Abstract
Cells produce tens of thousands of different lipid species, but the importance of this complexity in vivo is unclear. Analysis of individual tissues and cell types has revealed differences in abundance of individual lipid species, but there has been no comprehensive study comparing tissue lipidomes within a single developing organism. Here, we used quantitative shotgun profiling by high-resolution mass spectrometry to determine the absolute (molar) content of 250 species of 14 major lipid classes in 6 tissues of animals at 27 developmental stages raised on 4 different diets. Comparing these lipidomes revealed unexpected insights into lipid metabolism. Surprisingly, the fatty acids present in dietary lipids directly influence tissue phospholipid composition throughout the animal. Furthermore, Drosophila differentially regulates uptake, mobilization and tissue accumulation of specific sterols, and undergoes unsuspected shifts in fat metabolism during larval and pupal development. Finally, we observed striking differences between tissue lipidomes that are conserved between phyla. This study provides a comprehensive, quantitative and expandable resource for further pharmacological and genetic studies of metabolic disorders and molecular mechanisms underlying dietary response.
Keywords: Drosophila , lipidomics, membrane lipids, neutral lipids, nutrition
Introduction
Lipids are essential for life. They serve for energy storage, structural components of cell membranes and organelles, and are also important signaling molecules. Lipids are enormously diverse in their structure and composition. The lipidome, a full-lipid complement of the cell, tissue or organism, may comprise over 1000 lipid species for a single cell (van Meer, 2005; Wenk, 2010) and 10 000–100 000 species for tissues or organisms (Yetukuri et al, 2008). Part of this structural diversity arises from combinations of different fatty acids and functional headgroups that can be linked to each other in different ways (Dennis, 2009; Wenk, 2010). Within a given lipid class, different lipid species can be produced by the incorporation of a variety of fatty acids differing in chain length and unsaturation. How this immense lipid variability is deployed in vivo, and to what extent it can be altered in response to exogenous factors (such as nutrition) without jeopardizing the lipid homeostasis at the organism level, remains unclear (Shevchenko and Simons, 2010). While there are many examples of enrichment of specific lipids in particular tissues or cell types, the quantitative extent of lipid variability between tissues of a single organism has never been analyzed systematically. This is important because lipid populations display interesting collective behaviors that are not the property of any one lipid alone.
Mass spectrometry has been instrumental in identifying the vast number of different lipid species and is now recognized as a premier tool for lipidomics (reviewed in Harkewicz and Dennis, 2011). While some approaches require the prior separation of lipid classes or species by liquid chromatography (reviewed in Blanksby and Mitchell, 2010), the shotgun lipidomics approach is based on direct infusion of total lipid extracts into a mass spectrometer (reviewed in Han et al, 2012). Individual lipid species are then recognized either by accurately determined masses (reviewed in Schwudke et al, 2011), or by characteristic fragments produced by tandem mass spectrometry (reviewed in Gross and Han, 2011). With the advent of high-resolution mass spectrometry, shotgun lipidomics is now able to produce a rapid and comprehensive snapshot of the full-lipid composition, accounting for major lipid classes and major individual species within each class (Schwudke et al, 2007). This approach has been successfully applied for quantitative full-lipidome profiling in a variety of model organisms and cell systems, such as yeast (Ejsing et al, 2009) and viruses (Chan et al, 2008; Kalvodova et al, 2009; Gerl et al, 2012), mammalian cells (Sampaio et al, 2011) and blood plasma (Schuhmann et al, 2012). Typically, high-resolution full-organism shotgun profiling quantifies about 100–300 species from 10 to 15 classes, accounting for the bulk of the lipidome (Ejsing et al, 2009; Schuhmann et al, 2012).
Drosophila is rapidly emerging as a powerful model organism to study lipid metabolism. Its lipid metabolic pathways resemble those of vertebrates (Baker and Thummel, 2007; Leopold and Perrimon, 2007), but Drosophila is amenable to facile genetic manipulation. Drosophila lipid storage and mobilization mechanisms are well-characterized (Gronke et al, 2005; Guo et al, 2008). Molecules involved in membrane lipid biosynthesis and dietary lipid uptake can be easily accessed genetically (Pavlidis et al, 1994; Adachi-Yamada et al, 1999; Tschape et al, 2002; Herr et al, 2003; Huang et al, 2005; Kunte et al, 2006). Like vertebrates, Drosophila develop cardiomyopathy when fed diets high in saturated fats (Birse et al, 2010). Furthermore, the sterol auxotrophy of Drosophila simplifies the study of dietary sterol uptake (Carvalho et al, 2010; Niwa and Niwa, 2011). However, most previous studies have focused on only a few lipid classes when examining the consequences of different perturbations. Until recently, it has not been possible to address how genetic, nutritional or drug interference perturbs the whole lipidome.
Here, we present a systematic lipidomic effort to assess Drosophila lipid composition in different tissues, at different developmental stages and on different diets. We identify both qualitative and quantitative differences in tissue lipid composition. The diet has a direct and global effect on the lipidome; however, we also identify conserved differences in tissue lipid composition that are unaffected by the diet. We uncover specificity in the uptake, transport and tissue accumulation of different sterol species that is developmentally regulated. Finally, our data reveal unexpected shifts in the accumulation of membrane lipids and neutral lipids during larval growth and pupal development. Our study provides an essential framework and resource to understand lipid metabolism in Drosophila in molecular detail.
Results
Dissection of Drosophila lipidome by high mass resolution shotgun profiling
We wondered how the Drosophila lipidome varied in different tissues, on different diets, and at different developmental stages. To answer these questions, we applied quantitative shotgun profiling on high-resolution hybrid tandem mass spectrometers LTQ Orbitrap XL and (where specified) Q Exactive (Schwudke et al, 2007, 2011). Each sample contained <2 nmol of total lipids and at this level of sensitivity, the analysis of individual dissected tissues was possible. Total extracts were infused into a mass spectrometer in a fully automated fashion using a nanoflow robotic ion source. Using new pipette tip and spraying nozzle for each analysis precluded any danger of cross-contamination between the samples. Lipids were identified by matching their intact masses at the sub-p.p.m. accuracy, within the class-specific elemental composition constraints (Herzog et al, 2011). In a few ambiguous cases, peak assignments were validated by tandem mass spectrometry performed on the respective precursors. Lipid species were quantified by comparing the abundances of their precursors to the abundances of peaks of internal standard—synthetic non-naturally occurring lipids spiked into samples prior to extraction (Ejsing et al, 2006). We excluded from our analysis certain low abundant lipid classes that, for technical reasons, could not be robustly quantified (e.g., sterol esters, lyso-phospholipids, cardiolipins). To obtain a comprehensive profile of individual sterols, we developed a new method combining sterol sulfation (Sandhoff et al, 1999) with high-resolution mass spectrometry. Ambiguous assignments were resolved by an alternative derivatization protocol (Liebisch et al, 2006) that allowed us to distinguish sterol isomers. With these methods, we obtained a 1000-fold dynamic range and a limit of detection of 300 fmol for sterols and from 1 to 50 fmol for other lipids. The analysis of a total extract with three separate acquisitions required only 10 min.
We analyzed lipid extracts prepared from six major larval tissues: gut, lipoproteins, fat body, salivary gland, wing imaginal disk and brain. These tissues were dissected from feeding third instar larvae raised on four diets with different lipid compositions: a yeast-based diet, a plant-based diet and two lipid-depleted foods (LDFs) supplemented with yeast or plant sterols. We also analyzed lipid extracts from whole animals at different developmental stages, ranging from hatching of first instar larvae until the emergence of adults (a total of 27 time points). In total, we analyzed 54 different biological conditions, from which 222 lipid extracts were prepared and >1300 spectra were acquired and analyzed.
Altogether, we systematically quantified 250 species from 14 major lipid classes—triacylglycerols (TAG), diacylglycerols (DAG), phosphatidylethanolamines (PE), phosphatidylcholines (PC), phosphatidylinositols (PI), phosphatidylserines (PS), phosphatidylglycerols (PG), phosphatidic acids (PA), ether lipids (PE-O and PC-O), ceramides (Cer), phosphorylethanolamine ceramides (CerPE), hexosyl ceramides (HexCer) as well as 13 individual sterols. To the best of our knowledge, this is currently the most detailed assessment of the spatial and temporal distribution of lipids in Drosophila. Below, we highlight some of the most interesting and novel findings that emerge from these data. The full data set is available (Supplementary Information, Datasets 1, 2 and 3) and is open for future data mining efforts.
Larval tissues differ in their lipid composition
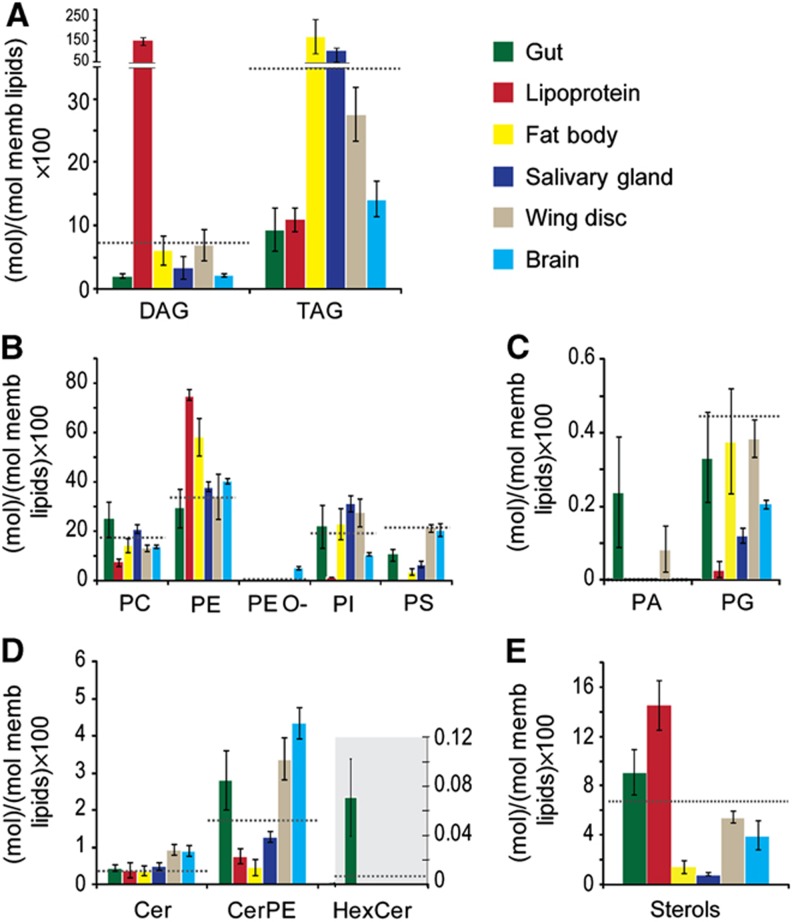
To characterize the composition of individual tissues, we examined the lipid profiles of gut, lipoproteins, fat body, salivary gland, wing disk and brain from larvae fed on yeast food (YF). We determined the absolute quantities of lipid species (in moles) and calculated the total abundance of each lipid class by summing up the abundances of individual species. To make lipid profiles comparable, we normalized the content of each individual lipid class to the total content of all membrane lipids—this includes phospholipids, sphingolipids and sterols, but not DAG or TAG. We then compared the normalized abundances of lipid classes between the tissues and the average lipid composition of stage-matched whole third instar larvae. We applied principal component analysis (PCA) to highlight the major compositional differences between tissues (Supplementary Figures S1 and S2), and then examined the altered lipids in detail (Figures 1 and 2). We noted striking differences in tissue lipid composition. Here, we highlight the most dramatic and interesting tissue-specific differences.
