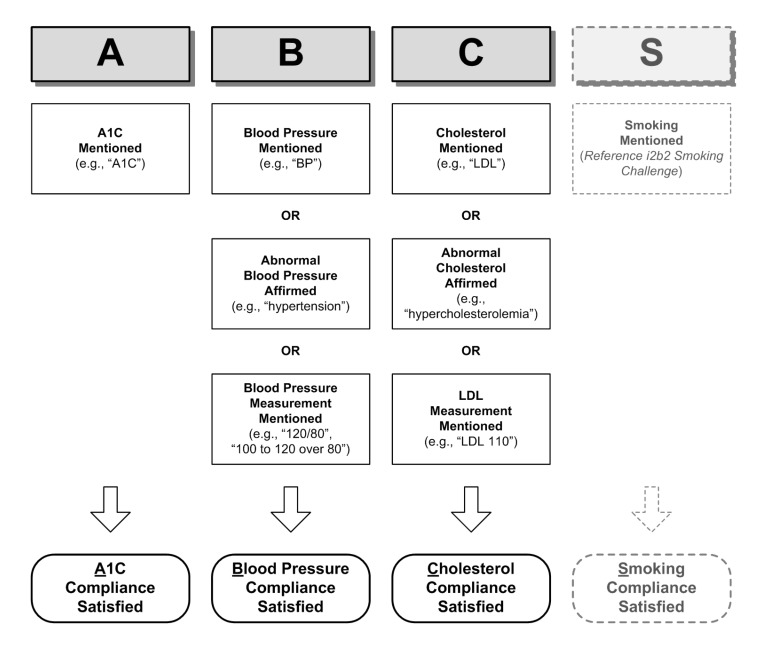
The ABCS protocol compliance classifier analyzes discharge summaries with a positive mention of diabetes to further assess compliance to ABCS protocol as follows:
Satisfied: assessment for the risk factor has been performed (eg, blood pressure was checked).
Unsatisfied: no mention of test, examination or risk factors (eg, physician makes no mention of blood pressure).
Figure 2 illustrates the rules utilized for three of the four ABCS protocol compliance assessment categories. The readers are referred to the i2b2 Smoking Competition20 for smoking assessment. As demonstrated in the assessments, discourse evidence such as direct mentioning of the examination, abnormal indications, as well as measurements encountered in the discourse can be used to determine compliance.
Figure 2.
ABCS protocol compliance assessment strategy for each indicator.
High Risk Factors Classifier
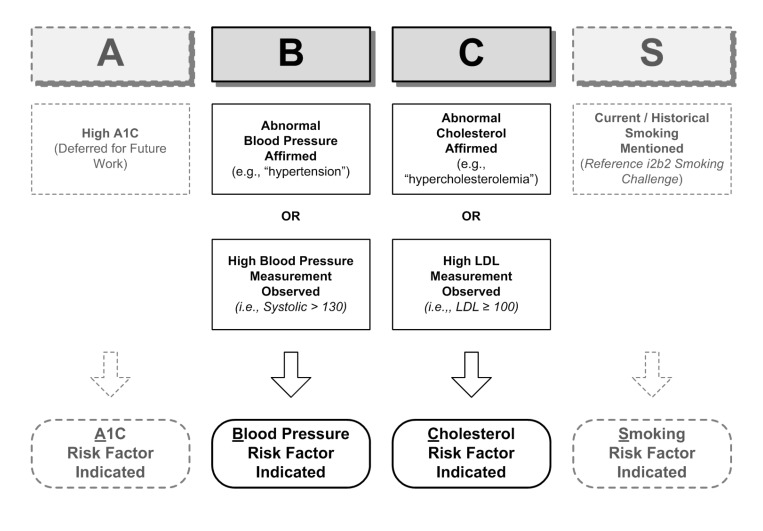
In contrast to the compliance classifier described above, the high risk factor classifier looks at values and assertions to determine the presence or absence of a high risk factor such as LDL >110 mg/dL. High risk factor assessment categories, based on ABCS indicators, are classified into one of two classes:
Indicated: high risk factor is present (eg, hyper-cholesterolemia).
Not Indicated: high risk factor is not present (eg, no mention of abnormal cholesterol or LDL measurement exceeding normal range).
Figure 3 illustrates the rules utilized for two of the four risk factor assessment categories. We used evidence from the clinical measurements mentioned in the text as well as clinical mention in the form of an assertion (eg, “Patient X has a history of hypertension”). In addition to the smoking risk factor, which is again deferred to the i2b2 Smoking Competition results, the A1c risk factor is deferred for future investigation, due to paucity of data used in this study.
Figure 3.
High-risk factors assessment strategy for each indicator.
