Abstract
Intraerythrocytic Babesia-like trophozoites were seen in postmortem kidney sections of a free-roaming cat in Hong Kong. DNA sequences of the 18S rRNA and mitochondrial cytochrome b genes had only 96.7% and 90.4% nucleotide identity with known Babesia sequences. We propose that this new species be named Babesia hongkongensis.
TEXT
Babesiosis is the commonest vectorborne canine infection in Hong Kong, with 48% and 33% of the stray and pet dogs being infected, respectively (14). Human babesiosis is usually caused by Babesia microti, B. divergens, and some newly described strains such as the WA1, EU1, -2, and -3, CA1, -2, -3, and -4, and KO1 types (9). We incidentally observed Babesia-like organisms in the erythrocytes in feline kidney sections during a previous study (15).
Postmortem kidney tissues and peripheral EDTA blood were collected from euthanized free-roaming cats between March 2009 and February 2011 for a previous study on a morbillivirus associated with feline tubulointerstitial nephritis (15). DNA was extracted from EDTA whole-blood and kidney samples by the use of an EZ1 minikit (Qiagen, Hilden, Germany). The DNA was eluted in 60 μl of elution buffer and was used as the template for PCR and sequencing.
Blood and kidney tissues were screened using primers listed in Table 1. The PCR mixture (25 μl) contained DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 3 mM MgCl2, and 0.01% gelatin), 200 μM each deoxynucleoside triphosphates (dNTPs), and 1.0 U of Taq polymerase (Applied Biosystems, Foster City, CA). The mixtures were amplified in 60 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min with a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystems, Foster City, CA). A Babesia gibsoni strain found in our previous study was used as a positive control. PCR products were gel purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). Both strands of the PCR products were sequenced twice using an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA). The sequences of the PCR products were compared with known sequences by BLAST analysis against the NCBI database.
Table 1.
Sequence of primers used in the study obtained by multiple alignments with CLUSTALW
| Primer category or species name | Target gene | Primer name (sequence) | Target length (bp) | Primer design |
|---|---|---|---|---|
| Piroplasms | 18S rRNA | P_18S1F (AAGATTAAGCCATGCATGTCTAA) | 1,612 | Consensus primers designed by multiple alignment of available 18S rRNA genes of known piroplasms and Hepatozoon spp. |
| P_18S1612R (AGTGATAAGGTTCACAAAACTT) | ||||
| Piroplasms | 18S rRNA | P_18S1F (AAGATTAAGCCATGCATGTCTAA) | Sequencing primers used for constructing the 18S rRNA gene of Babesia hongkongensis | |
