Abstract
A fundamental role for the endosymbiotic bacteria Wolbachia pipientis in the pathogenesis of Dirofilaria immitis infections has emerged in recent years. Diagnostic opportunities arising from this breakthrough have not yet been fully exploited. This study was aimed at developing conventional and real-time PCR assays to carry out a molecular survey in a convenience sample of cats living in an area where D. immitis is endemic and to evaluate the detection of bacterial DNA in blood as a surrogate assay for diagnosing filaria-associated syndromes in cats. COI and FtsZ loci were used as targets for D. immitis and Wolbachia PCR assays, respectively, and real-time TaqMan PCR assays were used only for Wolbachia. A convenience sample of 307 disease-affected or healthy cats examined at a University facility were PCR tested, and their medical records were investigated. Conventional nested PCR for Wolbachia amplified the endosymbionts of both D. immitis and D. repens, while real-time PCR was highly specific only for the former. Observed prevalences of 0.3 and 10.4% were found using conventional nested PCR assays for D. immitis and real-time PCR for Wolbachia, respectively. Similar prevalences were established using the Wolbachia nested PCR (98% concordance with real-time PCR). The group of Wolbachia-positive samples had a significantly higher proportion of subjects with respiratory signs (29.0% versus 9.7%; P = 0.002). The findings of this study indicate that a highly sensitive PCR assay can be used to detect the Wolbachia organism in the peripheral blood of cats with respiratory signs.
INTRODUCTION
Heartworm disease due to the nematode Dirofilaria immitis is globally widespread and is a leading cause of morbidity and mortality in dogs and cats in areas of endemicity. The prevalence in cats is consistent with the prevalence in dogs in a given area, even if in a lower proportion (5 to 20%) (10, 21). Nevertheless, feline prevalence data might be biased by a very low testing frequency estimated to be 0.06% versus a meaningful 33% in dogs (22). These testing frequencies clearly demonstrate that feline filariosis has not yet been perceived as an actual clinical problem by practitioners who erroneously believe that low infection rates occur in cats (9, 22). Certainly, until now, a combination of physical examination and ancillary methods have been recommended for establishing the likelihood of feline filariosis intra vitam (2, 7).
Filarial nematodes harbor Wolbachia endosymbionts. Wolbachia is an intracellular Gram-negative bacterium, belonging to the order Rickettsiales and found in 20 to 80% of the arthropod species and in the nematodes of the Onchocercidae family (4, 14, 36). In addition to the demonstrated symbiotic relationships regarding fecundity and long-term survival, Wolbachia has an important role in the pathogenesis of filarial infections of mammalians (19, 31). In cats, the peculiar pathobiology of filariosis characterized by the death of the majority of immature adults (L5 larvae) arriving in the lung vasculature has been advocated as the cause of the recently described heartworm-associated respiratory disease (HARD) (3). The reaction was partially ascribed to the release of Wolbachia antigens after the disintegration of the worms (24). Wolbachia surface proteins (WSPs) are highly immunogenic and elicit a strong inflammatory response (23). Some authors have speculated that a diagnosis of filariosis could be attained serologically by detecting anti-Wolbachia antibodies. Indeed, in cats, antibodies against WSPs are promptly produced as early as 2 months after infection and last for more than 8 months (26). Unfortunately, the increase in IgG persists months beyond the disappearance of adult nematodes and beyond the duration of D. immitis specific antibodies (17, 18). Thus, their diagnostic suitability is limited. Conversely, PCRs targeting Wolbachia were developed for detecting the bacteria either in tissues, nematodes (8, 20, 25, 32), or the blood of filaria-infected dogs (30). However, the studies reporting Wolbachia PCRs were not aimed at establishing diagnostic reliability but rather served as an insight into filarial pathogenesis. Thus, we hypothesized that the diagnostic expediency of PCR testing and, in particular of Wolbachia PCRs, was not fully exploited.
This study was aimed at developing conventional and real-time PCR assays to be used in carrying out a molecular survey in a convenience sample of cats from a D. immitis area of endemicity and for evaluating and comparing the detection of filarial and bacterial DNA in feline blood as surrogate assays for diagnosing feline filaria-associated syndromes in cats.