Classifier Framework
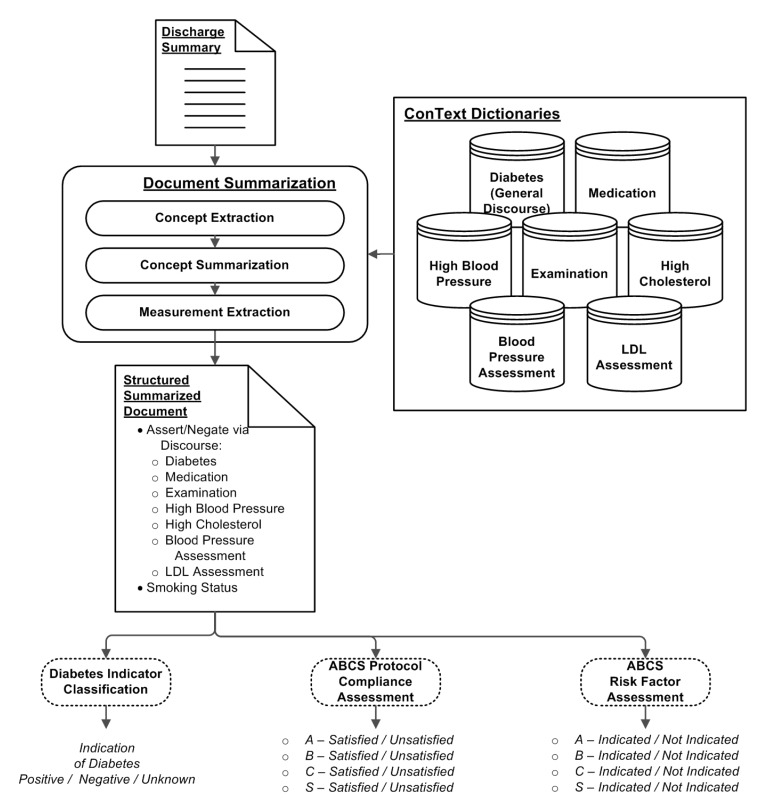
To perform each of the classification tasks described above, a clinical NLP framework using ConText was employed for each of the classifiers. As illustrated in figure 4, the analysis performed on a discharge document is broken down into multiple stages of: (1) Concept Extraction, (2) Concept Summarization, (3) Measurement Extraction (used only for the ABCS classifiers), and (4) Classification. In the Concept Extraction stage, an unstructured free-text document is transformed into a set of concept instances (eg, “diabetes”) that are encountered in the documents with various attributes associated with the concepts. The concept instances are summarized based on the collective extraction results to characterize the likely presence or absence of the concept (eg, has indications of diabetes). The resulting output from Concept Extraction is a structured representation of the discourse within the document. Additional measurement extraction is performed to support the ABCS protocol compliance and high risk factors classifiers through the extraction of relevant values, such as LDL and blood pressure measurements. Based on the structured document, a rule-based classification is performed, and the appropriate judgment class from above is assigned. The following subsections describe the details of each of these stages.
Figure 4.
Overview of discharge summary analysis for diabetes.
Concept Extraction
The initial step of classifying a discharge summary entails extracting “concepts of interest” at the utterance level from within the summary document. Employed in this initial step is ConText, a clinical NLP tool developed by Chapman et al12,13 that utilizes a regular expression approach to extract concepts of interest. ConText extracts concepts from a document such that each concept is associated with its experiencer (eg, patient, other), affirm/negate state (ie, has or does not have indications of a condition/concept), historical/currency context, and the sentence from which the concept was found. Examples of these concepts include “diabetes,” “glucose test,” “hypercholesterolemia,” and “hypertension.” These concepts are included in one or more dictionaries that are employed by ConText.
For this study, we defined seven different dictionaries employed by ConText, as shown in table 2. The first five dictionaries were used to identify affirmed or negated concepts within a document for their respective concept classes. Only concepts associated with the patient as the experiencer were considered. Through the compilation of the affirmative and negative concept mentions, five sets of affirmative/negative counts for each document were derived. The remaining two dictionaries were employed to identify candidate sentences that showed indications of blood pressure or LDL measurements associated with the patient, consistent with ABCS protocol compliance and high risk factor assessments.
Table 2.
Dictionaries utilized with ConText for concept extraction.
| Dictionary Name | Abbreviation | Associated Classification Task | Example Concept Term(s) | Number of Terms |
|---|---|---|---|---|
| Diabetes – General | gen | Indication | diabetes, dm | 21 |
| Diabetes – Medication | med | Indication | insulin, lispro, glucophage | 75 |
| Diabetes – Examination | exam | ABCS (Protocol Only) | A1c | 7 |
| High Blood Pressure | hbp | ABCS (Protocol + Risk) | hypertensive, hypertension | 3 |
| High Cholesterol | hc | ABCS (Protocol + Risk) | hypercholesterolemia | 3 |
| Blood Pressure Value Assessment | bp | ABCS (Protocol + Risk) | blood pressure, bp | 4 |
| LDL Assessment | ldl | ABCS (Protocol + Risk) | ldl | 5 |
LDL, low density liproprotein
The customized dictionaries were developed by domain experts consisting of a medical informatician and two medical doctors, including the principal author. The creation of the terminology lists included a review of the discourse in the “development” data set portion of the i2b2 data set used in this study, and supplemented by the domain experts.