Figure 1.
Lipid class profiles vary in different tissues. (A–E) The abundance of different lipid classes in tissues and lipoproteins of early wandering third instar larvae fed with YF. Each color represents one larval tissue, as indicated. Lipid class abundance is presented as moles per mole of total membrane lipid (phospholipids, sphingolipids and sterols—not including TAG or DAG). (A) Neutral lipids. (B) Major phospholipids. (C) Minor phospholipids. (D) Sphingolipids. (E) Sterols. Error bars indicate standard deviation. Dashed line indicates the lipid class amount in whole larval lipids from animals of the same stage.
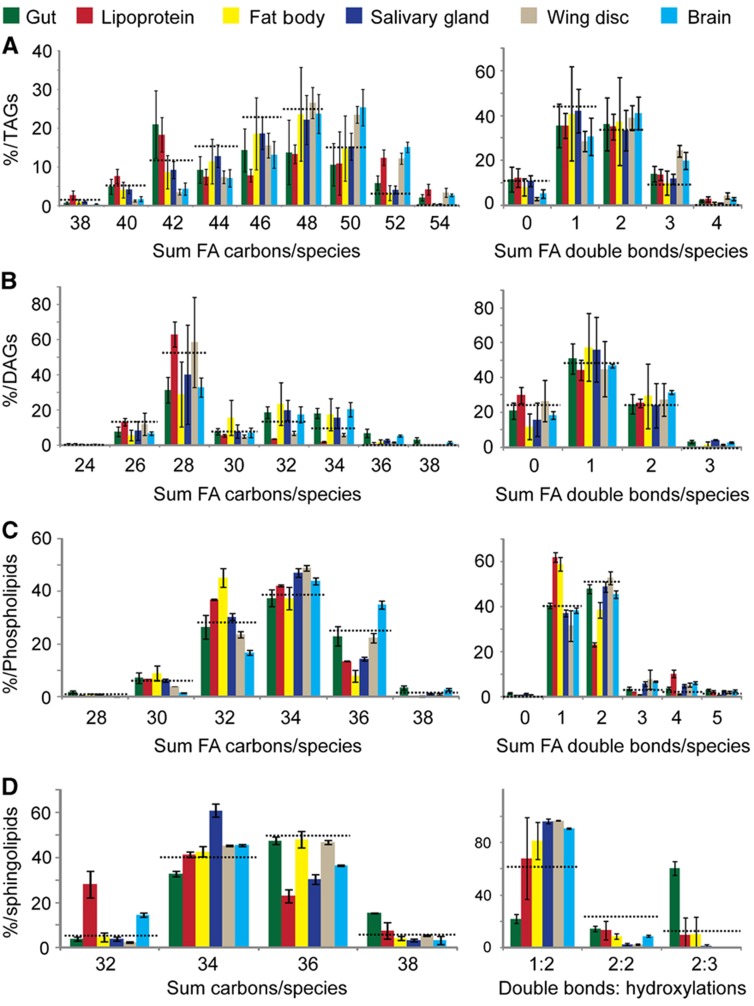
Figure 2.
Fatty acid chain length and unsaturation varies in different tissues. (A–D) The distributions of combined fatty acid chain length (left panels) and unsaturation (right panels) in (A) TAG (B) DAG (C) phospholipids (D) sphingolipids from different tissues of early wandering third instar larvae fed with YF. The abundance of each is presented as a percent relative to all lipids in its category. Dashed line indicates the corresponding profiles of whole larval lipids from animals of the same stage. Error bars indicate standard deviation.
Gut
The gut of Drosophila larvae differs from other tissues in its sphingolipid composition. We detected hexosyl ceramides only in the gut (Figure 1D). Furthermore, the gut contained a 4.8-fold higher fraction of 2:3 sphingolipids than the average from whole larvae (Figure 2D). 2:3 sphingolipids contain two double bonds in the long-chain base and hydroxylated fatty acid chains. Interestingly, these lipids do not seem to be generally characteristic of epithelial cells, since they are not elevated in either the wing disk or salivary gland (Figure 2D). Their function may be important to maintain a tight barrier against the harsh environment of the gut lumen; hydroxylated sphingolipids are known to promote tight lipid packing (Lofgren and Pascher, 1977) and are also found in mammalian membranes specialized to perform barrier functions (Uchida et al, 2007; Maldonado et al, 2008; Hama, 2009).
Lipoproteins
Polar lipid components of lipoprotein particles are present in strikingly different proportions than those observed for cell membranes. PE represents 76 mol% of all polar lipids in lipoproteins, but only between 30 and 40 mol% in most tissues we examined (35 mol% of total larval polar lipids) (Figure 1B). Sterols are also overrepresented in lipoproteins (14 mol%), compared with other tissues (7 mol% in total larval lipids) (Figure 1E). This may reflect the importance of lipoproteins as a source of sterols for peripheral tissues. While the most abundant neutral lipid class in most tissues is TAG, lipoproteins contain predominantly DAG (Figure 1A). The two fatty acid moieties in lipoprotein DAG comprise 26–28 carbon atoms, suggesting that they mostly contain medium chain fatty acids (12–14 carbon atoms) (Palm et al, 2012). In contrast, other tissues also contain long-chain DAGs, which have a total of>32 carbon atoms in their fatty acid moieties (Figure 2B).
Fat body and salivary gland
As expected, the fat body, which is the major lipid storage organ, contains much larger amounts of TAG than other tissues (Figure 1A). It is also enriched in PE (Figure 1B), compared with other tissues, although not as dramatically as lipoproteins. The most abundant PE species, 32:1 and 34:1 are the same as the major PE species in lipoproteins (Figure 2C; Supplementary Information, Dataset 2), supporting its role as the major source of lipoproteins (Panakova et al, 2005). We also note that fatty acids in fat body phospholipids tend to be shorter and more saturated than in other tissues (Figure 2C). For each glycero- and glycerophospholipid class, we calculated the average number of carbon atoms Cav and the average number of double bonds DBav by considering the relative abundances of corresponding lipid species:
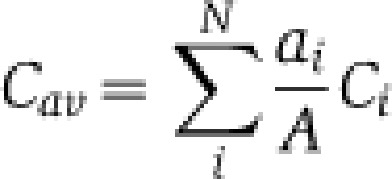 |
and
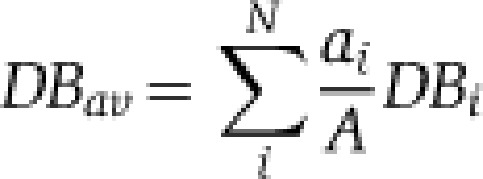 |
where ai is the abundance of i lipid species having Ci carbon atoms and DBi double bonds; A is the total abundance of all species of the lipid class and N is total number of species of this lipid class. To calculate the average carbon chain length and unsaturation of the fatty acid moieties, we divided Cav and DBav by the number of fatty acids present in each lipid species (two for DAGs and glycerophospholipids, and by three for TAGs). Cav and DBav are used to conveniently illustrate global changes in fatty acid length and unsaturation with a single number and they do not represent the individual fatty acids present in lipid species.
By this reckoning, the average fatty acid in fat body phospholipids contains 16.5 carbons and 0.7 double bonds (16.5:0.7, notations explained in MM). In contrast, the average fatty acids in the brain and wing disk were 17.0:1.0 and 17.2:0.9, respectively (Supplementary Table SI). This partly reflects the fact that the fat body has a lower proportion of PS, which contains longer and more unsaturated fatty acids than other phospholipid classes. But even within phospholipid classes, fatty acids are generally shorter and more saturated in the fat body (Supplementary Figure S3).
Interestingly, fat body and salivary gland, which are polyploid tissues, share several lipid composition features. Both contain relatively lower levels of lipids known to be enriched in plasma membranes, for example, sterols, PS and the sphingolipid CerPE (Figure 1). Intermediate levels of these lipids are present in the gut (Figure 1), which contains both diploid and polyploid cells. Plasma membrane represents a smaller fraction of total membrane as cell size increases. Thus, large cell size may account for differences in the proportions of these lipids.
Brain and wing disk
Brain and wing disk contain proportionally more sterol, CerPE and PS than polyploid tissues (as noted above). In addition, fatty acid moieties in phospholipids in brain and wing disk are longer and more unsaturated than those in other organs, and this phenomenon is even stronger in the brain than in the wing disk (Figure 2C; Supplementary Table SI). The same tendency was observed for the composition of storage lipids (TAG), which indicates the likely interconversion of phospholipids and TAGs. The brain exhibits additional unique features. It has less PI than any other organ, and it is the only organ analyzed that contains significant amounts of ether phospholipids (Figure 1B), similar to mammalian brain (Farooqui and Horrocks, 2001). These lipids contain mainly phosphoethanolamine as a headgroup, and comprise one fatty alcohol and one fatty acid moiety. They tend to be longer and more unsaturated than diacyl PE (Supplementary Figure S3)—this also contributes to the increased average length and unsaturation of fatty acids in the brain.
The diet influences membrane lipid composition in all tissues
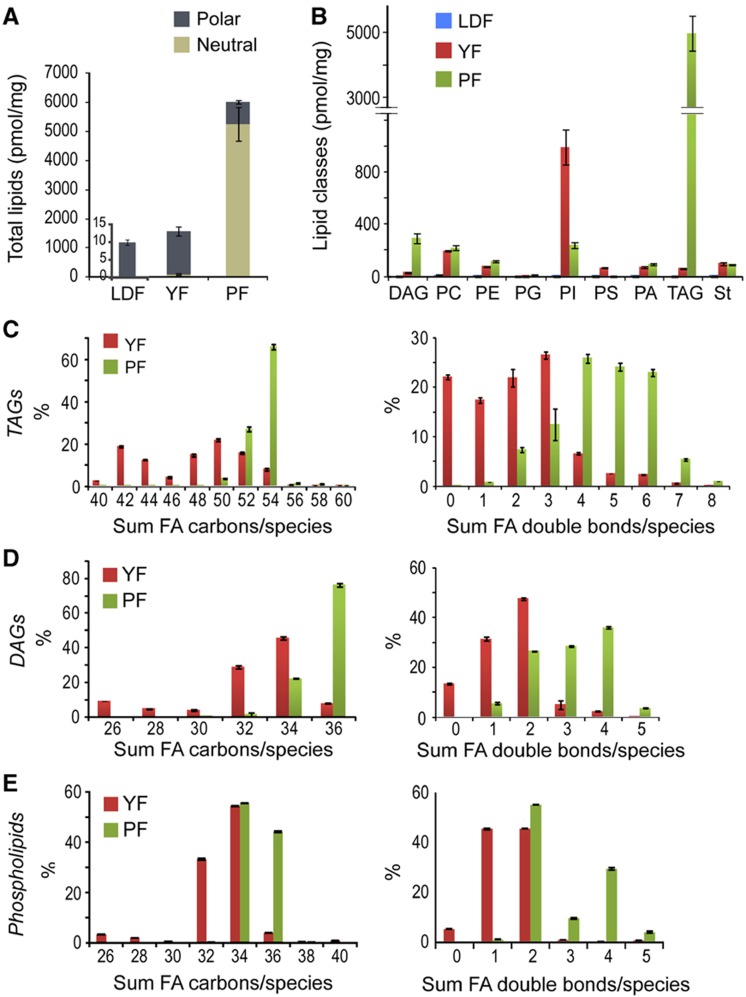
In mammals, ingested lipids are cleaved into fatty acids, monoacylglycerols, lyso-lipids and polar head groups before being absorbed by enterocytes (Zeisel, 1981; Iqbal and Hussain, 2009). How do metabolized dietary lipids affect the composition of organ lipidomes? To answer this, we fed Drosophila larvae with foods containing different lipids and quantified the lipid composition of individual tissues. In the wild, Drosophila feeds on yeast growing on rotting fruit. Here, to supply dietary lipids to flies, we used foods based on either yeast or plant material. Yeast food is enriched in PI and contains very little neutral lipid. In contrast, plant food (PF) contains mostly TAG, and a more heterogeneous composition of phospholipid classes (Figure 3B). In total, PF contains approximately four-fold more lipids than YF (Figure 3A). Overall, fatty acids in PF lipids are longer and more unsaturated than those in YF (Figure 3C–E). The full-lipidome compositions of the three foods were determined by mass spectrometry and are provided in Supplementary Information, Dataset 1. To investigate the contribution of endogenous lipid synthesis, we also fed larvae on LDFs supplemented with a plant or fungal sterol (which Drosophila cannot synthesize). We used PCA to compare the lipid class profiles (Supplementary Figure S4) and lipid species profiles (Supplementary Figures S5 and S6) from larvae grown on these four different foods, searching for systematic differences due to dietary lipids.