| P_18S522R (ATACGCTATTGGAGCTGGAATTA) | ||||
| P_18S500F (TAATTCCAGCTCCAATAGCGTAT) | ||||
| P_18S1071R (GTGTTGAGTCAAATTAAGCCGCA) | ||||
| P_18S1049F (TGCGGCTTAATTTGACTCAACAC) | ||||
| P_18S1612R (AGTGATAAGGTTCACAAAACTT) | ||||
| Babesia hongkongensis (proposed name of the new Babesia species described in this study) | 18S rRNA | BH_18S565F (CGTTTGGGCTTTTAGCTTT) | 173 | Screening primers designed specifically from the 18S rRNA gene of Babesia hongkongensis. |
| BH_18S737R (TTAACCATTACTAAGGTTCCCA) | ||||
| Piroplasms | mitochondrial cytochrome b (cytb) | P_cytbF (TGTTGCTCCCCAATAACTCATTT) | 359 | Consensus primers designed by multiple alignment of available cytb gene of Babesia bigemina, B. bovis, B. caballi, B gibsonii, and Theileria equi. |
| P_cytbR (AGGAATTTAAATTCTAATTGGAATT) |
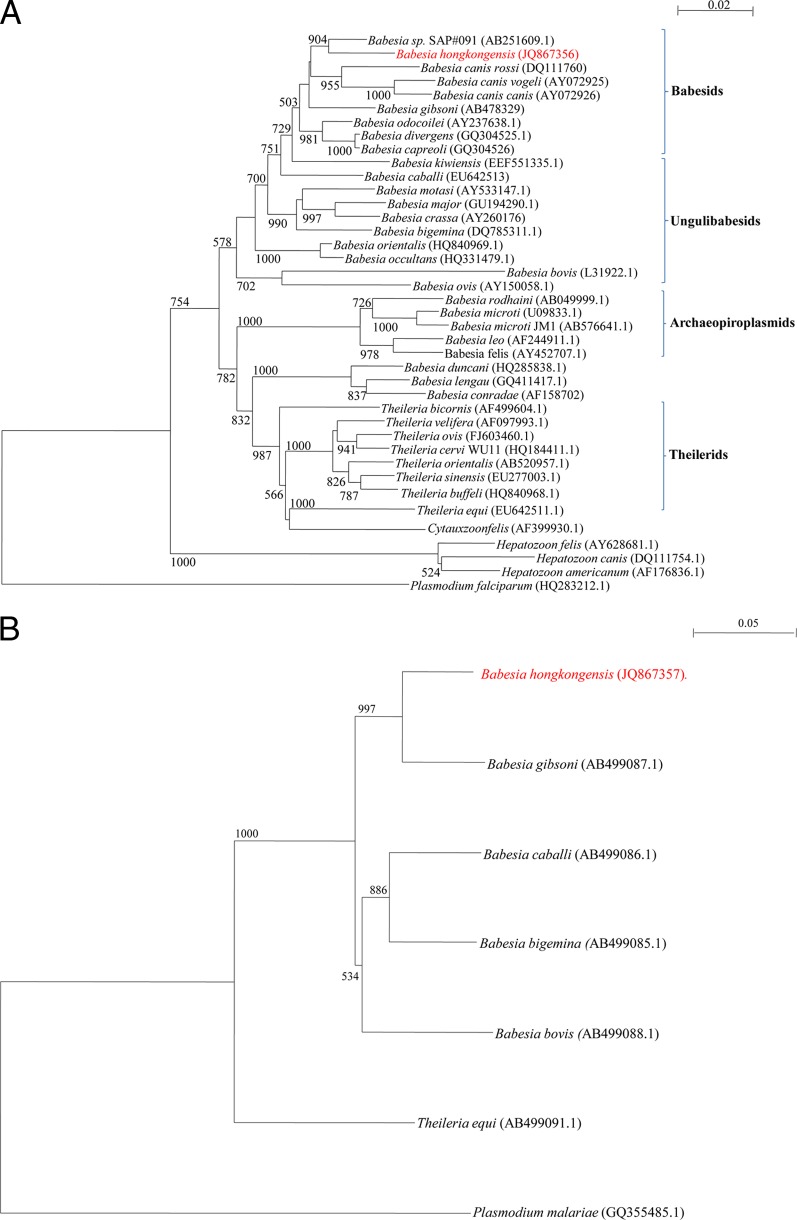
Phylogenetic tree was constructed by the neighbor-joining method using Kimura's two-parameter correction with ClustalX 1.83, with bootstrap values calculated from 1,000 trees. The 1,368 and 364 bp of amplicons from the 18S rRNA and mitochondrial cytochrome b genes of the new Babesia species detected in this study were included in the analysis, using Plasmodium spp. as the outgroup.
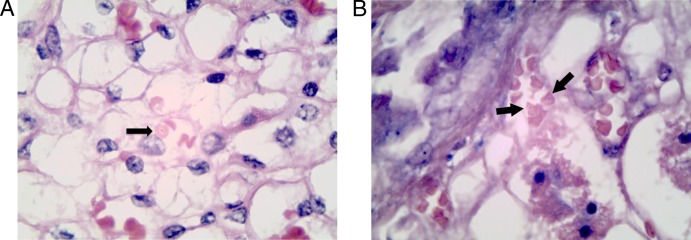
The infected cat was clinically asymptomatic antemortem, but a full autopsy was not performed. A total of 457 blood samples and 48 kidney samples were obtained. One of the 48 kidney sample sections showed intraerythrocytic Babesia-like trophozoites (Fig. 1A). The trophozoites were round to oval, with a light-blue cytoplasm and an eccentric purple nucleus. Single rings were slightly more often found to be located near the center of the erythrocyte. The organism resembles a small Babesia species, with ring forms measuring 1.4 to 1.6 μm in diameter. Similar trophozoites at various stages of development were also seen in Giemsa-stained sections of the kidney (Fig. 1B). No organisms were seen within the leukocytes in the sections.
Fig 1.
Photomicrographs of kidney sections showing Babesia-like organisms. (A) Single, small, round-to-oval intracellular organism with light-blue cytoplasm and an eccentric purple nucleus (signet ring-shaped) in an erythrocyte within the vasa recta of formalin-fixed renal tissue from a free-roaming cat in Hong Kong (hematoxylin and eosin [H&E] staining; original magnification, ×1,000). (B) Single, small, round-to-oval intracellular organism with light-blue cytoplasm in two erythrocytes within the vasa recta of formalin-fixed renal tissue (Giemsa staining; original magnification, ×1,000).