Given a sentence in a discharge summary and a concept of interest from a dictionary, ConText checks for the existence of the concept within the sentence. If the concept is found, ConText determines whether or not the utterance of the concept refers to the existence (ie, an affirmative mention) or absence (ie, a negated mention) of that concept. Also, ConText determines the experiencer to whom the concept refers, such as the patient or other (eg, family member). In this study, we were only interested in concepts that were attributed directly to the patient. Thus, family history, although embedded within a discharge summary, was not used as a contributing factor towards assessing indications of diabetes for the patient. Using ConText, findings associated with the patient and others can be differentiated, reducing the chance of false inclusion or exclusion of a patient status based on non-patient findings.
Upon processing a discharge summary (d) with the concepts of interest from each of the dictionaries listed in table 2, a set of these structured representations of concept instances (c ∈d) results. These concept instances (eg, utterance of “diabetes mellitus” [dm]) can then be employed for determining the existence or absence of the concept class (eg, “Diabetes–General”) from the patient’s discharge summary.
To better illustrate the utilization of ConText, consider the following example and the concept “diabetes”: a patient’s hemoglobin A1c levels actually came back within normal limits; however, the wide excursions of his blood sugar readings were consistent with possible dm. The resulting characterization from ConText would be:
c.class = gen [Diabetes–General]
c.term = dm
c.status = Affirmed
c.experiencer = Patient
c.history = Recent
where c is the concept utterance instance associated with the term “dm” encountered in the sentence.
Concept Summarization
After employing ConText to identify and characterize all utterances pertaining to the concepts of interest, a summarization step is employed to determine the status of the concept (ie, affirmed, negated, or indeterminate) within the discharge summary. Within a discharge report (d), all utterances pertaining to the same concept class (c.class) and associated with the patient are subsequently summarized based on the number of affirmed and negated utterances, as determined during ConText concept extraction. A function, fstatus(d, type), is defined such that:
where aff(d,type)=|{ c ∈d|c.class=type,c.status=AFFIRMED}| is the number of affirmed instances of the concept class type and neg(d,type)=|{c ∈d|c.class=type,c.status=NEGATED}| is the number of negated instances of the concept class type. The summarization is applied to all concept classes in table 2.
For each of the concept classes in this study, all concept term utterances associated with the class are treated as corresponding to a singleton entity instance. For example, diabetes (as characterized in the “Diabetes–General” dictionary) is treated as a singleton entity that exists or does not exist (or is otherwise indeterminate) for a given patient. Although special cases, such as gestational diabetes, can, in fact, be treated as separate instances over time, for this study, such a distinction is not made. Similarly, for the scope of this study, an exam was either performed or not performed; whether an A1c exam was performed multiple times or performed just a single time is not differentiated in this study. Concept classes (eg, tumors) that can pertain to multiple-instance entities (eg, multiple physical findings of tumor mentioned within a patient image report), where each entity is uniquely identified, are not addressed. Such tracking of multiple entity instances would require more extensive NLP analysis utilizing techniques, such as coreference resolution,21–24 which is outside the scope of this work.
Measurement Extraction
For blood pressure assessment and cholesterol assessment, an aggregate approach is employed that incorporates both the concept extraction and summarization mechanisms described above, as well as employing blood pressure value extraction and LDL measurement extraction techniques, modeled after Turchin’s25 regular expression blood pressure extraction patterns. Both extraction techniques utilized regular expression patterns to identify the respective measurement values, which were then used as a part of the ABCS classification process.
Classification of Diabetes Indication
Once summarization for each of the concept classes has been performed, the discharge summary is classified into its judgment category by utilizing a heuristic set of rules focusing on the General Diabetes Discourse and Diabetes Medication concept dictionaries. The heuristics are based on the premise that a diabetes “positive” discharge summary would include discourse that explicitly states that the patient has diabetes, or may implicitly indicate having diabetes through medications associated with the patient that are known or highly likely to be utilized for diabetes management. The rule-set employed in this study for classifying reports of interest for diabetes is shown in figure 1, where: fstatus(d, gen) and fstatus(d, med) are the summarized concepts for general diabetes terminology and diabetes medications, respectively.