Figure 3.
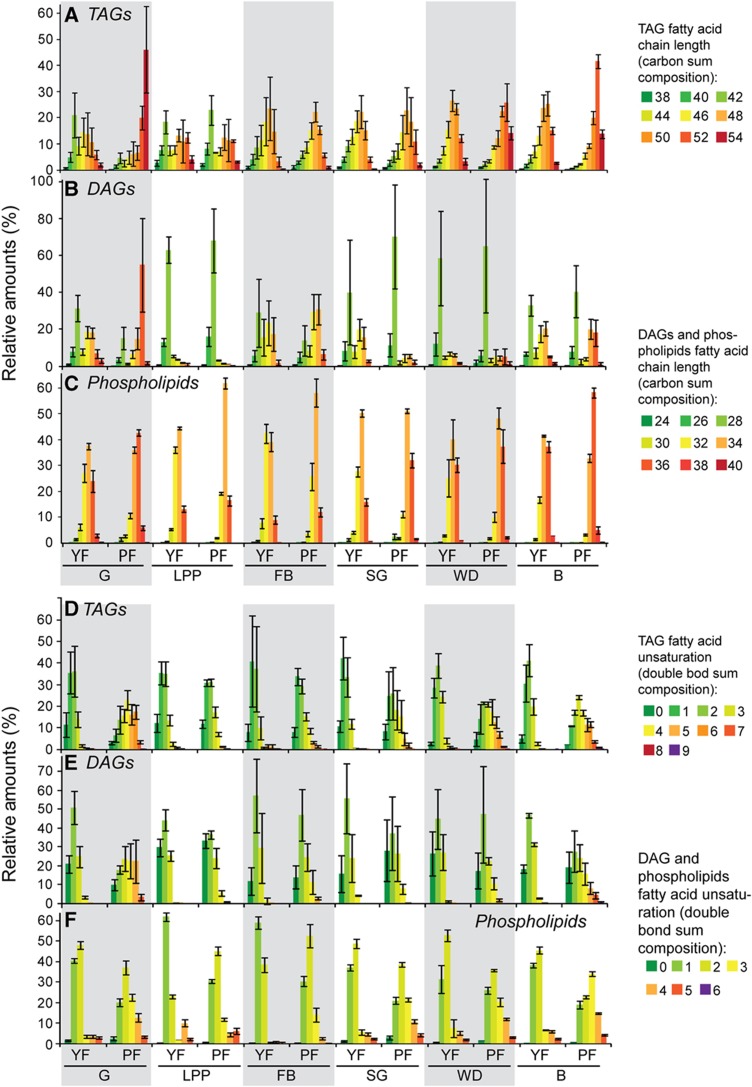
Lipid composition of different Drosophila foods. (A) The amount (pmol/mg) of neutral lipids (brown) and polar lipids (gray) present in LDF, YF and PF. Note that LDF is plotted on a different scale and its lipid content is only 1/150th that of YF. (B) The amount of different lipid classes present in LDF (blue), YF (red) and PF (green). (C–E) The relative abundance in YF (red) and PF (green) of lipids with different combined fatty acid chain lengths and unsaturation TAGs (C), DAGs (D) and phospholipids (E) are plotted separately. FA= fatty acid. Error bars indicate standard deviation.
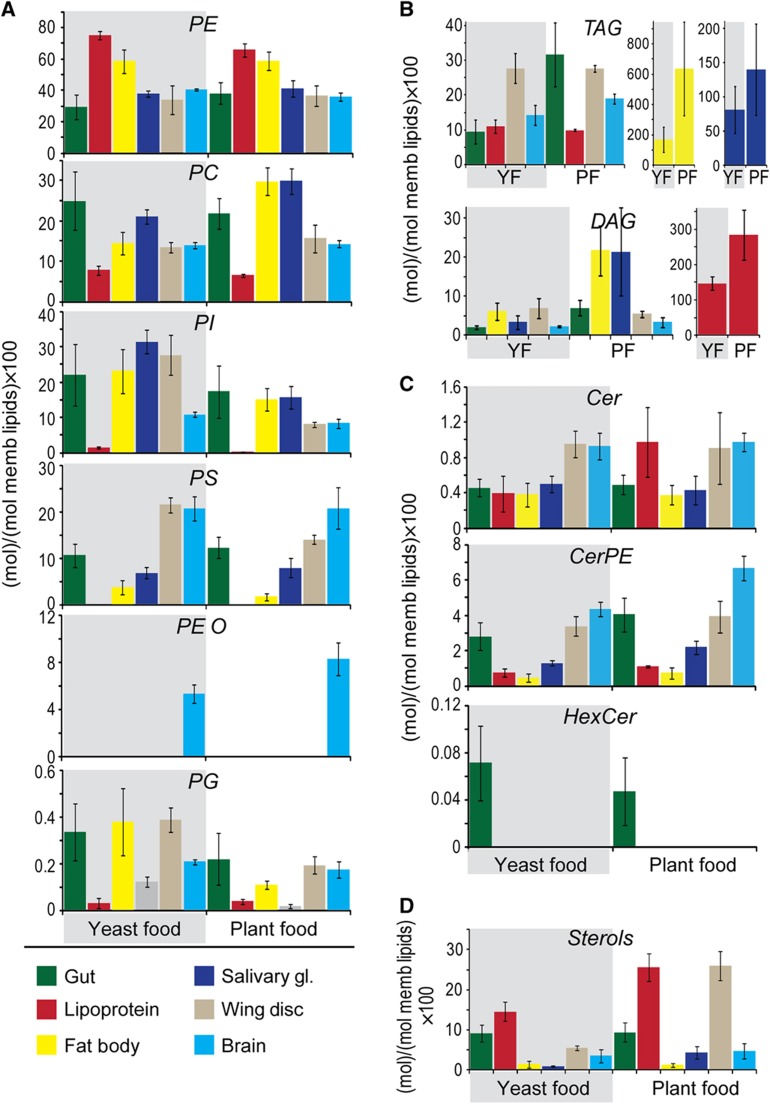
One of the most obvious changes in animals fed on PF is the dramatic increase in stored TAG in the fat body and gut (Figure 4B). Fat bodies from plant-fed larvae contain 3.7-fold more TAG than those fed on YF, and guts contain 3.3-fold more. These changes likely reflect the large amount of lipid in PF (Figure 3A and B). Consistent with this, circulating lipoproteins of plant-fed larvae contain much more medium chain DAG (Figures 4B and 5B). This could reflect the importance of this lipid as a major vehicle of fatty acid transport and suggests that dietary fatty acids may be shortened before export as medium chain DAG. TAG levels do not increase in wing disk or brain (Figure 4B), suggesting that accumulation of neutral lipids to these tissues is precisely controlled.
Figure 4.
Effect of diet on lipid class abundance in different tissues. (A–C) The amount of each lipid class in the different tissues and lipoproteins of early wandering third instar larvae fed either YF (shaded in gray), or PF (not shaded). Tissues are indicated with color, as shown. Lipid class amount is calculated as mol% with respect to total membrane lipids Membrane lipids include phospholipids, sphingolipids and sterols, but not TAG and DAG. (A) Phospholipids (B) Neutral lipids (C) Sphingolipids (D) Sterols. Error bars indicate standard deviation.
Figure 5.
Tissue lipids reflect the fatty acids present in the diet. (A–F) The distributions of combined fatty acid chain length (A–C) and unsaturation (D–F) in TAG (A, D) DAG (B, E) and phospholipids (C, F) from different tissues of early wandering third instar larvae fed with either YF or PF. Tissues are indicated at the bottom of each chart and are shaded in an alternating pattern to ease comparison: G=gut; LPP=lipoproteins; FB=fat body; SG=salivary gland; WD=wing disk and B=brain. Lipid species with equal fatty acid chain length or unsaturation were pooled together. Phospholipid classes were pooled in one group that represents the average profile of all phospholipids. Length and unsaturation were color coded from green to red according to the color scale as indicated. Green indicates shorter and less saturated fatty acids and red longer and more unsaturated fatty acids. Error bars indicate standard deviation.
We next compared phospholipid class profiles of animals fed with different diets. The relative abundance of specific phospholipid classes does change in some tissues; however, these changes cannot be directly attributed to the abundance of the corresponding phospholipid class in the diet. For example, PI represents 27.7 mol% of membrane lipids in the wing disks of yeast-fed larvae, but only 8.1 mol% in disks of plant-fed larvae. These changes cannot be a consequence of the high amounts of PI in YF; PI levels are also higher in wing disks when larvae are fed LDF, which does not contain PI (compare Figure 4A and Supplementary Figure S7A). Thus, the composition of lipid classes in the diet does not directly influence the amount of the corresponding phospholipid class in cell membranes.
In contrast, the dietary fatty acid composition does exert a direct effect on the fatty acids present in different tissues. PF has longer and less saturated fatty acids than YF; the abundance-averaged fatty acid in PF is 17.8:1.5, whereas in YF it is 16.5:0.7 (Figure 3; Supplementary Table SI). Tissue phospholipids of animals fed with YF resemble those of animals fed with no fatty acids at all. In contrast, the phospholipids of larvae fed a plant diet contain longer and more unsaturated fatty acids (Figure 5; Supplementary Table SI). For example, the average phospholipid fatty acid in the fat body changes from 16.5:0.7 on YF to 16.8:0.9 on PF. Phospholipid fatty acids in the brain change from 17.2:0.9 on YF to 17.6:1.3 on PF. This shift in the average length and unsaturation of phospholipid species is observed in all tissues examined and affects all phospholipid classes (Supplementary Information, Datasets 1 and 2). Thus, feeding with PF shifts the phospholipid fatty acid profile in all tissues, but tissue-specific differences in length and unsaturation are maintained. These data show that the diet exerts a direct and significant effect on the species of phospholipids present in cell membranes.
