Abstract
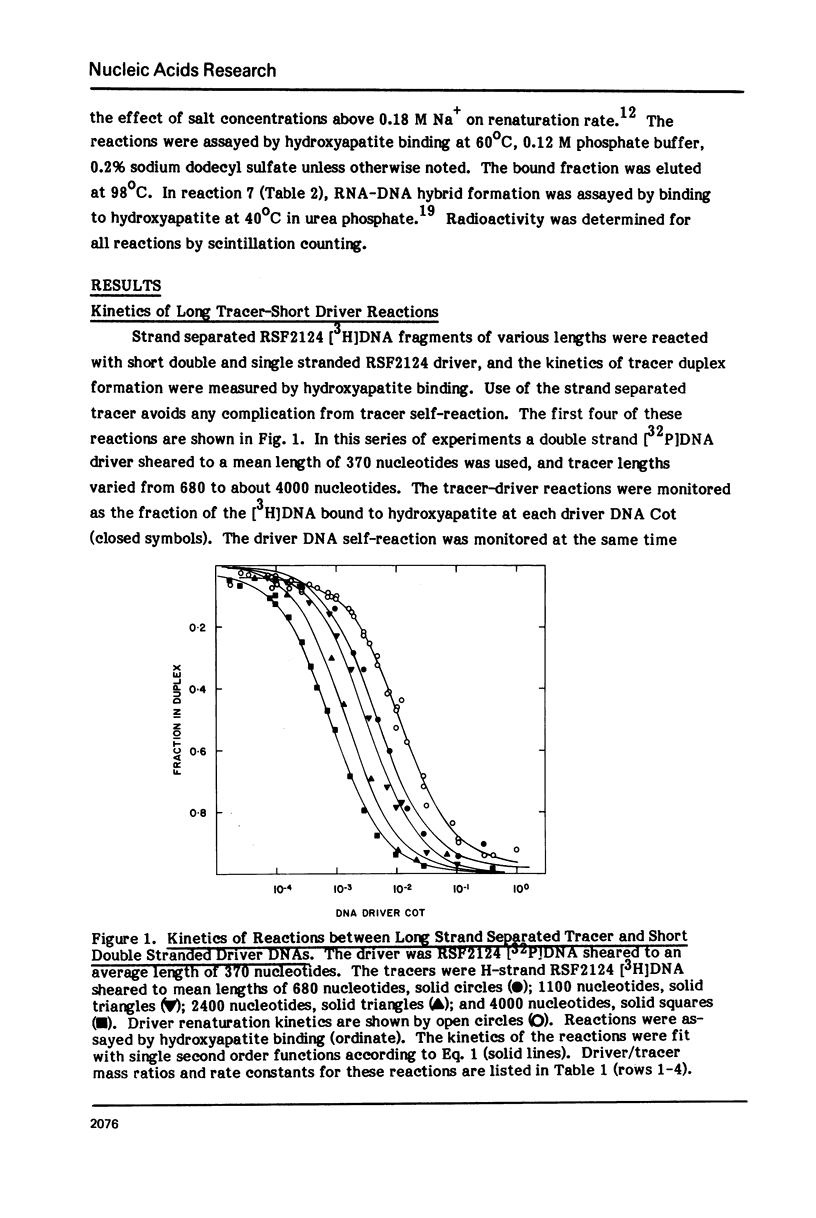
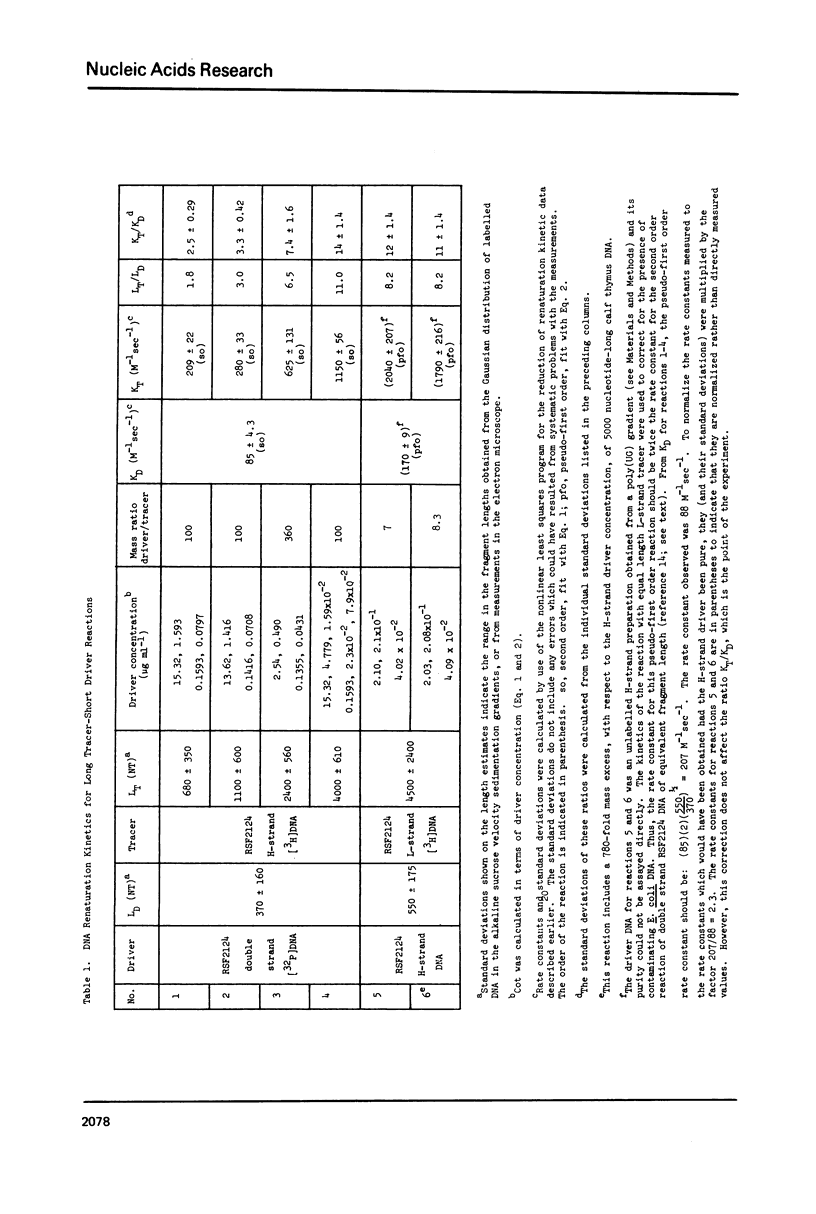
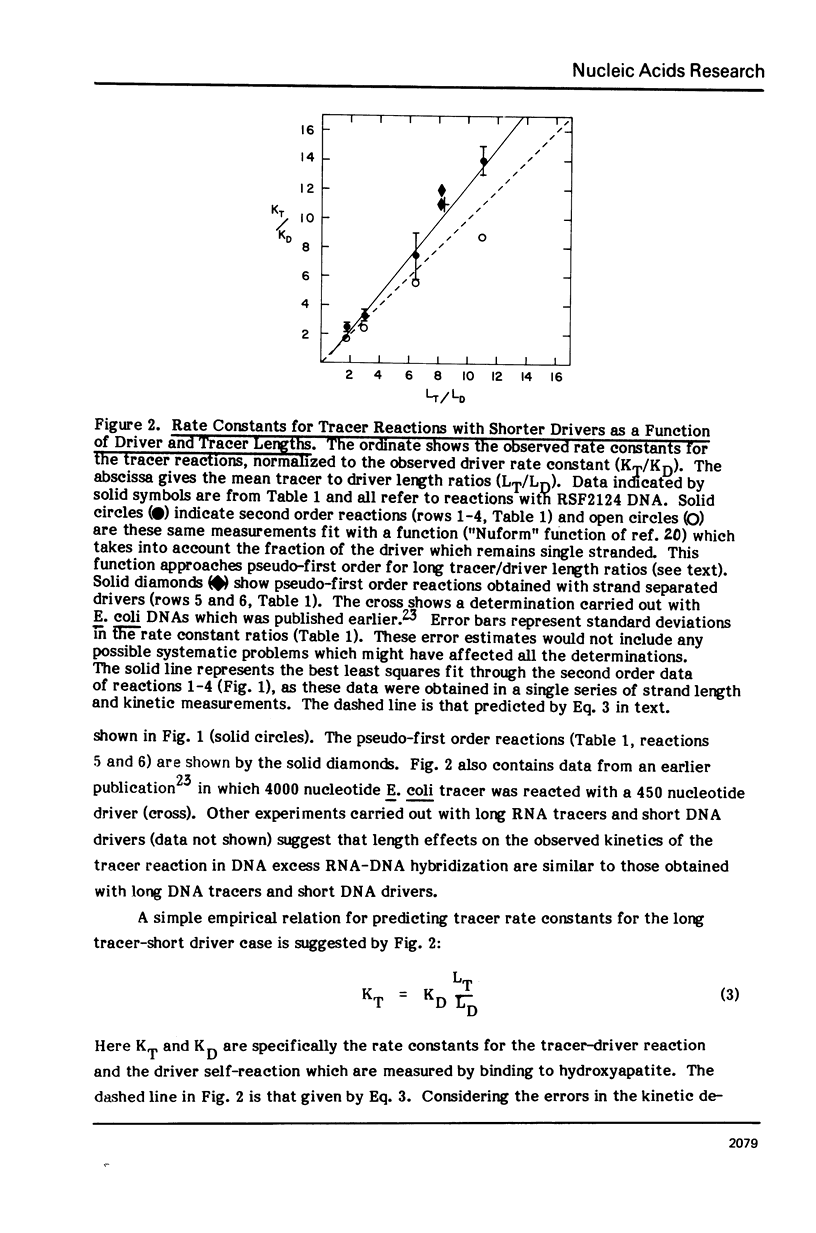
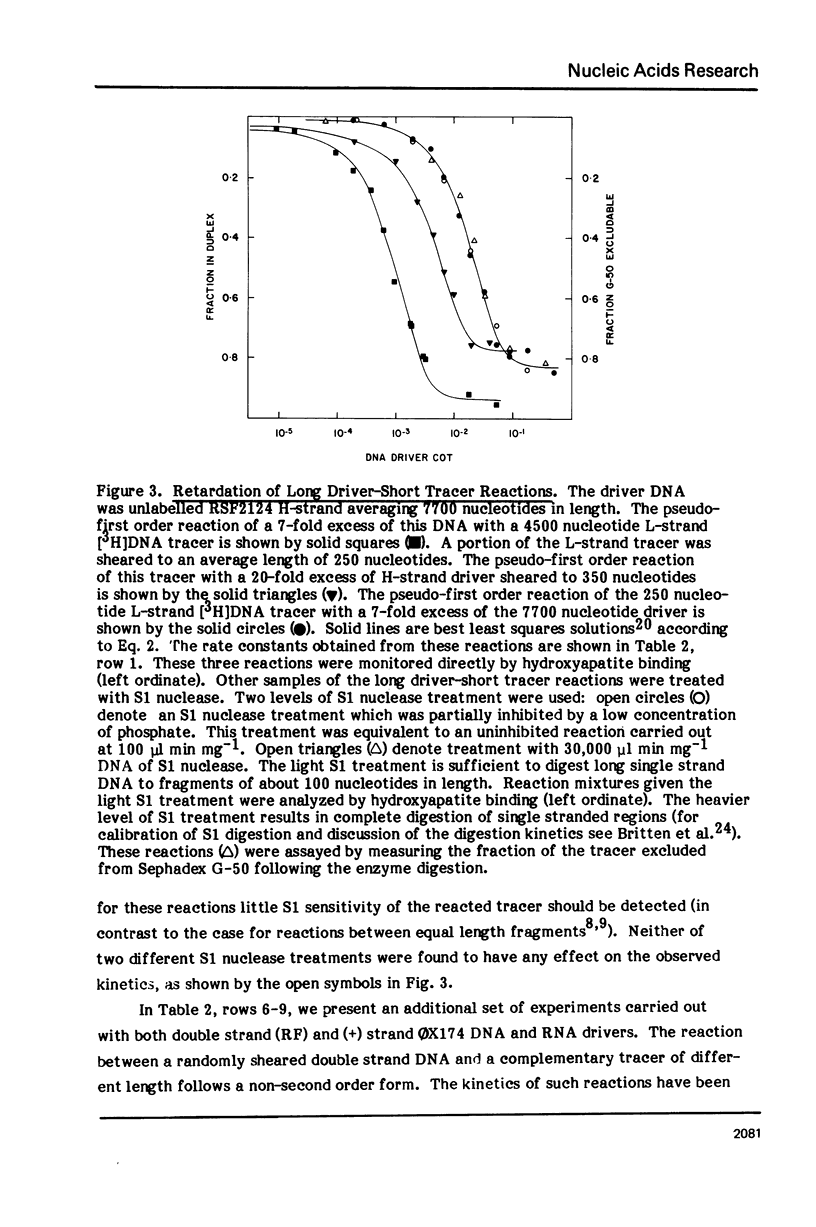
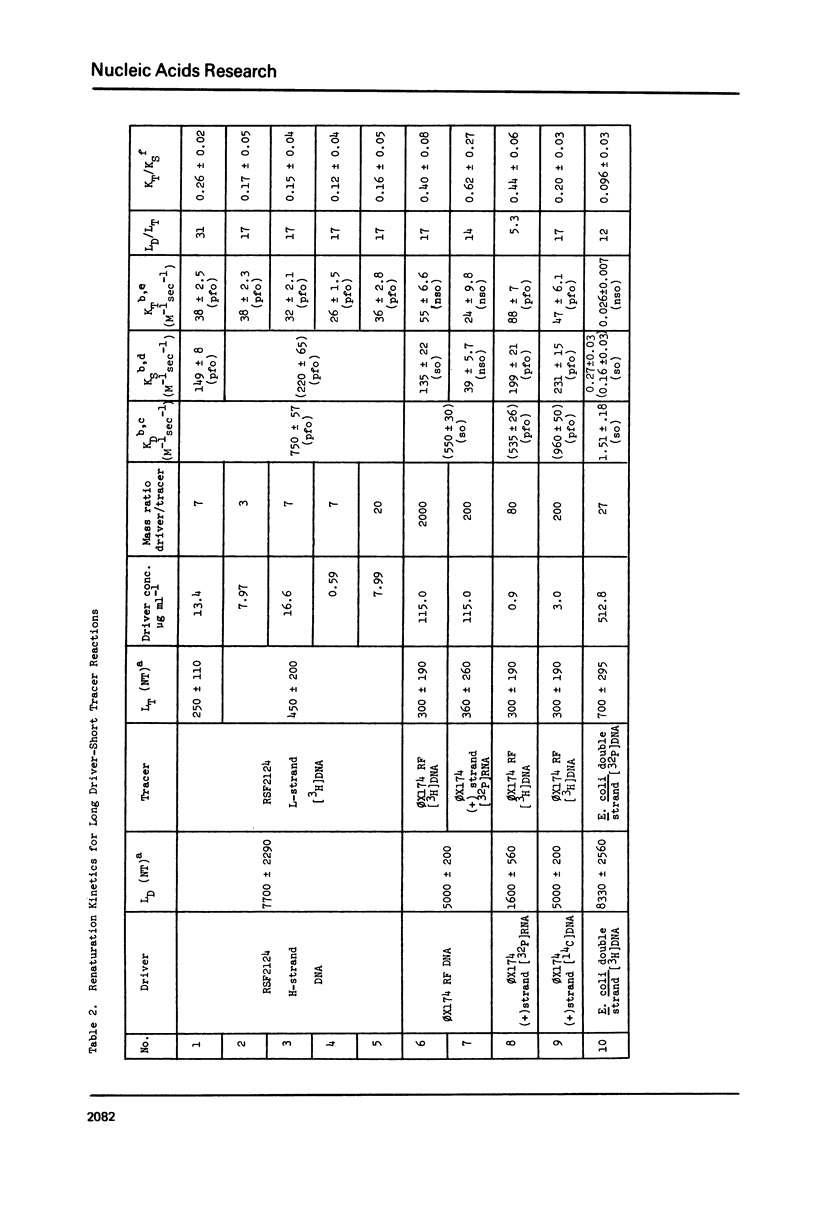
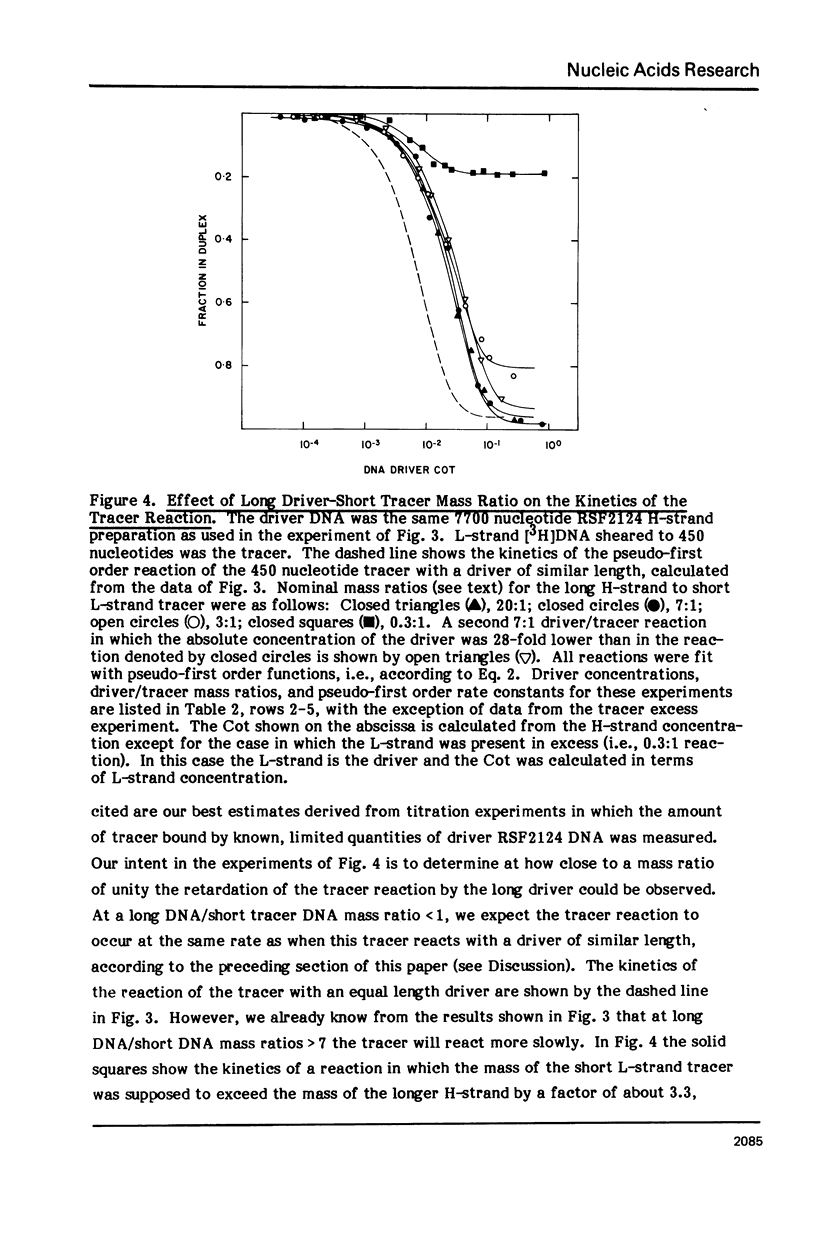
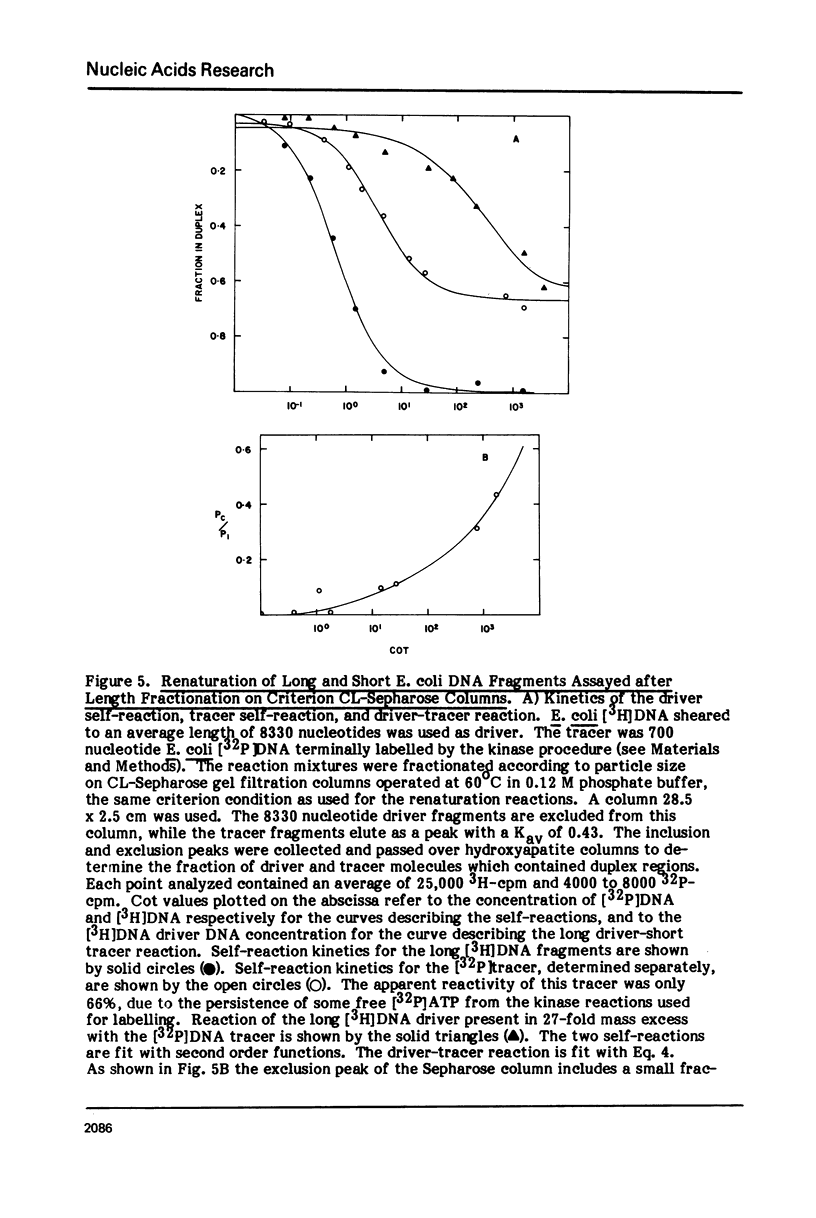
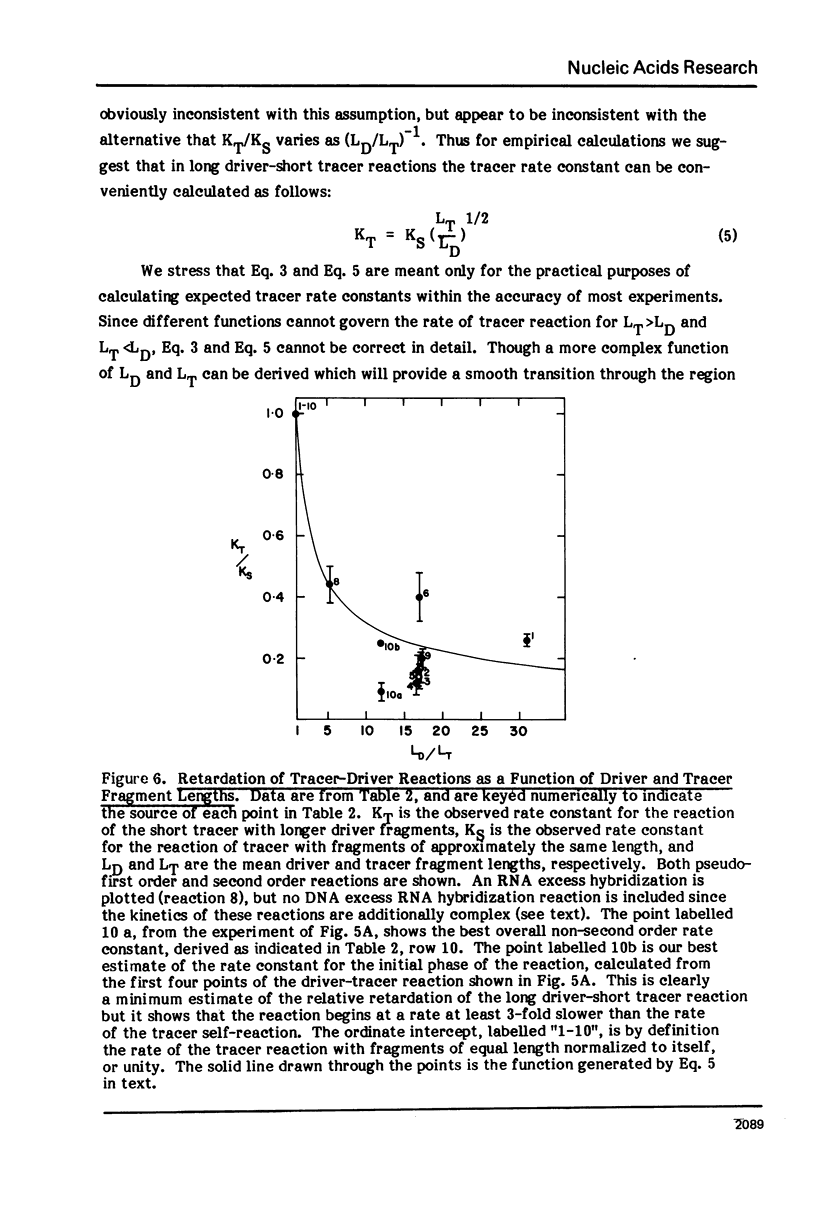
Measurements are described of the kinetics of nucleic acid strand pair reassociation where the complementary strands are of different lengths and are present in different concentrations. Rate constants for the reaction of labelled fragments ("tracer") with excess complementary strands ("driver") were determined, both for driver fragment length greater than tracer