Classification of ABCS Protocol Compliance and High Risk Factors Assessment
A similar strategy used in determining diabetes indication can also be applied to assess ABCS protocol adherence for patients who have been classified as “positive” for diabetes. Utilizing the same concept extraction and summarization mechanisms employed in diabetes classification, information can be extracted from the report to determine ABCS adherence.
In the ABCS protocol compliance classifiers, simple concept extraction is employed to determine if A1c, blood pressure, or cholesterol was mentioned, with the only restriction that the concepts are associated with the patient as the experiencer. For the abnormal findings, both concept extraction and summarization were employed to determine the presence of the respective abnormalities: fstatus(d, hbp)=AFFIRMED and fstatus(d, hc)=AFFIRMED. Regular expressions were used to extract the blood pressure and LDL measurements from the discharge summaries, if present. For the high risk factor assessment classifier, the values of the measurements were examined to determine if they were outside the norm.
Because of the paucity of known diabetes discharge summary reports from the existing data set (ie, only 56 positive diabetes cases), only preliminary results of the ABCS protocol assessment analysis are presented in this work. In this study, only ABC (ie, HbA1c, Blood Pressure, and Cholesterol) are evaluated. Strategies for smoking assessment are referred to the i2b2 Smoking Competition.20
Data Set
To evaluate the technique for classifying discharge summaries with regards to diabetes, a de-identified set of free-text patient discharge summary reports from the first i2b2 Shared Task (i2b2 Smoking Challenge)20 was utilized. The i2b2 data set consists of 889 de-identified discharge summaries.26 The discharge summaries were tagged by the committee of domain experts, assessing whether or not a report indicated diabetes. The review process consisted of two reviewers independently classifying the reports for diabetes with the third reviewer being used when a disagreement was encountered. The manual tagging of the reports was used as the gold standard for the classification evaluation. For this study, the reports were divided into two sets: 445 reports were used as the development set, and 444 reports were used as the test set to evaluate the technique. In the test set, there were 56 positive reports and 5 negative reports, with the remaining 382 reports not having any indications supporting or refuting the existence of diabetes, as determined by the committee.
Classifier Evaluations
Three implementations of the diabetes classifier described above were employed in the evaluation of the efficacy of the presented methodology. Each variant employs the same concept extraction and concept summarization approach described above. However, each exercises a different variation of the classification algorithm illustrated in figure 1:
Discourse Only: Utilizes only general diabetes discourse indicators in classification process; exercises only the “Diabetes Indicated?” decision point. No mention of diabetes results in an “Unknown Indication of Diabetes.”
Medication Only: Utilizes only diabetes medication indicators in classification process; exercises only the “Diabetes Medication Indicated?” decision point.
Combined: Incorporates both diabetes discourse and medication indicators; exercises complete algorithm.
Of the three implementations, the “Medication Only” classifier can only classify reports as “positive” or “unknown” for diabetes. The inability to classify to the “negative” judgment category is due to the fact that an absence or explicit non-use of a diabetes medication does not preclude the existence of the disease (eg, non-insulin-treated type 2 diabetes). The “Discourse Only” and “Combined” solutions can classify to all three judgment categories. The ABCS protocol compliance and high risk factor classifiers were also evaluated.
Evaluation Metrics
To evaluate each of the classifier’s ability to correctly classify a document, the F-measure metric was used, which combines precision (π) and recall (ρ) into a single value. Both micro-averaged and macro-averaged F-measures were computed in this study.27 The micro F-measure (Fμ) is a global computation of F-measure across all category decisions, and is computed using the equations below:
| (2) |
| (3) |
| (4) |
where π and ρ represent the aggregate precision and recall values, respectively.
The macro F-measure (FM) is an average of each category’s local F-measure, and is computed using the equations below:
| (5) |
| (6) |
| (7) |
| (8) |
where πi, ρi and Fi represent the precision, recall and F-measure values, respectively, for a given category, i (eg, “positive”, “negative”, or “unknown” for the diabetes classifier). Both computations are a function of the number of true positive (TPi), false positive (FPi), and false negative (FNi) for each category in set M, the possible classification outcomes.
Results
The discharge summary classifier was developed based on a Java implementation of ConText.
Diabetes Classification Results
Based on the described methodology, two evaluations were performed, coinciding with the objectives targeted by this study: (1) determining indications of diabetes, and (2) examining the discriminating potential of the “Discourse Only,” “Medication Only” and “Combined” approaches. The classifier was executed against the 444 discharge summary reports for indications of diabetes utilizing the presented methodology.
The initial evaluation was performed on the three variations of the diabetes indication classifier (ie, “Discourse Only,” “Medication Only” and “Combined”). The resulting precision, recall, and F-measure results, as well as the aggregate F-measure results, for the three possible classification outcomes are shown in table 3.
Table 3.