MATERIALS AND METHODS
Case material.
The present study utilized feline blood samples (EDTA anticoagulated) from a convenience-sampled population which had already been used in a different study (12). The samples had been obtained from cats seen at the Veterinary Teaching Hospital, University of Bologna, Bologna, Italy, and underwent routine blood testing at the Veterinary Clinical Pathology Service between 1 January and 31 December 2006. The samples were frozen daily at −20°C and stored until further analysis. Most of the samples had been collected from sick cats requiring hematological evaluation as part of a diagnostic profile, whereas a minority of samples were collected from apparently healthy cats which were at the hospital for presurgical testing or a pre-anesthetic check. The samples collected from cats during repeated presentation were discarded (n = 192). Only samples with a minimum blood volume of 100 μl after routine hematological testing were included. The medical records of the subjects included in the study were retrieved, and the examiner was blinded as to the previous molecular findings. Due to its retrospective nature, which examined a heterogeneous sample, the medical records were largely incomplete, and a definitive diagnosis had been obtained and/or reported in only a minority of cases. Nevertheless, in those cases without a definitive diagnosis, to complete the medical record, the clinician was forced to indicate the affected organ system on a definite field of the electronic medical records. This tool was used to categorize most of the subjects included in the study.
DNA extraction.
The DNA was extracted from whole blood samples after thawing at room temperature; the extraction was accomplished by using the QIAamp DNA blood minikit (Qiagen, Milan, Italy) according to the manufacturer's instructions. The DNA was eluted into 200 μl of Buffer AE and stored at −20°C until use. A subset of 25 samples was repurified from frozen blood using the NucleoSpin kit (Macherey & Nagel, Milan, Italy) according to the manufacturer's instructions.
Conventional PCR and PCR sensitivity.
Two different nested PCRs were set up for D. immitis and Wolbachia genomic DNA (gDNA) detection. The gDNAs of both D. immitis and Wolbachia were obtained from three dogs diagnosed with filariosis by means of the SNAP Filaria RT Antigene (IDEXX Laboratories, Milan, Italy) and microfilarial detection on blood smear observations. Further molecular characterization of the gDNA was accomplished with primer pairs DIDR_F1 and DIDR_R1, as described by Rishniw et al. (28).
The primer pairs were designed on the cytochrome oxidase subunit I (COI) gene of D. immitis (accession number gi:40255343), as well as on the cell division protein FtsZ of Wolbachia (gi:32562974). The prefixes Fil_COIclon and COIfel were assigned to the outer and inner primers for D. immitis, respectively. The prefixes Wol1 and Wol7 were assigned to the Wolbachia outer and inner primers, respectively. All of the primers used here are listed in Table 1.
Table 1.
Primers and probes used in this study
| Primer | Sequence (5′–3′) | Amplicon |
|---|---|---|
| Fil_COIclon_fwd | ATTGGTGGTTTTGGTAATTGGATGTTG | First-round PCR for D. immitis, 634 bp |
| Fil_COIclon_R | CAGAAGTCCCCAATACAGCAATCC | |
| COIfel_fwd | GGGTCCTGGGAGTAGTTGAAC | Second-round PCR for D. immitis, 406 bp |
| COIfel_R | TTCACTAACAATCCCAAACACCG | |
| Wol1_fwd | CCTGTACTATATCCAAGAATTACTG | First-round PCR for Wolbachia, 267 bp |
| Wol1_R | ACTATCCTTTATATGTTCCATAATTTC | |
| Wol7_fwd | GGTGGAAATGCTGTGAATAAC | Second-round PCR for Wolbachia, 147 bp |
| Wol7_R | AGCACCGAGCCCTTTAG | |
| Wolb3_fwd | TTGTTGTAGCAAATACGGATG | Real-time PCR, 127 bp |