It has been noted in yeast that the presence of different membrane sterols can influence the abundance of other lipid species, including phospholipids (Souza et al, 2011). Thus, we wondered whether changes in phospholipid length and unsaturation might be indirect consequences of the sterol composition of plant and YFs. To test this idea, we compared the phospholipid species synthesized by animals grown on LDF in the presence of either a plant sterol (stigmasterol) or a fungal sterol (ergosterol). Larvae fed with these two different sterols do not differ in the average length or unsaturation of phospholipid fatty acid moieties (Supplementary Figures S7 and S8; Supplementary Table SI). As noted above, larvae that must rely on endogenous fatty acid synthesis generate similar phospholipid species to those of larvae fed with YF (compare Figure 4 with Supplementary Figure S7 and Figure 5 with Supplementary Figure S8). Thus, larvae do not appear to adjust the fatty acids in their phospholipids to accommodate the structure of different sterols; rather, fatty acids available from the diet directly influence the composition of fatty acid moieties in cell membranes.
How do the long unsaturated fatty acid moieties present in dietary lipids reach the different peripheral tissues? We note that TAGs containing these fatty acids are dramatically elevated in guts of larvae fed with PF (Figures 4B and 5A and D). The lipoproteins that mobilize dietary lipids from the gut consist mainly of medium chain DAG, sterol and PE (Figure 1). Lipoprotein PE contains longer and more unsaturated fatty acids in larvae fed with PF (Figure 5C and F; Supplementary Table SI), while DAGs do not change—they still contain almost exclusively medium chain fatty acid moieties (Figure 5B and E). This suggests that lipoprotein PE is the major vehicle for the transport of longer unsaturated plant fatty acids to peripheral tissues. A plant diet also strongly affects fatty acid composition of stored TAG in most peripheral tissues. For example, the average fatty acid in the brain TAG is 16.1:0.6 when larvae are fed with YF, but 16.9:1.2 when larvae are fed with PF. In contrast, TAG species in the fat body are much less influenced by the fatty acid composition of the diet (Figure 5A and D; Supplementary Table SI), even though this tissue stores much more TAG when larvae are fed with the lipid-rich PF (Figure 4B). Thus, the fat body is not a passive depot for storage of dietary lipids; its ability to accumulate normal TAG stores on a lipid-free diet (compare Figure 4B with Figure 7B), suggests that endogenous synthesis supplies a large fraction of fat body TAG. In contrast, TAG stores in peripheral tissues tend to be directly derived from fatty acid moieties present in dietary lipids.
Figure 7.
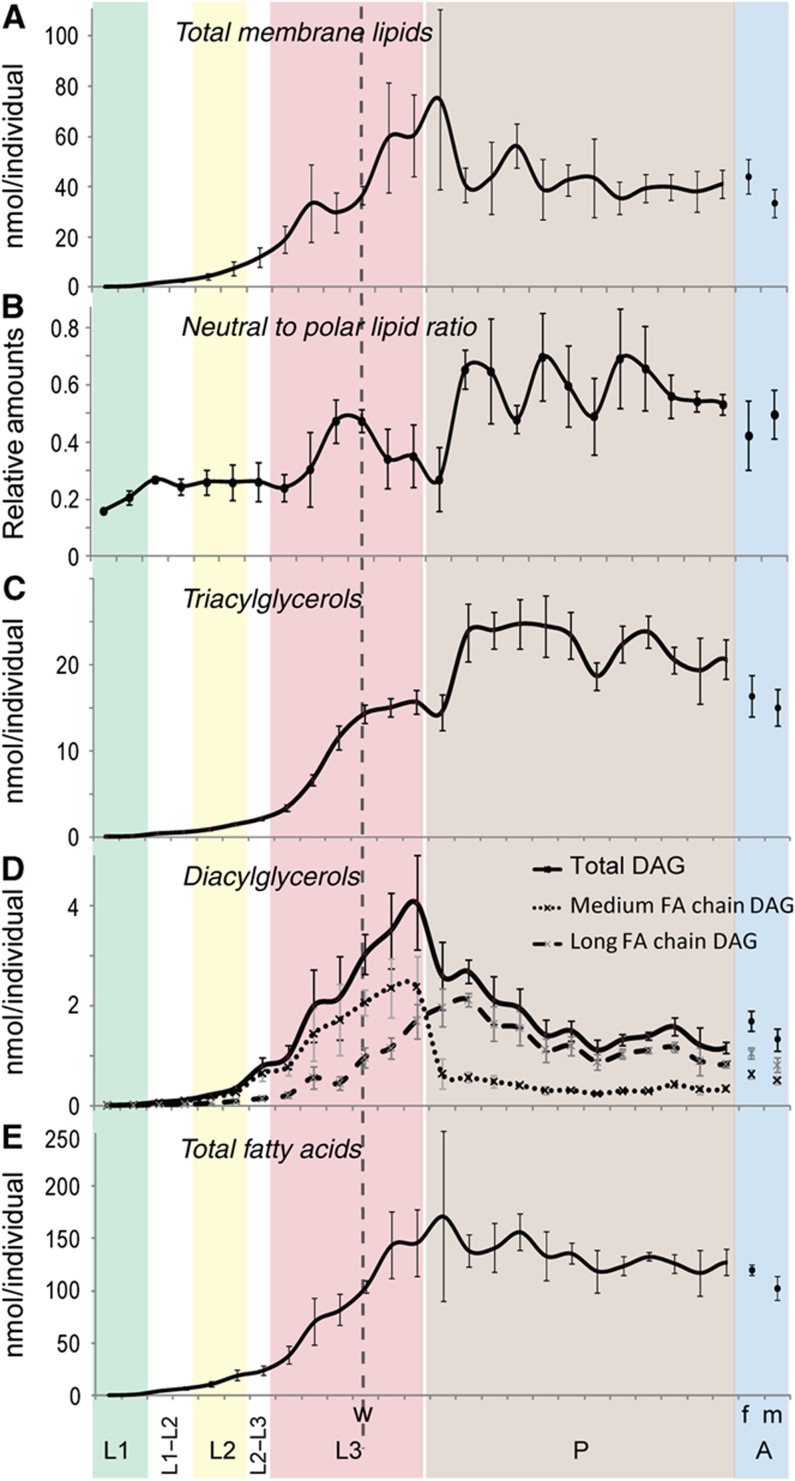
Changes in neutral and polar lipids during development. (A–E) The amounts of different types of lipids (nmol/animal) at different developmental stages from hatching to adulthood (time interval=8 h). (A) total membrane lipids, including phospholipids, sphingolipids and sterols, (C) TAG, (D) DAG and (E) total fatty acids. (B) The ratio of neutral lipids (DAGs+TAGs) to polar lipids (membrane lipids) at these different stages. L1=first instar, shaded green; L2=second instar, shaded yellow; L3=third instar, shaded red; P=pupae, shaded brown; A=adults, shaded blue, where m=males and f=females. The dashed line ‘w’ indicates the start of the wandering larval stage. Error bars indicate standard deviation.
Dietary sterols are mobilized onto lipoproteins with different efficiencies and show tissue-specific accumulation patterns
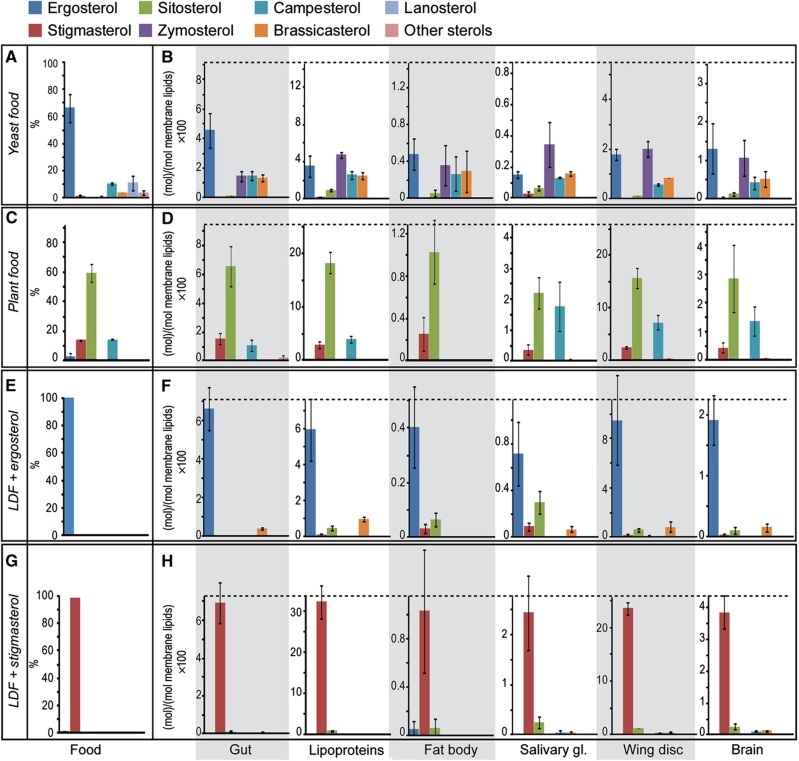
Drosophila requires bulk sterol in its membranes to develop normally (Carvalho et al, 2010; Niwa and Niwa, 2011). Yeast and plant-based foods each contain a mixture of different sterols (Figure 6A and C), and we wondered whether they could all be utilized equally. To investigate this, we compared the proportions of different sterols in the food with those that accumulated in different tissues. Ergosterol is the major sterol in YF, comprising close to 70% of all sterols (Figure 6A). In the gut, however, it represents only 50% of total sterols. Some sterols are completely excluded from the gut, most notably lanosterol. Other sterols present in the food are, conversely, relatively enriched in the gut—most dramatically, zymosterol (Figure 6B). This suggests that the gut either absorbs or effluxes different sterols with different efficiencies. Alternatively, some of these sterols might derive from maternal stores. To distinguish these possibilities, we examined the gut sterol profile of larvae fed with ergosterol alone on an otherwise lipid-depleted diet. These guts contain almost exclusively ergosterol and zymosterol is not detectable (Figure 6F). This rules out the possibility that zymosterol is derived from maternal sources, and further indicates that the gut does not convert ergosterol to zymosterol to any significant extent. Thus, the gut accumulates ergosterol relatively inefficiently compared with other sterols—particularly zymosterol.
Figure 6.
Differential uptake, mobilization and tissue accumulation of sterol species. (A, C, E, G) The proportions of different sterols in each food. (B, D, F, H) The amounts of different sterols present in each of the tissues of Drosophila early wandering third instar larvae fed on the corresponding food. The black dashed line indicates the total amount of sterol in each of the tissues. Different sterol species are shown in different colors, as indicated. Sterol amounts are represented as mol% with respect to membrane lipids (including phospholipids, sphingolipids, sterols but not DAG or TAG) (A, B) refer to YF; (C, D) refer to PF; (E, F) refer to LDF+ergosterol and (G, H) refer to LDF+stigmasterol. Error bars indicate standard deviation.
Lipoproteins of larvae fed with YF contain an even smaller proportion of ergosterol than the gut, whereas the proportion of zymosterol increases even more dramatically (Figure 6B). No further changes in sterol profiles are observed in membrane lipids of peripheral tissues, which generally resemble that of lipoproteins (Figure 6B). Thus, not only does the gut accumulate sterols with different efficiencies, it also prefers to mobilize specific sterols onto lipoproteins for delivery to peripheral tissues.