Precision [π], Recall [ρ], F-measure [F], aggregate macro F-measure [FM] and aggregate micro F-measure [Fμ] for diabetes indication classifiers based on three classes (Positive [+], Negative [−], Unknown [? ]).
| Indication Class | Metrics | Discourse Only | Medication Only | Combined |
|---|---|---|---|---|
| Positive [+] | π+ (TP+ + FP+) | 0.9455 (52+3) | 0.9783 (45+1) | 0.9310 (54+4) |
| ρ+ (TP+ + FN+) | 0.9286 (52+4) | 0.8036 (45+11) | 0.9643 (54+2) | |
| F+ | 0.9369 | 0.8824 | 0.9474 | |
|
| ||||
| Negative [−] | π− (TP− + FP−) | 0.8333 (5+1) | N/A (0+0) | 0.8333 (5+1) |
| ρ− (TP− + FN−) | 1.0000 (5+0) | 0.000 (0+5) | 1.000 (5+0) | |
| F− | 0.9091 | 0.0000 | 0.9091 | |
|
| ||||
| Unknown [?] | π? (TP? + FP?) | 0.9922 (380+3) | 0.9598 (382+16) | 0.9974 (379+1) |
| ρ? (TP? + FN?) | 0.9922 (380+3) | 0.9974 (382+1) | 0.9896 (379+4) | |
| F? | 0.9922 | 0.9782 | 0.9934 | |
|
| ||||
| FM | 0.9461 | 0.6202 | 0.9500 | |
| Fμ | 0.9842 | 0.9617 | 0.9865 | |
A second analysis was performed where “negative” and “unknown” judgment categories are combined into a single category, to compare “Medication Only” to “Discourse Only” and “Combined,” because “Medication Only” approach cannot definitively identify “negative” cases. Combining these judgment categories focuses the metric on high precision for positive cases, thus treating the remaining cases as “non-positive.” This statistical approach may be more appropriate for studies where falsely positive cases may more significantly bias a study than falsely negative cases. The results of the second analysis are shown in table 4.
Table 4.
Precision [π], Recall [ρ], F-measure [F], aggregate macro F-measure [FM] and aggregate micro F-measure [Fμ] for diabetes indication classifiers based on two classes (Positive [+] and Combined Negative/Unknown[!]).
| Indication Class | Metrics | Discourse Only | Medication Only | Combined |
|---|---|---|---|---|
| Positive [+] | π+ (TP+ + FP+) | 0.9455 (52+3) | 0.9783 (45+1) | 0.9310 (54+4) |
| ρ+ (TP+ + FN+) | 0.9286 (52+4) | 0.8036 (45+11) | 0.9643 (54+2) | |
| F+ | 0.9369 | 0.8824 | 0.9474 | |
|
| ||||
| Combined Negative/Unknown [!] | π! (TP! + FP!) | 0.9897 (385+4) | 0.9724 (387+11) | 0.9948 (384+2) |
| ρ! (TP! + FN!) | 0.9923 (385+3) | 0.9974 (387+1) | 0.9897 (384+4) | |
| F! | 0.9910 | 0.9847 | 0.9922 | |
|
| ||||
| FM | 0.9640 | 0.9335 | 0.9698 | |
| Fμ | 0.9842 | 0.9730 | 0.9865 | |
ABCS Protocol Compliance and High Risk Factor Assessment Results
To obtain a preliminary assessment of detecting ABCS protocol compliance as well as risk assessment, two sets of analyses were performed using the above methodology. The ABCS protocol compliance classifier was executed against the 56 known diabetes discharge summary reports (as determined by our gold standard) for ABC compliance utilizing the methodology described above. The resulting precision, recall, and F-measure results, as well as the aggregate F-measure results, for the three possible classification outcomes are shown in table 5.
Table 5.
Precision [π], Recall [ρ], F-measure [F], aggregate macro F-measure [FM] and aggregate micro F-measure [Fμ] for ABCS protocol compliance assessment classifiers based on (Satisfied [+] and Not Satisfied [−]).
| Indication Class | Metrics | A1c Mentioned | Blood Pressure Mentioned | Cholesterol Mentioned |
|---|---|---|---|---|
| Positive [+] | π+ (TP+ + FP+) | 1.0000 (5+0) | 0.9535 (41+2) | 1.0000 (9+0) |
| ρ+ (TP+ + FN+) | 1.0000 (5+0) | 0.9318 (41+3) | 1.0000 (9+0) | |
| F+ | 1.0000 | 0.9425 | 1.0000 | |
|
| ||||
| Negative [−] | π− (TP− + FP−) | 1.0000 (51+0) | 0.7692 (10+3) | 1.0000 (47+0) |
| ρ− (TP− + FN−) | 1.0000 (51+0) | 0.8333 (10+2) | 1.0000 (47+0) | |
| F− | 1.0000 | 0.8000 | 1.0000 | |
|
| ||||
| FM | 1.0000 | 0.8713 | 1.0000 | |
| Fμ | 1.0000 | 0.9107 | 1.0000 | |
A further evaluation was performed to determine high risk factors, again using the presented methodology. Due to paucity of data, specifically the limited number of diabetes-related discharge summaries with abnormal A1c measurements in our data set, only high blood pressure and high cholesterol were evaluated. The associated results are shown in table 6.