fragment length and for the reverse case. Second order reactions and pseudo-first order reactions utilizing strand separated drivers and tracers were studied. The nucleic acids which served for this investigation were phiX174 DNA and RNA, plasmid RSF2124 DNA and E. coli DNA. Approximate empirical expressions relating driver and tracer fragment lengths with the observed rate constants were obtained for practical use. In long tracer-short driver reactions the observed rate constant for the tracer reaction increases proportionately with tracer length. In long driver-short tracer reactions the rate of tracer reaction is retarded. The latter result is unexpected and appears to represent a departure from standard interpretations of the renaturation reaction.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrell B. G., Air G. M., Hutchison C. A., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976 Nov 4;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Bishop J. O., Beckmann J. S., Campo M. S., Hastie N. D., Izquierdo M., Perlman S. DNA-RNA hybridization. Philos Trans R Soc Lond B Biol Sci. 1975 Nov 6;272(915):147–157. doi: 10.1098/rstb.1975.0077. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: empirical equations describing DNA reassociation. Proc Natl Acad Sci U S A. 1976 Feb;73(2):415–419. doi: 10.1073/pnas.73.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Eden F. C., Painchaud D. M., Davidson E. H. Evolutionary divergence and length of repetitive sequences in sea urchin DNA. J Mol Evol. 1976 Dec 31;9(1):1–23. doi: 10.1007/BF01796119. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. E., Britten R. J., Davidson E. H. Sequence organization in Xenopus DNA studied by the electron microscope. J Mol Biol. 1975 Aug 5;96(2):317–333. doi: 10.1016/0022-2836(75)90351-4. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Klein W. H., Britten R. J. Structural genes adjacent to interspersed repetitive DNA sequences. Cell. 1975 Mar;4(3):217–238. doi: 10.1016/0092-8674(75)90170-1. [DOI] [PubMed] [Google Scholar]