PF contains a different complement of sterols; sitosterol, comprising about 60%, is the most abundant (Figure 6C). No alteration in the proportion of different plant sterols is observed in the gut or in lipoproteins (Figure 6D), indicating that these plant sterols are absorbed and mobilized with comparable efficiencies. Interestingly, however, these sterols show different tissue-specific enrichments. Although campesterol is present in circulating lipoproteins, it is undetectable in the fat body (Figure 6D). In contrast, campesterol is relatively enriched in salivary gland, wing disk and brain, compared with its abundance in lipoproteins (Figure 6D). This cannot reflect maternally contributed campesterol, because animals fed with either stigmasterol or ergosterol alone do not accumulate campesterol in these tissues (Figure 6F and H). Taken together, these data suggest that Drosophila larvae can specifically control the uptake, mobilization and tissue accumulation of different sterols.
Strikingly, animals fed on LDF containing single sterols show dramatic differences in total sterol accumulation in all tissues except the gut. Lipoproteins from ergosterol-fed larvae have five-fold less sterol as a fraction of total polar lipids than those from stigmasterol-fed larvae (Figure 6F and H). Furthermore, different tissues of ergosterol-fed larvae accumulate between 1/3 and 1/2 the amount of sterol as animals fed stigmasterol (Figure 6F and H). Since ergosterol fulfills the bulk membrane sterol requirement in Drosophila (Carvalho et al, 2010), this suggests that the sterol structure may influence the amount of sterol required for membrane homeostasis.
Developmental changes in lipid composition
Growth and development of Drosophila take place over ∼10 days when larvae are raised under standard laboratory conditions. After hatching, larvae grow rapidly and undergo two molts. Larval tissues grow by becoming polyploid, whereas the imaginal tissues, which will give rise to the adult, remain diploid as they proliferate. When larvae have reached a threshold size, the critical weight for pupariation, they stop feeding and begin to wander (Beadle et al, 1938; Nijhout and Williams, 1974). Although they do not feed during this time, the imaginal tissues continue to grow (Nijhout and Grunert, 2010). In response to hormonal signals, the animals then pupariate (Mirth and Riddiford, 2007). Larval tissues undergo histolysis (Jiang et al, 1997; Rusten et al, 2004), and the imaginal tissues develop their final adult shape over the course of several days (Aldaz et al, 2010). The development of Drosophila therefore comprises a phase of feeding and rapid growth, followed by a prolonged period of tissue remodeling in the absence of additional nutrients. We wondered how changes in growth and metabolism are reflected in lipid composition. To address this question, we performed timed collections of larvae and pupae raised on YF and determined their lipid composition over the course of development (Supplementary Information, Dataset 3). Figures 7 and 8 show the amount of each lipid class (in nmoles/animal) at each developmental stage. We also calculated the sum of all membrane lipids (including phospholipids, sphingolipids and sterols) at each stage and plotted the total content of membrane lipids per organism over time (Figure 7A).
Figure 8.
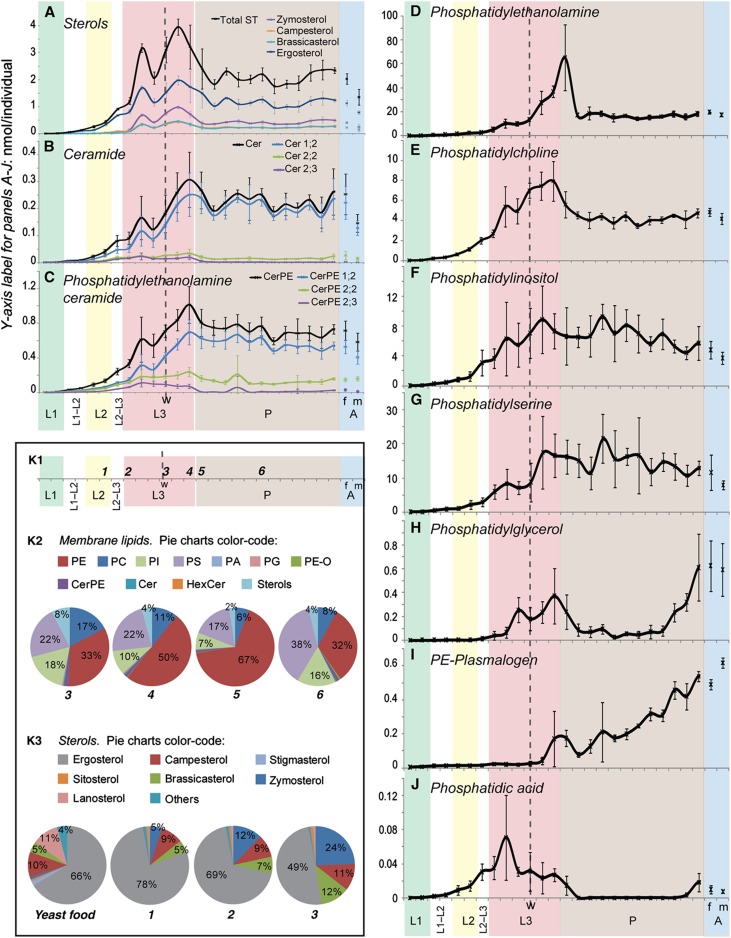
Changes in the amounts of different lipid classes during development. (A–J) Amounts (nmol per individual) of different polar lipid classes (indicated) at different developmental stages from hatching to adulthood (time interval=8 h). L1=first instar, shaded green; L2=second instar, shaded yellow; L3=third instar, shaded red; P=pupae, shaded brown; A=adults, shaded blue, where m=males and f=females. The dashed line ‘w’ indicates the start of the wandering larval stage. Error bars indicate standard deviation. (K) Membrane lipid composition at selected developmental time points. (K1) The developmental time points 1–6 that are represented in the pie charts of (K2) and (K3). (K2) Membrane lipids, and highlights the dramatic changes in PE relative to the other membrane lipids that occur during and after pupariation (time points 3–6). (K3) The proportions of different sterol species in YF, and in larvae at time points 1, 2 and 3. It highlights changes in ergosterol and zymosterol accumulation during larval stages.
Changes in membrane lipids and TAG during larval development
The total content of membrane lipids increases during larval growth, with the exception of a clear pause that occurs in the third instar just prior to the time when larvae stop feeding and start to wander. After the larvae start to wander, the amount of membrane lipid per larva increases again, despite the fact that larvae are not feeding. As they near pupariation, the membrane lipid increase decelerates (Figure 7A).
Like membrane lipids, TAGs increase as larvae grow, but do so with different dynamics (Figure 7C). For example, during the pause in membrane lipid accumulation that occurs prior to wandering, larvae keep accumulating TAG and their neutral to polar lipid ratio increases from 0.26 to 0.46 (Figure 7A–C). This suggests that a metabolic shift occurs prior to wandering causing larvae to favor lipid storage over new membrane synthesis and tissue growth. During wandering, as total membrane lipid levels increase, the amount of TAG is not reduced, despite the fact that these animals do not feed (Figure 7A and C). This indicates that the fatty acids stored in TAG at earlier stages are not being depleted for the new phospholipid synthesis that occurs during wandering. Furthermore, there is no net consumption of TAG to support energy production at this time.
Changes in membrane lipids and TAG during pupal development
After pupariation, as larval tissues undergo histolysis, the amount of membrane lipid drops by about half, then remains roughly constant for the rest of pupal life (Figure 7A). At the same time, TAG levels almost double (Figure 7C). This suggests that fatty acids liberated from phospholipids during histolysis may be stored in TAG—consistent with this, no significant changes in total fatty acids are observed at this time (Figure 7E). Furthermore, this transition correlates with a transient increase in long-chain DAG, an intermediate in the conversion of phospholipid to TAG (Figure 7D).
Although all phospholipids eventually decrease during the larval-to-pupal transition, they do so with different dynamics. All phospholipids except PE start to decrease slightly before pupariation. As the levels of these phospholipids drop, PE transiently increases before dropping just after pupariation (Figure 8D–J and K2). During this time, total fatty acids do not change significantly (Figure 7E). This suggests that phospholipids may be converted to PE before being incorporated into TAG.
After the increase in TAG that occurs during tissue histolysis, TAG levels remain high throughout the rest of pupal development (Figure 7B). The observation that TAG stores are not strongly depleted during pupal life indicates that β-oxidation of fatty acids derived from TAG is not a significant source of energy for pupal metabolism.
Abundance of specific sterols changes during larval development
As noted above, YF contains a mixture of sterols—ergosterol being the most abundant (66%) (Figure 8K3, pie chart YF). The most abundant sterol in early larvae is ergosterol, which comprises 78% of total sterols—a higher percentage than is found in the food (Figure 8A K3, pie chart 1). However, at the beginning of the third instar, sterols that represent only a minor fraction in YF start to accumulate disproportionately with respect to ergosterol (Figure 8A). Ergosterol contributes only 49% of total sterols in wandering third instar larvae. In contrast, zymosterol, which is almost undetectable in food, represents 24% of larval sterols by this stage. Campesterol and brassicasterol also more modestly increase their relative representation at this stage (Figure 8K3, pie chart 3). This is consistent with the proportions observed in lipoproteins and in different tissues at the late third instar (Figure 6B), indicating that these changes do not reflect excessive accumulation of zymosterol in only one tissue. As argued in the previous section, disproportionate uptake and mobilization of minor sterol components might be responsible for changing tissue sterol profiles. Taken together, these data show that the mobilization mechanism that enriches specific sterol species is under developmental control.
Other stage-specific changes
Ether lipids increase dramatically during pupal development (Figure 8I). Since we detected ether lipids only in the brain (Figure 4A), this may reflect the rewiring and growth of the brain that takes place at this time. Although adult males are smaller than females and contain less lipid overall, they contain more ether lipids than females. Vertebrate testes are rich in ether lipids (Diagne et al, 1984; Reisse et al, 2001), and it would be interesting to investigate whether this is also the case in Drosophila.
Finally, we note that levels of the mitochondrial cardiolipin precursor, PG, decrease over 10-fold during pupal stages before increasing again to even higher levels just before adults emerge. Adult flies contain much more PG than at any larval stage (Figure 8H). This suggests that the rate of oxidative metabolism is very low throughout pupal development, consistent with the lack of utilization of TAG at this stage. It also suggests that adults have a higher rate of oxidative metabolism than larvae.
Discussion
Over the last 10 years, the capacity of mass spectrometry to identify and quantify lipid species has increased dramatically, and the power of this approach is now beginning to be applied to genetic model organisms. For example, LC—MS/MS has recently been used to profile 78 lipids from whole adult Drosophila, and to quantify the absolute abundance of 28 PE and PC species (Hammad et al, 2011). In this study, we took advantage of the recent developments in high mass resolution shotgun lipidomics (Schwudke et al, 2007, 2011; Ejsing et al, 2009), which enable direct identification of lipid species without recourse to MS/MS, to establish an unprecedented coverage of the Drosophila lipidome. With <10 min required for a single sample, we were able to systematically quantify 250 species from 14 major lipid classes. While previous analyses required lipid extracts prepared from multiple animals, the technology used in the present study is sensitive enough to allow us to quantify the lipids in individual organs of Drosophila third instar larvae—one analysis requires the equivalent of ½ of a gut or five brains of third instar larvae. The speed of this technique also made it possible to analyze large numbers of samples—for example, we quantified lipid profiles at 27 developmental time points from hatching until adulthood, and examined six different tissues of larvae fed with four different diets.