| Wolb3_R | GCTGCACCTTTACCAATATC | |
| WOLB_TQ3 | [6FAM]AAGGCACCAGCACCGAGC[BHQ1] | |
| F_wol_is1 | ACTATATCCAAGAATTACTGTTGCGTTGTTGATGG | Mimic for first-round PCR for Wolbachia, 220 bp |
| R_wol_is1 | CTTTATATGTTCCATAATTTCATGCTGATACAATAAAGG | |
| F_wol_is7 | GGAAATGCTGTGAATAACACCACTGGTATTGTCATGGACTCTG | Mimic for first-round PCR for Wolbachia, 309 bp |
| R_wol_is7 | CACCGAGCCCTTTAGGCTCTTCTCCAGGGAGGACGA | |
| Fwd_Fil_COIclon_is | TGGTTTTGGTAATTGGATGTTGTCCTGGGAGTAGTTGAAC | Mimic for first-round PCR for D. immitis, 280 bp |
| R_Fil_COIclon_is | AGTCCCCAATACAGCAATCCCTAACAATCCCAAACACCG | |
| Fwd_COIfel_is | TCCTGGGAGTAGTTGAACATGATCTGTCTCTCTTTTCTCCCC | Mimic for second-round PCR for D. immitis, 238 bp |
| R_Fil_COIfel_is | CTAACAATCCCAAACACCGGTACACAAAAAGGTTACATGGAAAGC |
The first-round PCR produced the expected size bands of 634 and 267 bp for D. immitis and Wolbachia, respectively. Therefore, the PCR products were purified using commercial kits (Perfectprep purification; Eppendorf, Milan, Italy). The purified amplicons were sequenced using a BigDye Terminator v1.1 kit (Applied Biosystems, Milan, Italy), purified with Centri-Sep columns (Applied Biosystems), and electrophoresed on an ABI Prism 310 sequencer after denaturation with HiDi Formamide (Applied Biosystems) at 95°C for 5 min. Sequences covering almost 90% of the amplicons were 100% homologous with D. immitis and its Wolbachia endosymbiont. Consequently, the first-round PCRs were cloned into the pCR-4 TOPO (Invitrogen, Milan, Italy) plasmid vectors. The plasmids were used to transform TOP10 chemically competent E. coli (Invitrogen, Milan, Italy) and were purified using a plasmid purification kit (Sigma-Aldrich, Milan, Italy).
The plasmid copy number was determined by spectrophotometry. The plasmids were then linearized with SphI endonuclease (New England Biolabs/Euroclone, Milan, Italy), and serial 10-fold dilutions were prepared for a PCR sensitivity assay. The precise serial dilutions of the linearized plasmid were also used to calibrate the real-time PCR assay for Wolbachia detection.
For the assessment of the specificity, in addition to the sequencing of three Wolbachia-positive samples, PCR assays were used to assess canine and feline genomic templates. In particular, Fil_COIclon and COIfel primers were assayed against canine gDNA containing Acanthocheilonema (Dipetalonema) reconditum and Dirofilaria repens canine gDNA, while conventional and real-time Wolbachia PCRs were checked in PCR assays using Ehrlichia canis, Anaplasma platys (34), Bartonella henselae, and D. repens canine and feline gDNA that were previously positive in routine testing as templates.
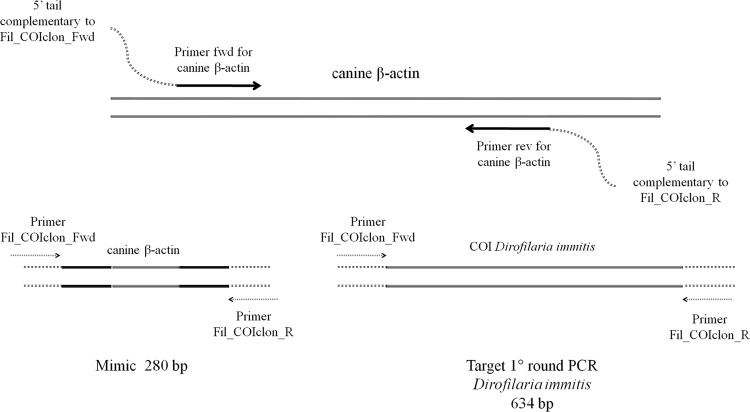
To reduce the risk of contamination, the two nested PCR assays were set up with an internal control known as a mimic. The mimics were obtained as described by Ballagi-Pordány and Belák (1) with only slight modifications. Briefly, mimics are PCR amplicons that include the sequences recognized by the primers at each end of a different target (off-target) so that the sequence length between each target primer site is different from that of the target sequence. Mimics are easily and cheaply obtained by amplifying the off-target with a primer pair composed of a 3′ end specific for the off-target and a 5′ oligonucleotide tail made up of sequences of the target. Unlike the original method, in the present study the 5′ oligonucleotide tail was 2 to 3 bp shorter than the target primers in order to facilitate the recognition of the target sequence of the gDNA with respect to the mimics (Fig. 1). The PCR products were diluted 10−10 in molecular-biology-grade water and used in the PCR mixtures.