Table 6.
Precision [π], Recall [ρ], F-measure [F], aggregate macro F-measure [FM] and aggregate micro F-measure [Fμ] for high-risk factor classifiers (Indicated [+] and Not Indicated [−]).
| Indication Class | Metrics | Blood Pressure Risk | Cholesterol Risk |
|---|---|---|---|
| Positive [+] | π+ (TP+ + FP+) | 0.9429 (33+2) | 0.7500 (3+1) |
| ρ+ (TP+ + FN+) | 0.9167 (33+3) | 0.6000 (3+2) | |
| F+ | 0.9296 | 0.6667 | |
|
| |||
| Negative [−] | π− (TP− + FP−) | 0.8571 (18+3) | 0.9615 (50+2) |
| ρ− (TP− + FN−) | 0.9000 (18+2) | 0.9804 (50+1) | |
| F− | 0.8780 | 0.9709 | |
|
| |||
| FM | 0.9038 | 0.8188 | |
| Fμ | 0.9107 | 0.9464 | |
Discussion
Utilizing ConText for identifying diabetes “positive” discharge summaries shows promising results with macro and micro F-scores of 0.9500 and 0.9865 when employing a three-classclassifier (ie, “positive,” “negative” and “unknown” indication classes), and macro and micro F-scores of 0.9698 and 0.9865 when combining “negative” and “unknown”, into a single category.
Both “Discourse Only” and “Combined” classifier results showed comparable F-score results with the “Combined” classifier having a slight improvement in F-score relative to “Discourse Only,” as shown in table 3. The improvement shown in the “Combined” classifier stemmed from the incorporation of medication evidence, resulting in two additional correctly classified “positive” discharge summary reports, without incurring appreciable addition of misclassified reports.
In our error analysis of the “Medication Only” classifier, only one false positive was observed in the “positive” indication class. This false positive was caused by an observed diabetes medication (ie, insulin), which was being employed for treatment of hyperkalemia. Although rare, the false positive error points out the potential for a diabetes medication observation to be misleading without consideration of other details in the discourse, such as the discourse of “hyperkalemia,” a condition in which insulin is used without the presence of diabetes. This high precision outcome resulted from the notion that reports including diabetes medications would have a high likelihood of indicating a patient’s history of diabetes.
The F-scores for the “Medication Only” classifier were lower than either the “Discourse Only” and “Combined” classifiers. The absence of diabetes medication from the discharge summary does not strictly indicate an absence of diabetes as a disease condition, since hyperglycemia may also be managed by other interventions such as exercise and diet. Thus, medication evidence cannot be independently employed to classify discharge summaries for diabetes. However, combined with other discourse evidence, medication can supplement the classification process because of its high precision when diabetes medication discourse is encountered in a report.
Other contributors of misclassification that were encountered in the evaluation included an observation of diabetes discourse that was incorrectly associated to a patient, when actually it was in reference to a family member’s condition. In this case, the report would be incorrectly identified as positively indicating diabetes, when no other evidence supports a positive classification. Another error stemmed from a verbose list of principal diagnoses in one of the documents that included a negation of an unrelated condition (eg, “without remission”) along with the diagnosis of diabetes. Due to the structure of the discourse (eg, the list of diagnoses were uttered in a single sentence), the “without” negation was inadvertently associated with the both the “remission” and the “diabetes” utterances.
Although encumbered by limited data, ABCS protocol compliance and high risk factor classifiers show promising results using the concept extraction and summarization methodology with all but two F-measures above 0.9; the remaining two metrics were above 0.8.
One observation to note regarding the ABCS protocol compliance results is the apparent lack of A1c and cholesterol reporting for diabetes patients, in contrast to blood pressure reporting. Although this observation might suggest a lack of ABCS protocol compliance, it may also suggest that evaluating ABCS protocol compliance should be performed across multiple reports over a range of time, since A1c and cholesterol tests may not be ordered during a given encounter, if they had been performed during a prior, recent visit.
Conclusions
In this study, we have demonstrated that it is possible to extract and classify concepts directly tied to diabetes from medical text with high precision and recall to support not only diabetes detection, but also protocol compliance and risk factor assessments. Natural language processing using ConText in conjunction with heuristic rules can help us to process a great number of medical records for preliminary screening at minimal cost and potentially augment better quality-of-care by tagging medical records for concepts of interest; such relatively simple to implement rule-based approaches can perform well, especially in circumstances where training data is limited, a potential shortcoming for statistical approaches.