Taken together, this study established the high-resolution shotgun methodology as a valuable tool for molecular characterization of the Drosophila lipidome. Lipid abundance is expressed in absolute values (moles), rather than fold-changes; thus the data set can be expanded by including additional lipid classes or conditions. The robustness, reproducibility, simplicity and speed of shotgun lipidomics allowed us to generate a comprehensive resource in which fundamental biological questions can now be addressed regarding the role of lipids in development. Its multidimensional interpretation can potentially address a variety of specific problems far beyond those considered in this work.
Phosphoethanolamine lipids dominate the Drosophila lipidome
It has been noted previously that Drosophila membranes contain more PE than PC (Fast, 1966)—in mammalian cells, this ratio is reversed (Sampaio et al, 2011). Furthermore, Drosophila synthesizes CerPE rather than the phosphorylcholine-containing sphingomyelin found in mammalian membranes. Here, we show that this tendency to replace choline with ethanolamine extends to ether lipids as well. Furthermore, just as mammalian lipoproteins dramatically enrich the most abundant mammalian glycerophospholipid, PC (Maldonado et al, 2002), Drosophila lipoproteins contain almost exclusively PE. Interestingly, different head group dominance has evolved in yeast, which build both phospholipids and sphingolipids that contain phosphoinositols (Ejsing et al, 2009). Thus, the Drosophila lipidome appears to be PE-centric, in contrast to PC-centric mammals and PI-centric yeast.
Is there an advantage to membrane biophysical properties in having the most abundant phospholipid headgroup also present in ether lipids and sphingolipids? It would be interesting to examine whether these are compensated by differences in fatty acid chain lengths in these different organisms. Alternatively, as yet unidentified constraints in the architecture of lipid biosynthetic pathways may account for this phenomenon. Each of these organisms has evolved functional membranes based on highly divergent components, but with conserved design principles. How different combinations of lipids are selected to ensure membrane integrity and key biophysical properties will be an important topic for the future.
Tissue specificity
Our experiments revealed both quantitative and qualitative differences in lipid composition between the tissues we examined: gut, salivary gland, brain, imaginal disk, fat body and lipoproteins. Some lipids are almost exclusively present in a subset of these tissues. For example, we could detect ether phospholipids only in the brain, and hexosyl ceramides with hydroxylated fatty acid moieties only in the gut. Interestingly, these lipids are also enriched in the corresponding mammalian tissues, suggesting that their tissue-specific functions are conserved across phyla. In addition to tissue-specific lipids, we also observed quantitative differences in the proportions of lipid classes and species between tissues. For example, Drosophila brain phospholipids and neutral lipids contain longer and more unsaturated fatty acids than those of other tissues. Although no systematic comparisons of mammalian tissue lipidomes has yet been made, many investigators have noted the striking abundance of long polyunsaturated fatty acids in mammalian brain phospholipids (Salem et al, 2001). It will be interesting to see whether other quantitative differences in tissue membrane lipid composition are conserved as well.
Epithelial cells share many architectural features, and lipid-based trafficking pathways are thought to have an important function in epithelial polarization (Nelson and Yeaman, 2001). Although this might have suggested that epithelial tissues would share some lipidomic features, our data show that the lipidomes of different epithelial tissues (disks, salivary glands and gut) are no more similar to each other than to non-epithelial tissues such as brain and fat body (see Figure 1 and Supplementary Figure S2). Although changes in membrane lipid composition clearly occur during epithelial polarization (Sampaio et al, 2011), these may reflect the specific functions of a given epithelial tissue, rather than epithelial architecture in general. For example, glycosylated sphingolipids with hydroxylated fatty acids increase in abundance during MDCK cell polarization (Sampaio et al, 2011), and they are important for normal gut differentiation in Caenorhabditis elegans (Zhang et al, 2011). In Drosophila, these lipids are abundant in the gut, but not in other epithelial tissues. They may improve barrier function, which is particularly important in kidney and gut but less so in organs like the wing imaginal disk that are not exposed to harsh environments at their apical surfaces.
Dietary fatty acids directly influence membrane lipid composition
Cells should be capable of exerting exquisite control over the structures of lipids they incorporate into cell membranes by de novo lipid synthesis and interconversion. However, lipid constituents—fatty acids and a variety of headgroups—are also available from the diet. Are all lipids autonomously synthesized to precise cellular specifications, or might the abundance of different dietary lipids directly influence what is incorporated into cell membranes? Certainly, it is clear that a few fatty acids are only available from dietary sources—for example, vertebrates rely on nutritional sources of certain long unsaturated fatty acids (omega-3 and omega-6 fatty acids) (Burr and Burr, 1930; Bezard et al, 1994). These fatty acids are incorporated into cell membranes when available (Little et al, 2007), and their deficiency is associated with impaired brain function (Holman et al, 1982; Yehuda et al, 1999; Uauy and Dangour, 2006). However, the extent to which the balance of other dietary fatty acids influences membrane lipid composition in different organs was unclear (Farquhar and Ahrens, 1963; Yeo et al, 1989). Our data reveal a striking and direct effect of dietary fatty acid composition on all phospholipid classes in every organ we examined. When fed a plant diet rich in long unsaturated fatty acids, the fatty acids present in tissue phospholipids tend to be longer and more unsaturated than in animals fed with yeast, or with a lipid-depleted diet. Thus, although Drosophila can synthesize de novo all the fatty acids it needs to survive, they incorporate different fatty acids into membrane lipids if they are provided by the diet. We also observed indirect, possibly compensatory, effects on the proportion of different lipid classes in larvae fed with a plant diet. Nevertheless, the lipid composition of different tissues remains distinct on different diets. This flexibility in lipid composition is surprising, given the central role of membrane lipid composition in cellular function. It suggests that a wide range of different lipid compositions is consistent with normal tissue function. Tissues may sense and regulate important bulk properties of membranes, rather than the concentration of individual lipids.
In humans, a diet rich in saturated fatty acids and cholesterol is associated with metabolic syndrome and heart disease (Riccardi et al, 2004; Tanasescu et al, 2004; Eckel et al, 2005). The mechanisms underlying this linkage are not entirely clear; disturbed cardiac phospholipid metabolism and increased triglyceride storage have been implicated in cardiomyopathy (Borradaile and Schaffer, 2005; Wende and Abel, 2010), but it is not understood why consumption of saturated fatty acids leads to these problems. Our data suggest a novel mechanism for future exploration—might a diet high in saturated fats change membrane lipid composition throughout the body, contributing to pathogenesis?
Control of sterol accumulation
Cholesterol represents the vast majority of sterols in mammalian cells. Genetic mutations in genes involved in cholesterol biosynthesis or phytosterol efflux result in the accumulation of non-cholesterol sterols and cause a variety of pathologies (Bhattacharyya and Connor, 1974; Porter, 2002; Yu et al, 2004). Thus, mammalian cells seem to be highly adapted to the use of cholesterol. In contrast, insects are auxotrophic for sterols (Hobson, 1935; Clayton, 1964) and have relaxed the structural requirements for sterols in their membranes—many different sterols support adult development in Drosophila (Carvalho et al, 2010). Here we have shown that, despite this flexibility, Drosophila does have strong preferences for particular sterols. When presented with a mixture of different sterols, Drosophila larvae accumulate them with different efficiencies overall, and specific sterols accumulate preferentially in particular tissues. Mammals also exert control over sterol uptake at the intestine by effluxing non-cholesterol sterols back into the gut lumen via ABCG5 and ABCG8 transporters (Duan et al, 2004). The Drosophila genome encodes orthologous proteins (CG11069 and CG31121) that could contribute to this process (Tarr et al, 2009). Interestingly, Drosophila also controls the distribution of specific sterols at two additional levels: mobilization from the intestine into circulation, and tissue-specific accumulation of particular sterols. It would be interesting to know whether mammals might also have such mechanisms. Because of its sterol auxotrophy, Drosophila will be a powerful model for the study of nutritional sterol uptake and mobilization.
Drosophila fed with single sterols do not interconvert them significantly. Interestingly, individual tissues accumulate vastly different levels of total sterol depending on which sterol is available. For example, flies fed with ergosterol have three-fold less sterol in their wing imaginal disks than those fed with stigmasterol. Ergosterol-fed animals develop to adulthood, at least when supplemented with ecdysone (Carvalho et al, 2010). Thus, membranes that contain these low levels of ergosterol function normally. Model membrane studies have shown that ergosterol can increase lipid packing order at lower concentrations than stigmasterol (Bernsdorff and Winter, 2003). This may suggest that the amount of sterol in cell membranes is controlled by monitoring biophysical membrane properties, rather than sterol concentration itself. This would allow wider flexibility in the use of different dietary sterols.
Fat storage and utilization
Studying how the lipidome changes during larval and pupal development has suggested novel and testable hypotheses about metabolic shifts during these stages. For example, during the middle of the third larval instar, membrane lipid accumulation slows down while neutral lipid storage increases. This suggests that lipid metabolism shifts from the generation of new membrane to the buildup of energy stores. Interestingly, neutral lipid stores do not decrease when larvae stop feeding, or during the long phase of pupariation when feeding is impossible. Thus, fats are not used to provide energy or building materials during these periods. Consistently, PG is low during pupal stages and increases only shortly before adult emergence. Strikingly, levels of PG exactly parallel those of oxygen consumption reported in other studies (Merkey et al, 2011).
A lipidomic resource for Drosophila
This work provides a comprehensive lipidomics resource built on clearly understandable analytical values (moles of individual species, in contrast to fold-changes or arbitrary values indirectly associated with lipid abundances) and opens new perspectives for in-depth, hypothesis-driven research. Importantly, the resource is independent of the analytical platform and lends itself to further expansion once new lipidomics data become available in the same (absolute concentrations of lipid species) format. Our comparison of the lipid composition of different tissues, developmental stages and diets has suggested new hypotheses about organ function, membrane homeostasis, lipid transport and energy metabolism. This lipidomic resource provides a new framework within which to manipulate and understand these processes.
Materials and methods
Fly stocks
Oregon R flies are available from the Bloomington Stock Center. All experiments were performed at 25°C and constant humidity.
Drosophila diets
Drosophila larvae were fed on four different diets: YF, PF and on a LDF supplemented either with ergosterol or with stigmasterol. YF is based on a Bloomington Stock Center recipe http://fly.bio.indiana.edu/Fly_Work/media-recipes/germanfood.htm and contains per liter: 8.7 g agar, 80 g yeast (brewers), 20 g yeast extract, 20 g peptone, 30 g sucrose, 60 g dextrose, 0.5 g CaCl2(2)H2O, 0.5 g MgSO4(6)H2O, 6.3 ml propionic acid, 0.02% nipagen. It has 790 kcal/l, of which 66% are derived from sugar, 31% from protein and 3% from fat. 1 liter PF contains: 7 g agar, 76 g soy, 160 g cornmeal, 160 g malt, 0.4 g vegetable oil, 44 g treacle, 12.6 ml propionic acid, 2.61 g nipagen. It has 1570, kcal/l, of which 68% are derived from sugar, 26% from protein and 6% from fat. LDF: made as in Carvalho et al (2010). It has 810 kcal/l, of which 75% are derived from sugar, 25% from protein and 0% from fat. Sterols were added in a 1-mM ethanol solution to a final concentration of 6.2 μg sterol per 1 ml of LDF. Lipid compositions of the foods are shown in Figure 3 and in Supplementary Information, Dataset 1.