Fig 1.
Schematic representation of the mimic technique. (Top) Method for obtaining the mimic amplicons. (Bottom) Use of the mimic amplicons in the first D. immitis PCR round.
The PCR mixture for all PCR assays included 2.5 μl of 10× PCR buffer (Invitrogen), 1.5 mM magnesium chloride, 300 nM (each) forward and reverse primers, 250 nM deoxynucleoside triphosphates (10 mM dNTP mix, PCR grade; Invitrogen), 1 U of recombinant Taq polymerase (Invitrogen), and 2 μl of template brought up to 25 μl with mimics diluted 10−10 in molecular-biology-grade water (Eppendorf). Each PCR run included negative controls represented by molecular-biology-grade water. The PCRs were carried out using an EP-gradient S thermal cycler (Eppendorf). The first D. immitis PCR round included an initial denaturation at 95°C for 4 min, followed by 40 cycles of 94°C for 30 s, 56.5°C for 30 s, and 72°C for 45 s, with a final extension step of 72°C for 5 min. The nested D. immitis PCR round differed only with respect to an annealing temperature of 54.5°C and an extension step of 30 s. The Wolbachia first-round PCR consisted of an initial denaturation at 95°C for 4 min, followed by 40 cycles of 94°C for 30 s, 51°C for 30 s, and 72°C for 30 s, and final treatment of 72°C for 5 min. The Wolbachia second round differed only with respect to an annealing temperature of 57°C. In both cases, the second round template was represented by a 1:10 dilution of the first-round PCR mixture. The PCR products were evaluated after electrophoresis on 1.5% agarose gel and gel staining with ethidium bromide.
All conventional PCRs were carried out in duplicate. The last dilution yielding a positive result was assumed to be the limit of detection.
Real-time TaqMan PCR.
A TaqMan assay was designed using Primer Express v3 software (Applied Biosystems) within the Wolbachia cloned sequences (Table 1), purchased (Proligo; Sigma-Aldrich), and used to re-assay all of the samples. The real-time PCRs were carried out with a mixture composed of 10 μl of PCR mix 2× (Maxima probe master mix; Fermentas, Milan, Italy), 900 nM concentrations (each) of forward and reverse primers, 300 nM TaqMan probe, 2 μl of template, and molecular-biology-grade water to reach a final volume of 20 μl. The real-time PCRs were carried out with a four-step protocol: initial denaturation at 95°C for 10 min, followed by 45 cycles of 92.5°C for 15 s, 54°C for 15 s, and 54°C for 10 s with signal acquisition and finally at 72°C for 25 s in a StepOne thermal cycler (Applied Biosystems). The calibration was carried out by assessing each sample in triplicate. The findings of both conventional and real-time PCR assays were considered positive when at least one of the two replicates yielded a specific amplicon or a fluorescent signal.
Statistical analysis.
A chi-square test was used to evaluate the differences in presenting clinical signs, and the odds ratio with a 95% confidence interval (CI) was calculated. A P value of <0.05 was considered statistically significant.
RESULTS
Sequencing.
All positive controls, three randomly chosen samples positive for Wolbachia, and the sole sample positive for D. immitis were sequenced. All confirmed 100% homology with the reference sequences.
Nested PCR assay sensitivity and specificity.