One challenge in the current design of this study is distinguishing between different types of diabetes, for example, the differentiation between diabetes mellitus type 2 and gestational diabetes. In free-text discourse, understanding the true meaning of the utterance “diabetes” requires word sense disambiguation. Beyond word sense disambiguation, disambiguation of the underlying motivation for an event is also a challenge requiring further exploration to identify ways of resolving such ambiguities, as well as understanding the prevalence of potential false-positive classification (eg, misclassifying insulin usage as an indication of diabetes) resulting from these ambiguous concepts.
Although effective against discharge summaries, the true value of disease screening can only be realized when we use NLP-based systems in conjunction with other sources of information such as procedure lists, problem lists, diagnosis codes, etc. Most of the factual and objective pieces of information can be derived from rule-based systems, but these systems lack the ability to be inferential, which is a hallmark of a domain expert’s judgment. Hence, we recommend these tools be used as preliminary screening devices to assist the gold standard manual processes, such as chart reading performed by people with required expertise.
The preliminary screening of textual medical records, like discharge summaries and clinical reports, facilitates medical and epidemiological study by providing statistically relevant data for analysis. The findings observed and reported for elements of a disease concept are of key importance in control and prevention of such diseases.28 Although rule-based solutions, such as this work, do not necessarily require extensive training sets (like statistical methods); testing against a larger, diverse corpus of data can elucidate unusual presentations of diabetes, ABCS protocol compliance attributes, and high-risk factor indicators that are not accurately handled by the current classifiers.
Future work includes testing against a broader set of data, as they become available. Future work should also explore employing this approach on higher fidelity (eg, physician’s notes) and more voluminous data sets to better assess and refine the current methodology.
Acknowledgments
The authors thank Wendy Chapman for her assistance with ConText. Deidentified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr. Özlem Uzuner, i2b2 and SUNY. Authors also thank Danielle Kahn in assisting us with the literature search on use of NLP for disease screening and classification.
Footnotes
Funding Source: CDC Funded Research
References
- 1.Centers for Disease Control and Prevention National Diabetes Fact Sheet: national estimates and general information on diabetes and prediabetes in the United States, 2011. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2011 [Google Scholar]
- 2.Dall T, Mann SE, Zhang Y, Martin J, Chen Y, Hogan P. Economic costs of diabetes in the U.S. in 2007. Diabetes Care 2008; 31:1–20 [Google Scholar]
- 3.Saydah SH, Geiss LS, Tierney E, Benjamin SM, Engelgau M, Brancati F. Review of the performance of methods to identify diabetes cases among vital statistics, administrative, and survey data. Annals Epidemio 2004; 14:507–516 [DOI] [PubMed] [Google Scholar]
- 4.Turchin A, Kohane IS, Pendergrass ML. Identification of patients with diabetes from the text of physician notes in the electronic medical record. Diabetes Care 2005; 28:1794–1795 [DOI] [PubMed] [Google Scholar]
- 5.Turchin A, Pendergrass ML, Kohane IS. DITTO — a tool for identification of patient cohorts from the text of physician notes in the electronic medical record. AMIA Annu Symp Proc 2005: 744–748 [PMC free article] [PubMed] [Google Scholar]
- 6.Solti I, Gennari JH, Payne T, Solti M, Tarczy-Hornoch P. Natural language processing of clinical trial announcements: exploratory-study of building an automated screening application. AMIA Annu Symp Proc 2008:1142 [PubMed] [Google Scholar]
- 7.Li L, Chase HS, Patel CO, Friedman C, Weng C. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Annu Symp Proc 2008:404–408 [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung BM, Ong KL, Cherny SS, Sham PC, Tso AW, Lam KS. Diabetes prevalence and therapeutic target achievement in the United States, 1999 to 2006. AM J Med 2009; 122:443–453 [DOI] [PubMed] [Google Scholar]
- 9.American Diabetes Association Standards of Medical Care in Diabetes—2011. Diabetes Care 2011; 34:S11–S61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.American Diabetes Association Standards of Medical Care in Diabetes—2009. Diabetes Care 2009; 32:S13–S61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Brief Bioinform 2005; 6:57–71 [DOI] [PubMed] [Google Scholar]
- 12.Chapman WW, Chu D, Dowling JN. ConText: An algorithm for identifying contextual features from clinical text. Czech Republic: BioNLP Workshop of the Association for Computational Linguistics; 2007. 81–88 [Google Scholar]
- 13.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009; 42:839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001; 34:301–310 [DOI] [PubMed] [Google Scholar]