Feeding experiments and sample preparation
To synchronize larval development, we allowed flies to lay eggs on yeast apple juice plates for 1 h, discarded the first batch of embryos, and then collected for another 1.5 h. We placed groups of embryos into vials containing YF and PF, or single embryos into individual wells containing 1 ml of LDF supplemented with 6.2 μg of sterol. Raising these animals in isolation avoids cannibalism as an undesirable source of lipids. Embryos were prepared as in Carvalho et al (2010). Larvae were allowed to develop until the third instar, at the beginning of the wandering stage. Before tissue dissection, YF and PF-fed larvae were collected and placed on LDF for 2 h in order to empty their guts of dietary unabsorbed lipids. Larvae were kept on ice-cold ammonium bicarbonate (150 mM) while the gut, fat body, salivary gland, wing disk and brain were dissected. The hemolymph (lipoproteins) was collected in ice-cold PBS. The hemolymph protein content was measured using a BCA assay (Thermo Scientific). Larval tissues and hemolymph were fast-frozen in liquid nitrogen and stored at −80°C until lipid extraction.
Developmental experiments
Embryos were collected for 1.5 h, as described for feeding experiments, and placed on petri dishes containing YF. The animals were collected from hatching to adulthood at intervals of 8 h. During larval development, collected larvae were fed for 2 h in LDF before freezing to empty the guts of unabsorbed dietary lipids. Organisms were then washed in water, fast-frozen in liquid nitrogen and stored at −80°C until lipid extraction.
Annotation of lipid species
Glycerophospholipid, DAG and TAG species were annotated as: 〈lipid class〉 〈no. of carbons in all fatty acids〉:〈no. of double bonds in all fatty acids〉. Sphingolipid species were annotated as: 〈lipid class〉 〈no. of carbons in the long-chain base and fatty acid moieties〉:〈no. of double bonds in the long-chain base and fatty acid moieties〉; 〈no. of hydroxyl groups in the long-chain base and fatty acid moieties〉. Sterols were annotated as: 〈ST〉 〈no. of carbons additional to cholesterol〉:〈no. of double bonds〉:〈no. of hydroxyl groups additional to the hydroxyl group at position 3〉.
Lipid extraction
Samples were homogenized in 150 mM ammonium bicarbonate using a pestle attached to a cordless motor. For each replicate, per sample we used: 3 guts, 3 fat bodies, 4 pairs of salivary glands, 20 wing disks, 5 brains, 30 first instar larvae, 15 second instar larvae and 3 third instar larvae, pupae or adults. From each homogenate, we removed a volume containing ∼2 nmol of lipid material to process for extraction (this corresponded to ½ gut, ½ fat body, two pairs of salivary glands, 20 wing disks, 5 brains, 15 first instar larvae, 8 second instar larvae, 1 early third instar larvae, 1/2 of third instar larvae from wandering to pupae and 1/4 of pupae or adult, 2–5 μl hemolymph). Each series of experiments comprised three replicates derived from the progeny of three different groups of parents that were raised separately. Replicate samples were processed in parallel.
Sample volume was adjusted to 200 μl. For absolute quantification, internal standards were added to control for lipid-class dependent differences in extraction and ionization (Koivusalo et al, 2001; Ejsing et al, 2006, 2009). The internal standard mix contained PG 17:0 and TAG 36:0, 10 pmol; Cer 17:0, GalCer 12:0 and DAG 17:0, 20 pmol; PA 17:0, 25 pmol; PS 17:0 and PC 18:3, 40 pmol; PI 17:0, CerPE 12:0, Chol-D7 and PE 17:0, 50 pmol. Lipids were extracted using a modified Folch extraction protocol (Carvalho et al, 2010): 265 μl of methanol was added to the aqueous phase and vortexed; 730 μl of chloroform was added and the samples were vortexed for 1 h; after centrifugation, the organic phase was collected and dried under vacuum to avoid lipid oxidation. The whole extraction procedure including sample preparation was performed at 4°C in order to prevent lipid degradation. All lipid standards were purchased from Avanti Polar Lipids (Alabaster, Alabama). Solvents were purchased from Sigma-Aldrich (Taufkirchen, Germany).
Mass spectrometry
Mass spectrometric analyses were performed on a LTQ Orbitrap XL instrument (Thermo Fisher Scientific, Bremen) equipped with a robotic nanoflow ion source TriVersa NanoMate (Advion BioSciences, Ithaca, NY) using chips with the diameter of spraying nozzles of 4.1 μm. The ion source was controlled by Chipsoft 8.3.1 software. Ionization voltages were +1.25 kV and −0.9 kV in positive and negative modes, respectively; back pressure was set at 0.95 psi in both modes. The temperature of ion transfer capillary was 125°C; tube voltages were 90 V (MS+) and −150 V (MS−). Acquisitions were performed at the mass resolution Rm/z 400=100 000. AGC control was set at 106 ions and maximum injection time was set to 50 ms.
Dried total lipid extracts were re-dissolved in 100 μl of chloroform:methanol 1:2. For the analysis, 10 μl of samples were loaded onto 96-well plate (Eppendorf, Hamburg) of the TriVersa NanoMate ion source and sealed with aluminum foil. Each sample was analyzed for 4 min in positive ion mode where PE, PC PC-O, TAG, CerPE and DAG were detected and quantified. This was followed by an acquisition in negative ion mode for 5 min where PA, PI, PS, PG, PE, PEO-, Cer, HexCer were detected and quantified. Finally, after derivatization (see below for details), the sample was analyzed in negative ion mode for sterol detection and quantification.
Sterol quantification method was adapted from Sandhoff et al (1999). Briefly, dried samples were sulfated with sulfur trioxide pyridine complex in pyridine (Sigma-Aldrich, Munich, Germany), sonicated and incubated at room temperature. Then barium acetate (Sigma-Aldrich) was added, samples sonicated and incubated 10 min at room temperature and then 1 hour at 4°C. Sulfated sterols were quantified in MS mode on a LTQ Orbitrap XL mass spectrometer using cholesterol-D7 (Avanti Polar Lipids) as internal standard. Since signal response varies between different sterols (Sandhoff et al, 1999), correction factors determined in separate experiments with available sterol standards were applied for the groups of structurally related molecules (Supplementary Figure S9): 0.75 for stigmasterol, campesterol, sitosterol and brassicasterol; and 0.61 for ergosterol and stigmasta-5,7,22-trienol. For low abundant lanosterol and zytosterol, no correction factor was applied because we lack the appropriate structurally related standards.
Lipids were identified by LipidXplorer software (Herzog et al, 2011) by matching m/z of their monoisotopic peaks to the corresponding elemental composition constraints. Molecular Fragmentation Query Language (MFQL) queries compiled for all the aforementioned lipid classes are available at the LipidXplorer wiki site: https://wiki.mpi-cbg.de/wiki/lipidx/index.php/Main_Page. Mass tolerance was 5 p.p.m. and intensity threshold was set according to the noise level reported by Xcalibur software (Thermo Scientific).
The mass spectrometry data associated with this publication may be downloaded from ProteomeCommons.org/tranche/ using the following hash: 8ic85vEi++Xsn9nEWzWklQGrdqsKM4Hpoe7bne0yS6D0FpdTCC6uuFOdpg0WGTlfZdN9dT32CqOBlcCKeNCGJ49U16wAAAAAAANtZQ==
Supplementary Material
Supplementary Figs S1-9, Supplementary table S1
Datasets 1–3
Acknowledgments
Work in the AS laboratory was supported by TRR 83 Grant from Deutsche Forschungsgemeinschaft (DFG) and Virtual Liver Network Grant (Code/0315757) from Bundesministerium f. Bildung u. Forschung (BMBF). Work in the SE laboratory was supported by a grant through the European Science Foundation Euromembranes network (EA4/5-1), and by a grant from the Deutsche Forschugsgemeinschaft (EA4/2–4). We thank Kai Simons, Teymuras Kurzchalia (MPI CBG, Dresden) and Vijay Raghavan (NCBS, Bangalore) for helpful advice and critical comments on the manuscript.