Three hundred randomly selected archival blood specimens were screened for Babesia 18S rRNA PCR. Only the aforementioned cat's specimen was positive. The Babesia-positive cat's blood and the kidney tissue were both PCR positive using consensus 18S rRNA primers for Babesia.
Nearly the full ∼1,700 bp of the 18S rRNA gene of the new Babesia species were built by consensus primer PCR and sequencing (Table 1). The DNA sequences from the kidney section and peripheral blood were identical. BLAST analyses of the sequence did not fully match with any of the sequences in GenBank. It was most closely related (94.3% to 96.7% nucleotide identity with 98% to 100% coverage) to various Babesia sequences found in feral raccoon and dogs (Table 2). By using the ClustalW option of BioEdit, we aligned 1,368 bp of the B. hongkongensis 18S rRNA gene sequence to 38 sequences (Table 2) of other members of the Piroplasmida and Hepatozoon spp. representative of the 5 groups identified within this order as previously defined (8). A representative tree is shown in Fig. 2A. B. hongkongensis falls into a distinct branch of the Babesiidae. The phylogenetic tree is consistent with the topology of previously reported analyses based on 18S rRNA gene sequences of piroplasmids (8). Internal branches of the trees were statistically supported by high bootstrap values. Phylogenetic analysis of a 364-bp region of the mitochondrial cytochrome b gene (Fig. 2B) was most closely related to B. gibsoni. Although only a few Babesia mitochondrial cytochrome b gene sequences have been reported to date, our strain's sequence has only 90.4% identity with that of B. gibsonii. As with the 18S rRNA gene sequence analysis, the Ungulibabesids and Theilerides were grouped separately. We propose that this new Babesia strain, genetically and geographically distinct from all other previously described species, be tentatively named Babesia hongkongensis sp. nov.
Table 2.
Taxonomy, GenBank accession numbers, hosts, geographical regions of isolation, and percentages of sequence identity of Babesia hongkongensis sp. nov. with the 38 piroplasms and Hepatozoon spp. used as operational taxonomic units in the phylogenetic analysis
| Species | GenBank accession no. | Host | Geographical region of isolation | Yr of isolation | Percent sequence identity with B. hongkongensis |
|---|---|---|---|---|---|
| Babesia sp. SAP#091 | AB251609.1 | Feral raccoon | Japan | 2009 | 96.7 |
| Babesia capreoli | GQ304526 | Deer | France | 2011 | 95.1 |
| Babesia divergens | GQ304525.1 | Deer | France | 2011 | 95.1 |
| Babesia odocoilei | AY237638.1 | Reindeer | United States | 2004 | 95.1 |
| Babesia gibsoni | AB478329 | Dog | Japan | 2010 | 95.3 |
| Babesia canis rossi | DQ111760 | Dog | Japan | 2005 | 94.6 |
| Babesia canis canis | AY072926 | Dog | Europe | 2002 | 94.5 |
| Babesia canis vogeli | AY072925 | Dog | Europe | 2002 | 94.3 |
| Babesia caballi | EU642513 | Horse | South Africa | 2009 | 94.3 |
| Babesia kiwiensis | EF551335.1 | Brown kiwi | Australia | 2008 | 94.5 |
| Babesia major | GU194290.1 | Cattle | France | 2009 | 93.5 |
| Babesia orientalis | HQ840969.1 | Water buffalo | China | 2011 | 93.6 |
| Babesia bigemina | DQ785311.1 | Cattle | Spain | 2007 | 93.6 |
| Babesia crassa | AY260176 | Sheep | Germany | 2004 | 92.9 |
| Babesia motasi | AY533147.1 | Sheep | Spain | 2004 | 93.2 |
| Babesia occultans | HQ331479.1 | Hyalomma ticks | Tunisia | 2011 | 93.4 |
| Babesia ovis | AY150058.1 | Goat | Spain | 2006 | 90.7 |
| Babesia duncani | HQ285838.1 | Human | United States | 2011 | 88.8 |
| Babesia leo | AF244911.1 | Lion | South Africa | 2004 | 88.6 |
| Babesia felis | AY452707.1 | Cat | South Africa | 2004 | 88.8 |
| Babesia lengau | GQ411417.1 | Cheetah | South Africa | 2010 | 88.4 |
| Babesia conradae | AF158702 | Dog | United States | 2008 | 88.1 |
| Babesia rodhaini | AB049999.1 | Mouse | Japan | 2008 | 87.8 |
| Babesia microti | U09833.1 | Mouse | United States | 1994 | 87.7 |
| Babesia microti JM1 | AB576641.1 | Monkey | Japan | 2011 | 87.4 |
| Babesia bovis | L31922.1 | Cattle | Mexico | 2001 | 85.1 |
| Theileria velifera | AF097993.1 | Cattle | Tanzania | 1999 | 89.9 |
| Theileria ovis | FJ603460.1 | Goat | China | 2011 | 89.9 |
| Theileria cervi WU11 | HQ184411.1 | Sika deer | China | 2010 | 89.9 |