- 15.Mishra NK, Cummo DM, Arnzen JJ, Bonander J. A rule-based approach for identifying obesity and its comorbidities in medical discharge summaries. J Am Med Inform Assoc 2009; 16:576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambert KH, Cohen AM. A system for classifying disease comorbidity status from medical discharge summaries using automated hotspot and negated concept detection. J Am Med Inform Assoc 2009; 16:590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo Clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008; 15:25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamon T, Grabar N. Linguistic approach for identification of medication names and related information in clinical narratives. J Am Med Inform Assoc 2010; 17:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen AN, Lawley MJ, Hansen DP, Bowman RV, Clarke BE, Duhig EE, Colquist S. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc 2010; 17:440–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzuner Ö. Second i2b2 workshop on natural language processing challenges for clinical records. AMIA Annu Symp Proc 2008:1252–1253 [PubMed] [Google Scholar]
- 21.Lappin S, Leass HJ. An algorithm for pronominal anaphora resolution. Computational Linguistics 1994; 20:535–561 [Google Scholar]
- 22.Ge N, Hale J, Charniak E. A statistical approach to anaphora resolution. Paper presented at the Sixth Workshop on Very Large Corpora; August 16, 1998; Montreal, Quebec, CA [Google Scholar]
- 23.Son RY, Taira RK, Kangarloo H. Inter-document coreference resolution of abnormal findings in radiology documents. Stud Health Technol Inform 2004; 107:1388–1392 [PubMed] [Google Scholar]
- 24.Son RY, Taira RK, Kangarloo H, Cardenas AF. Context-sensitive correlation of implicitly related data: an episode creation methodology. IEEE Trans Inf Technol Biomed 2008; 12:549–560 [DOI] [PubMed] [Google Scholar]
- 25.Turchin A, Kolatkar NS, Grant RW, Makhni EC, Pendergrass ML, Einbinder JS. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. J Am Med Inform Assoc 2006; 13:691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007; 14:550–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y. A study of thresholding strategies for text categorization. SIGIR ’01 Proceedings of the 24th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval New Orleans, LA, 2001:137–145 [Google Scholar]
- 28.Solt I, Tikk D, G·l V, Kardkovács ZT. Semantic classification of diseases in discharge summaries using a context-aware rule-based classifier. J Am Med Inform Assoc 2009; 16:580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilke RA, Berg RL, Peissig P, Kitchner T, Sijercic B, McCarty CA, McCarty DJ. Use of an electronic medical record for the identification of research subjects with diabetes mellitus. Clin Med Res 2007; 5:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voorham J, Denig P. Computerized extraction of information on the quality of diabetes care from free text in electronic patient records of general practitioners. J Am Med Inform Assoc 2007; 14:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc 2000; 7:593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001; 34:301–310 [DOI] [PubMed] [Google Scholar]
- 33.Chapman WW, Cooper GF, Hanbury P, Chapman BE, Harrison LH, Wagner MM. Creating a text classifier to detect radiology reports describing mediastinal findings associated with inhalational anthrax and other disorders. J Am Med Inform Assoc 2003; 10:494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilcox AB, Hripcsak G. The role of domain knowledge in automating medical text report classification. J Am Med Inform Assoc 2003; 10:330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapman WW, Christensen LM, Wagner MM, Haug PJ, Ivanov O, Dowling JN, Olszewski RT. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med 2005; 33:31–40 [DOI] [PubMed] [Google Scholar]
- 36.Zeng Q, Goryachev S, Weiss S, Sordo M, Murphy S, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedlin J, McDonald CJ. Using a natural language processing system to extract and code family history data from admission reports. AMIA Annu Symp Proc 2006:925 [PMC free article] [PubMed] [Google Scholar]
- 38.Clark C, Good K, Jezierny L, Macpherson M, Wilson B, Chajewska U. Identifying smokers with a medical extraction system. J Am Med Inform Assoc 2008;15:36–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Childs LC, Enelow R, Simonsen L, Heintzelman NH, Kowalski KM, Taylor RJ. Description of a rule-based system for the i2b2 challenge in natural language processing for clinical data. J Am Med Inform Assoc 2009; 16:571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farkas R, Szarvas G, Heged?s I, Alm·si A, Vincze V, Orm·ndi R, Busa-Fekete R. Semi-automated construction of decision rules to predict morbidities from clinical texts. J Am Med Inform Assoc 2009; 16:601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Spasic I, Keane JA, Nenadic G. A text mining approach to the prediction of disease status from clinical discharge summaries. J Am Med Inform Assoc 2009; 16:596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ware H, Mullett CJ, Jagannathan V. Natural language processing framework to assess clinical conditions. J Am Med Inform Assoc 2009; 16:585–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murff HJ, FitzHenry F, Matheny ME, Gentry N, Kotter KL, Crimin K, Dittus RS, Rosen AK, Elkin PL, Brown SH, Speroff T. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011; 306:848–855 [DOI] [PubMed] [Google Scholar]