Author contributions: MC and JLS planned and performed the experiments and collected the majority of the data, with some contribution by WP. MB developed the PF recipe. AS and SE planned the experiments. MC, JLS, WP, SE and AS analyzed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adachi-Yamada T, Gotoh T, Sugimura I, Tateno M, Nishida Y, Onuki T, Date H (1999) De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs. Mol Cell Biol 19: 7276–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaz S, Escudero LM, Freeman M (2010) Live imaging of Drosophila imaginal disc development. Proc Natl Acad Sci USA 107: 14217–14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KD, Thummel CS (2007) Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab 6: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle GW, Tatum EL, Clancy CW (1938) Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biol Bull 75: 447–462 [Google Scholar]
- Bernsdorff C, Winter R (2003) Differential properties of the sterols cholesterol, ergosterol, b-sitosterol, trans-7-dehydrocholesterol, stigmasterol and lanosterol on DPPC bilayer order. J Phys Chem B 107: 10658–10664 [Google Scholar]
- Bezard J, Blond JP, Bernard A, Clouet P (1994) The metabolism and availability of essential fatty acids in animal and human tissues. Reprod Nutr Dev 34: 539–568 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya AK, Connor WE (1974) Beta-sitosterolemia and xanthomatosis. A newly described lipid storage disease in two sisters. J Clin Invest 53: 1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, Ocorr K, Bodmer R, Oldham S (2010) High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab 12: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanksby SJ, Mitchell TW (2010) Advances in mass spectrometry for lipidomics. Annu Rev Anal Chem 3: 433–465 [DOI] [PubMed] [Google Scholar]
- Borradaile NM, Schaffer JE (2005) Lipotoxicity in the heart. Curr Hypertens Rep 7: 412–417 [DOI] [PubMed] [Google Scholar]
- Burr GO, Burr MM (1930) On the nature and role of the fatty acids essential in nutrition. J Biol Chem 86: 587–621 [Google Scholar]
- Carvalho M, Schwudke D, Sampaio JL, Palm W, Riezman I, Dey G, Gupta GD, Mayor S, Riezman H, Shevchenko A, Kurzchalia TV, Eaton S (2010) Survival strategies of a sterol auxotroph. Development 137: 3675–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR (2008) Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol 82: 11228–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton RB (1964) The Utilization of sterols by insects. J Lipid Res 15: 3–19 [PubMed] [Google Scholar]
- Dennis EA (2009) Lipidomics joins the omics evolution. Proc Natl Acad Sci USA 106: 2089–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagne A, Fauvel J, Record M, Chap H, Douste-Blazy L (1984) Studies on ether phospholipids. II. Comparative composition of various tissues from human, rat and guinea pig. Biochim Biophys Acta 793: 221–231 [DOI] [PubMed] [Google Scholar]
- Duan LP, Wang HH, Wang DQ (2004) Cholesterol absorption is mainly regulated by the jejunal and ileal ATP-binding cassette sterol efflux transporters Abcg5 and Abcg8 in mice. J Lipid Res 45: 1312–1323 [DOI] [PubMed] [Google Scholar]
- Eckel RH, Grundy SM, Zimmet PZ (2005) The metabolic syndrome. Lancet 365: 1415–1428 [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Moehring T, Bahr U, Duchoslav E, Karas M, Simons K, Shevchenko A (2006) Collision-induced dissociation pathways of yeast sphingolipids and their molecular profiling in total lipid extracts: a study by quadrupole TOF and linear ion trap-orbitrap mass spectrometry. J Mass Spectrom 41: 372–389 [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA 106: 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui AA, Horrocks LA (2001) Plasmalogens: workhorse lipids of membranes in normal and injured neurons and glia. Neuroscientist 7: 232–245 [DOI] [PubMed] [Google Scholar]
- Farquhar JW, Ahrens EH Jr. (1963) Effects of dietary fats on human erythrocyte fatty acid patterns. J Clin Invest 42: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast PG (1966) A comparative study of the phospholipids and fatty acids of some insects. Lipids 1: 209–215 [DOI] [PubMed] [Google Scholar]
- Gerl MJ, Sampaio JL, Urbano S, Kaldova L, Verbavatz JM, Binnington B, Lindemann D, Lingwood CA, Shevchenko A, Schroeder C, Simons K (2012) Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol 192: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP (2005) Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab 1: 323–330 [DOI] [PubMed] [Google Scholar]
- Gross RW, Han X (2011) Lipidomics at the interface of structure and function in systems biology. Chem Biol 18: 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV (2008) Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453: 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H (2009) Fatty acid 2-Hydroxylation in mammalian sphingolipid biology. Biochim Biophys Acta 1801: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad LA, Cooper BS, Fisher NP, Montooth KL, Karty JA (2011) Profiling and quantification of Drosophila melanogaster lipids using liquid chromatography/mass spectrometry. Rapid Commun Mass Spectrom 25: 2959–2968 [DOI] [PubMed] [Google Scholar]
- Han X, Yang K, Gross RW (2012) Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev 31: 134–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkewicz R, Dennis EA (2011) Applications of mass spectrometry to lipids and membranes. Annu Rev Biochem 80: 301–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr DR, Fyrst H, Phan V, Heinecke K, Georges R, Harris GL, Saba JD (2003) Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development 130: 2443–2453 [DOI] [PubMed] [Google Scholar]
- Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A (2011) A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol 12: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson RP (1935) On a fat-soluble growth factor required by blow-fly larvae: identity of the growth factor with cholesterol. Biochem J 29: 2023–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman RT, Johnson SB, Hatch TF (1982) A case of human linolenic acid deficiency involving neurological abnormalities. Am J Clin Nutr 35: 617–623 [DOI] [PubMed] [Google Scholar]
- Huang X, Suyama K, Buchanan J, Zhu AJ, Scott MP (2005) A Drosophila model of the Niemann-Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development 132: 5115–5124 [DOI] [PubMed] [Google Scholar]
- Iqbal J, Hussain MM (2009) Intestinal lipid absorption. Am J Physiol Endocrinol Metab 296: E1183–E1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Baehrecke EH, Thummel CS (1997) Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683 [DOI] [PubMed] [Google Scholar]
- Kalvodova L, Sampaio JL, Cordo S, Ejsing CS, Shevchenko A, Simons K (2009) The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J Virol 83: 7996–8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P (2001) Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J Lipid Res 42: 663–672 [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB (2006) Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab 3: 439–448 [DOI] [PubMed] [Google Scholar]
- Leopold P, Perrimon N (2007) Drosophila and the genetics of the internal milieu. Nature 450: 186–188 [DOI] [PubMed] [Google Scholar]
- Liebisch G, Binder M, Schifferer R, Langmann T, Schulz B, Schmitz G (2006) High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim Biophys Acta 1761: 121–128 [DOI] [PubMed] [Google Scholar]
- Little SJ, Lynch MA, Manku M, Nicolaou A (2007) Docosahexaenoic acid-induced changes in phospholipids in cortex of young and aged rats: a lipidomic analysis. Prostaglandins Leukot Essent Fatty Acids 77: 155–162 [DOI] [PubMed] [Google Scholar]
- Lofgren H, Pascher I (1977) Molecular arrangements of sphingolipids. The monolayer behaviour of ceramides. Chem Phys Lipids 20: 273–284 [DOI] [PubMed] [Google Scholar]
- Maldonado EN, Alderson NL, Monje PV, Wood PM, Hama H (2008) FA2H is responsible for the formation of 2-hydroxy galactolipids in peripheral nervous system myelin. J Lipid Res 49: 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado EN, Casanave EB, Aveldano MI (2002) Major plasma lipids and fatty acids in four HDL mammals. Comp Biochem Physiol A Mol Integr Physiol 132: 297–303 [DOI] [PubMed] [Google Scholar]
- Merkey AB, Wong CK, Hoshizaki DK, Gibbs AG (2011) Energetics of metamorphosis in Drosophila melanogaster. J Insect Physiol 57: 1437–1445 [DOI] [PubMed] [Google Scholar]
- Mirth CK, Riddiford LM (2007) Size assessment and growth control: how adult size is determined in insects. Bioessays 29: 344–355 [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Yeaman C (2001) Protein trafficking in the exocytic pathway of polarized epithelial cells. Trends Cell Biol 11: 483–486 [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Grunert LW (2010) The cellular and physiological mechanism of wing-body scaling in Manduca sexta. Science 330: 1693–1695 [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Williams CM (1974) Control of moulting and metamorphosis in the tobacco hornworm, Manduca sexta (L.): growth of the last-instar larva and the decision to pupate. J Exp Biol 61: 481–491 [DOI] [PubMed] [Google Scholar]
- Niwa R, Niwa YS (2011) The Fruit Fly Drosophila melanogaster as a Model System to Study Cholesterol Metabolism and Homeostasis. Cholesterol 2011: 176802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, Eaton S (2012) Lipoproteins in Drosophila melanogaster—assembly, function, and influence on tissue lipid composition. PLoS Genet 8: e1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panakova D, Sprong H, Marois E, Thiele C, Eaton S (2005) Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 435: 58–65 [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Ramaswami M, Tanouye MA (1994) The Drosophila easily shocked gene: a mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell 79: 23–33 [DOI] [PubMed] [Google Scholar]
- Porter FD (2002) Malformation syndromes due to inborn errors of cholesterol synthesis. J Clin Invest 110: 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisse S, Rothardt G, Volkl A, Beier K (2001) Peroxisomes and ether lipid biosynthesis in rat testis and epididymis. Biol Reprod 64: 1689–1694 [DOI] [PubMed] [Google Scholar]
- Riccardi G, Giacco R, Rivellese AA (2004) Dietary fat, insulin sensitivity and the metabolic syndrome. Clin Nutr 23: 447–456 [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H (2004) Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell 7: 179–192 [DOI] [PubMed] [Google Scholar]
- Salem N Jr., Litman B, Kim HY, Gawrisch K (2001) Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids 36: 945–959 [DOI] [PubMed] [Google Scholar]
- Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A (2011) Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci USA 108: 1903–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhoff R, Brugger B, Jeckel D, Lehmann WD, Wieland FT (1999) Determination of cholesterol at the low picomole level by nano-electrospray ionization tandem mass spectrometry. J Lipid Res 40: 126–132 [PubMed] [Google Scholar]
- Schuhmann K, Almeida R, Baumert M, Herzog R, Bornstein SR, Shevchenko A (2012) Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J Mass Spectrom 47: 96–104 [DOI] [PubMed] [Google Scholar]
- Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A (2007) Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem 79: 4083–4093 [DOI] [PubMed] [Google Scholar]
- Schwudke D, Schuhmann K, Herzog R, Bornstein SR, Shevchenko A (2011) Shotgun lipidomics on high resolution mass spectrometers. Cold Spring Harb Perspect Biol 3: a004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Simons K (2010) Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol 11: 593–598 [DOI] [PubMed] [Google Scholar]
- Souza CM, Schwabe TM, Pichler H, Ploier B, Leitner E, Guan XL, Wenk MR, Riezman I, Riezman H (2011) A stable yeast strain efficiently producing cholesterol instead of ergosterol is functional for tryptophan uptake, but not weak organic acid resistance. Metab Eng 13: 555–569 [DOI] [PubMed] [Google Scholar]
- Tanasescu M, Cho E, Manson JE, Hu FB (2004) Dietary fat and cholesterol and the risk of cardiovascular disease among women with type 2 diabetes. Am J Clin Nutr 79: 999–1005 [DOI] [PubMed] [Google Scholar]
- Tarr PT, Tarling EJ, Bojanic DD, Edwards PA, Baldan A (2009) Emerging new paradigms for ABCG transporters. Biochim Biophys Acta 1791: 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschape JA, Hammerschmied C, Muhlig-Versen M, Athenstaedt K, Daum G, Kretzschmar D (2002) The neurodegeneration mutant lochrig interferes with cholesterol homeostasis and Appl processing. EMBO J 21: 6367–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy R, Dangour AD (2006) Nutrition in brain development and aging: role of essential fatty acids. Nutr Rev 64: S24–S33; discussion S72-S91 [DOI] [PubMed] [Google Scholar]
- Uchida Y, Hama H, Alderson NL, Douangpanya S, Wang Y, Crumrine DA, Elias PM, Holleran WM (2007) Fatty acid 2-hydroxylase, encoded by FA2H, accounts for differentiation-associated increase in 2-OH ceramides during keratinocyte differentiation. J Biol Chem 282: 13211–13219 [DOI] [PubMed] [Google Scholar]
- van Meer G (2005) Cellular lipidomics. EMBO J 24: 3159–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende AR, Abel ED (2010) Lipotoxicity in the heart. Biochim Biophys Acta 1801: 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR (2010) Lipidomics: new tools and applications. Cell 143: 888–895 [DOI] [PubMed] [Google Scholar]
- Yehuda S, Rabinovitz S, Mostofsky DI (1999) Essential fatty acids are mediators of brain biochemistry and cognitive functions. J Neurosci Res 56: 565–570 [DOI] [PubMed] [Google Scholar]
- Yeo YK, Philbrick DJ, Holub BJ (1989) Altered acyl chain compositions of alkylacyl, alkenylacyl, and diacyl subclasses of choline and ethanolamine glycerophospholipids in rat heart by dietary fish oil. Biochim Biophys Acta 1001: 25–30 [DOI] [PubMed] [Google Scholar]
- Yetukuri L, Ekroos K, Vidal-Puig A, Oresic M (2008) Informatics and computational strategies for the study of lipids. Mol Biosyst 4: 121–127 [DOI] [PubMed] [Google Scholar]
- Yu L, von Bergmann K, Lutjohann D, Hobbs HH, Cohen JC (2004) Selective sterol accumulation in ABCG5/ABCG8-deficient mice. J Lipid Res 45: 301–307 [DOI] [PubMed] [Google Scholar]
- Zeisel SH (1981) Dietary choline: biochemistry, physiology, and pharmacology. Annu Rev Nutr 1: 95–121 [DOI] [PubMed] [Google Scholar]
- Zhang H, Abraham N, Khan LA, Hall DH, Fleming JT, Göbel V (2011) Apicobasal domain identities of expanding tubular membranes depend on glycosphingolipid biosynthesis. Nat Cell Biol 13: 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs S1-9, Supplementary table S1
Datasets 1–3