- Galau G. A., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: rate of hybridization of excess RNA with DNA, compared to the rate of DNA renaturation. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1020–1023. doi: 10.1073/pnas.74.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galau G. A., Smith M. J., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: retarded rate of hybridization of RNA with excess DNA. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2306–2310. doi: 10.1073/pnas.74.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Renaturation of bacteriophage phiX174 DNA-RNA hybrid: RNA length effect and nucleation rate constant. J Mol Biol. 1973 Jul 15;77(4):495–500. doi: 10.1016/0022-2836(73)90218-0. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Davidson E. H., Britten R. J. A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 1977 Jun;4(6):1727–1737. doi: 10.1093/nar/4.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller R. H., Thomas T. L., Lee A. S., Klein W. H., Niles W. D., Britten R. J., Davidson E. H. Clones of individual repetitive sequences from sea urchin DNA constructed with synthetic Eco RI sites. Science. 1977 Apr 8;196(4286):197–200. doi: 10.1126/science.847467. [DOI] [PubMed] [Google Scholar]
- Smith M. J., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: reactivity of single-stranded tails in DNA-DNA renaturation. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4805–4809. doi: 10.1073/pnas.72.12.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. J., Hough B. R., Chamberlin M. E., Davidson E. H. Repetitive and non-repetitive sequence in sea urchin heterogeneous nuclear RNA. J Mol Biol. 1974 May 5;85(1):103–126. doi: 10.1016/0022-2836(74)90132-6. [DOI] [PubMed] [Google Scholar]
- So M., Gill R., Falkow S. The generation of a ColE1-Apr cloning vehicle which allows detection of inserted DNA. Mol Gen Genet. 1975 Dec 30;142(3):239–249. doi: 10.1007/BF00425649. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Excluded volume effects on the rate of renaturation of DNA. Biopolymers. 1971;10(4):601–613. doi: 10.1002/bip.360100402. [DOI] [PubMed] [Google Scholar]