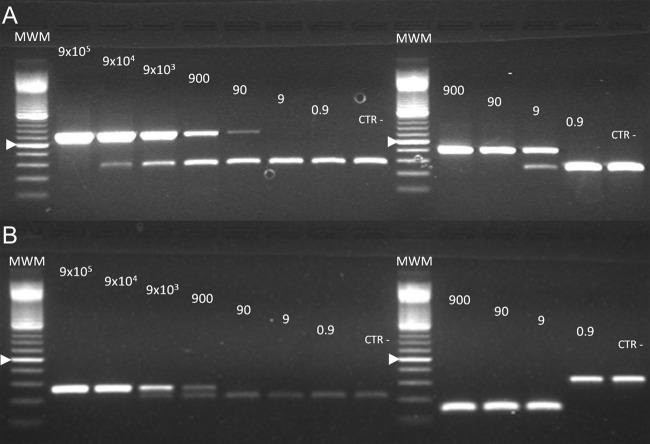
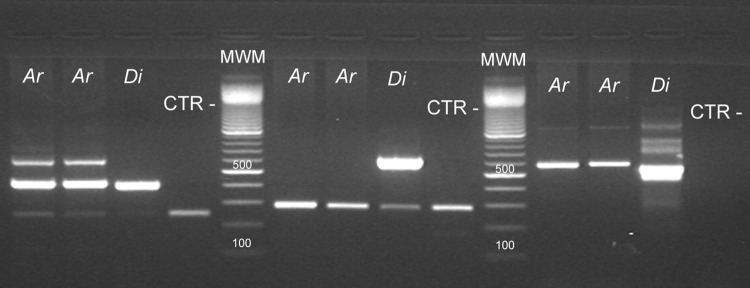
Both nested PCR assays showed similar sensitivity performances. Assay sensitivities of ∼900 copies/reaction for the first round and of 9 copies/reaction for the second round were obtained in both conventional PCR assays (Fig. 2). As expected, the internal standard did not show interference since similar assay sensitivity was demonstrated with or without the inclusion of mimics in the PCR mixtures (Fig. 2 and 3). The D. immitis nested PCR did not yield PCR products when canine gDNAs positive for Acanthocheilonema (Dipetalonema) reconditum (Fig. 4) and D. repens were amplified. The Wolbachia nested PCR did not yield products when canine gDNAs PCR positive for E. canis or A. platys or when feline gDNAs positive for B. henselae by PCR were used as templates. Conversely, nested PCR amplified Wolbachia from D. repens-positive gDNA samples.
Fig 2.
Sensitivity of the Dirofilaria Immitis nested PCR (A) and Wolbachia nested PCR (B) assays. Precise serial dilutions of the linearized pCR-4 TOPO vector containing the D. immitis COI and Wolbachia FtsZ sequences were used as a PCR template. The amounts of target are indicated above each lane. (A) Lanes 2 to 9, first PCR round. Upper bands of the expected 634-bp size represent the Fil_COIclon amplicons, whereas the lower 280-bp bands represent the mimic internal standard. Lanes 11 to 15, second PCR round. Upper bands of the expected 406-bp size represent the COIfel amplicons, whereas the lower 237-bp bands represent the mimic internal standard. (B) Lanes 2 to 9, first PCR round. Upper bands of the expected 267-bp size represent the Wol1 amplicons, whereas the lower bands of 220 bp represent the mimic internal standard. Lanes 11 to 15, second PCR round. Lower bands of the expected 147-bp size represent the Wol7 amplicons, whereas the upper 309-bp bands represent the mimic internal standard. MWM, molecular weight marker, 100-bp ladder. The 500-bp band is indicated by an arrowhead.
Fig 3.