| Theileria sinensis | EU277003.1 | Bos grunniens | China | 2008 | 89.8 |
| Thseileria orientalis | AB520957.1 | Cattle | Australia | 2011 | 89.6 |
| Theileria buffeli | HQ840968.1 | Water buffalo | China | 2011 | 89.6 |
| Theileria bicornis | AF499604.1 | Black rhinoceros | South Africa | 2003 | 88.8 |
| Theileria equi | EU642511.1 | Horse | South Africa | 2009 | 88.7 |
| Cytauxzoon felis | AF399930.1 | Cat | United States | 2002 | 87.4 |
| Hepatozoon felis | AY628681.1 | Cat | Spain | 2006 | 87.4 |
| Hepatozoon americanum | AF176836.1 | Dog | United States | 2001 | 86.1 |
| Hepatozoon canis | DQ111754.1 | Dog | Japan | 2005 | 85.3 |
| Plasmodium falciparum | M19172.1 | Human | Africa | 1993 | 79.4 |
Fig 2.
Phylogenetic study of Babesia hongkongenesis. (A) Phylogenetic analysis of nucleotide sequences of the 1,368-bp fragment of the 18S rRNA gene of Babesia hongkongensis sp. nov. identified from a free-roaming cat in the present study. The tree was constructed by the neighbor-joining method using Kimura-2 correction and bootstrap values calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 50 nucleotides. Plasmodium falciparum (HQ283212.1) was used as the outgroup. (B) Phylogenetic analysis of nucleotide sequences of the 364-bp fragment of the mitochondrial cytochrome b gene of Babesia hongkongensis sp. nov. identified from the free-roaming cat in the present study. The tree was constructed by the neighbor-joining method using Kimura-2 correction and bootstrap values calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 20 nucleotides. Plasmodium malariae (GQ355485.1) was used as the outgroup.
Feline babesiosis has been described in domestic cats and wild felines (lions, leopards, panthers, cougars, and cheetahs) and is caused by B. felis, B. cati, B. leo, B. canis presentii, B. canis canis, B. canis vogeli, B. pantherae, B. herpalluri, and B. microti-like spp. (Theileria annae) (2–6, 10, 11, 12). Few studies have addressed the prevalence of Babesia in domestic and urban free-roaming cats. In Pakistan, a prevalence of 3.14% was found in pet cats as detected by light microscopy (1). Using PCR to supplement light microscopy, Babesia was found in 1.4% of stray cats in Bangkok (12). Molecular studies contribute to the identification of new species which may have similar microscopic appearances and to diagnosing novel infections caused by environmental species which may initially be misdiagnosed as babesiosis (16). Accurate species identification is important in that different species may have different clinical manifestations and antiparastic drug susceptibilities (2).
The prevalence of B. hongkongensis appears to be low (0.3%) among free-roaming cats. Its prevalence and pathogenicity in pet cats have to be explored. Pet ownership is common is most countries. In Hong Kong, 12.6% of the households were keeping pet animals, with 22.3% of them having cats (7). If B. hongkongensis causes disease in pet animals, this could represent a significant veterinary problem.
Thorough examination of the peripheral blood is important to confirm the absence of a schizogony cycle in leukocytes, which would classify the organism as a Theileria (13). However, our current phylogenetic analysis suggests that this is unlikely, since it is closely clustered with other Babesia species. The vector for B. hongkongensis is unknown, but all known Babesia species utilize hard ticks as the arthropod vector for transmission. Further study should be performed to understand the epidemiology, life cycle, host vector, pathogenicity, and drug susceptibility of this new feline Babesia species. The pathogenicity and zoonotic potential of this new feline Babesia species remain to be determined by further studies.
Nucleotide sequence accession numbers.
Partial nucleotide sequences of the 18S rRNA and mitochrondrial cytochrome b genes obtained in this study have been deposited in the GenBank sequence database under accession numbers JQ867356 to JQ867357.
ACKNOWLEDGMENTS
We thank Alan Chi-Kong Wong, Siu-Fai Leung, Chik-Chuen Lay, Thomas Sit, K. F. Chan, Michelle L. Yeung, Byung Mo Hwang, Suet Yee Ng, Patrick I. T. Lau, and Steven D. Benton from the HKSAR Department of Agriculture, Fisheries, and Conservation (AFCD) for facilitation and support and members of the Animal Management Centres of AFCD.
This work is partly supported by the Tung Wah Group of Hospitals Fund for Research in Infectious Diseases, the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau; the Providence Foundation Limited in memory of the late Lui Hac Minh; and the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health. We are grateful for the generous support of Carol Yu, Richard Yu, Hui Hoy, and Hui Ming in the genomic sequencing platform on emerging infectious disease research.
We declare no conflict of interest.
Footnotes
Published ahead of print 30 May 2012
REFERENCES
- 1. Ahmad SS, Khan MS, Khan MA, Ahmad N. 2011. Prevalence of babesiosis in cats in Lahore, Pakistan. J. Animal Plant Sci. 21(2 Suppl.):354–357 [Google Scholar]
- 2. Ayoob AL, Hackner SG, Prittie J. 2010. Clinical management of canine babesiosis. J. Vet. Emerg. Crit. Care (San Antonio) 20:77–89 [DOI] [PubMed] [Google Scholar]
- 3. Ayoob AL, Prittie J, Hackner SG. 2010. Feline babesiosis. J. Vet. Emerg. Crit. Care (San Antonio) 20:90–97 [DOI] [PubMed] [Google Scholar]
- 4. Baneth G, et al. 2004. Infection with a proposed new subspecies of Babesia canis, Babesia canis subsp. presentii, in domestic cats. J. Clin. Microbiol. 42:99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosman AM, Oosthuizen MC, Peirce MA, Venter EH, Penzhorn BL. 2010. Babesia lengau sp. nov., a novel Babesia species in cheetah (Acinonyx jubatus, Schreber, 1775) populations in South Africa. J. Clin. Microbiol. 48:2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosman AM, Venter EH, Penzhorn BL. 2007. Occurrence of Babesia felis and Babesia leo in various wild felid species and domestic cats in Southern Africa, based on reverse line blot analysis. Vet. Parasitol. 144:33–38 [DOI] [PubMed] [Google Scholar]
- 7. Census and Statistics Department Hong Kong Special Administrative Region 2005. Thematic household survey report—report No. 26. Census and Statistics Department, Hong Kong Special Administrative Region. http://www.censtatd.gov.hk/products_and_services/products/publications/statistical_report/social_data/index_cd_B1130226_dt_detail.jsp
- 8. Criado-Fornelio A, Martinez-Marcos A, Buling-Saraña A, Barba-Carretero JC. 2003. Molecular studies on Babesia, Theileria and Hepatozoon in southern Europe. Part II. Phylogenetic analysis and evolutionary history. Vet. Parasitol. 114:173–194 [DOI] [PubMed] [Google Scholar]
- 9. Gray J, Zintl A, Hildebrandt A, Hunfeld KP, Weiss L. 2010. Zoonotic babesiosis: overview of the disease and novel aspects of pathogen identity. Ticks Tick Borne Dis. 1:3–10 [DOI] [PubMed] [Google Scholar]
- 10. Luaces I, et al. 2005. First report of an intraerythrocytic small piroplasm in wild Iberian lynx (Lynx pardinus). J. Wildl. Dis. 41:810–815 [DOI] [PubMed] [Google Scholar]
- 11. Penzhorn BL, Kjemtrup AM, López-Rebollar LM, Conrad PA. 2001. Babesia leo n. sp. from lions in the Kruger National Park, South Africa, and its relation to other small piroplasms. J. Parasitol. 87:681–685 [DOI] [PubMed] [Google Scholar]
- 12. Simking P, Wongnakphet S, Stich RW, Jittapalapong S. 2010. Detection of Babesia vogeli in stray cats of metropolitan Bangkok, Thailand. Vet. Parasitol. 173:70–75 [DOI] [PubMed] [Google Scholar]
- 13. Uilenberg G. 2006. Babesia—a historical overview. Vet. Parasitol. 138:3–10 [DOI] [PubMed] [Google Scholar]
- 14. Wong SS, et al. 2011. Comparative evaluation of a point-of-care immunochromatographic test SNAP 4Dx with molecular detection tests for vector-borne canine pathogens in Hong Kong. Vector Borne Zoonotic Dis. 11:1269–1277 [DOI] [PubMed] [Google Scholar]
- 15. Woo PC, et al. 2012. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc. Natl. Acad. Sci. U. S. A. 109:5435–5440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan CL, et al. 2012. Colpodella spp.-like parasite infection in woman, China. Emerg. Infect. Dis. 18:125–127 [DOI] [PMC free article] [PubMed] [Google Scholar]