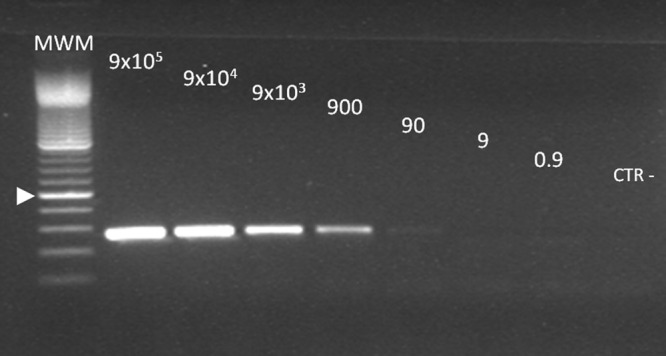
Sensitivity of the first-round Wolbachia nested PCR without the mimic internal standard. This figure should be compared to Fig. 2, lanes 2 to 9. A pattern almost identical to that seen in Fig. 2 was observed, indicating the same limit of detection of 900 copies/reaction.
Fig 4.
Molecular identification of D. immitis. Amplicons obtained using different filarial gDNA templates. Lanes 1 to 4, PCRs using primer of D. immitis COIfel; lanes 6 to 9, PCRs using a primer of D. immitis Fil_COIclon; lanes 11 to 14, PCRs using primers DIDR_F and DIDR_R. Ar, Acanthocheilonema (Dipetalonema) reconditum templates; Di, D. immitis templates; CTR−, no DNA control. The lowest bands of lanes 1 to 4 and lanes 6 to 9 represent the mimic internal standard PCR product. MWM, molecular weight marker, 100-bp ladder. The 500-bp band is indicated by an arrowhead.
Retrospective survey using conventional nested PCR.
One of 307 samples was positive when using the D. immitis nested PCR. The sample was already positive after the first round of PCR. Of the 307 samples, 34 were positive when using the Wolbachia nested PCR. All except one sample were negative after the first round and positive only after the second round. The only sample that was positive when using the first round was referred to as the D. immitis-positive sample. Thus, approximately less than 900 and more than 9 copies/reaction of Wolbachia could be estimated in all cases without circulating D. immitis. The observed prevalences using conventional nested PCR were 0.3% (0.0 to 1.8% [95% CI]) and 11.1% (7.8 to 15.1% [95% CI]) for D. immitis and Wolbachia, respectively.
Retrospective survey using real-time PCR and concordance with conventional PCR.
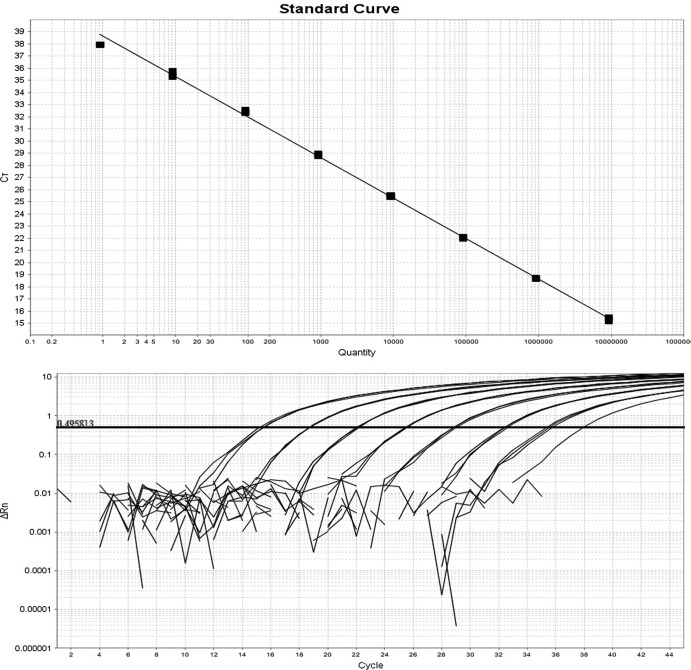
Real-time PCR was linear over 8 orders of magnitude from 9 × 106 to 9 target/reaction with a coefficient R2 of 0.998. The mean coefficient of variation of each triplicate calibration point was 0.42% (range, 0.22 to 0.92%), and there was a PCR efficiency of 97.521 (Fig. 5). The limit of detection was 9 targets/reaction, although 1 of 3 replicates at 0.9 copies/reaction yielded a signal. If the 0.9 target calibration point was included, the calculated efficiency was 99.212 with an R2 of 0.999. Real-time TaqMan PCR did not originate a signal when canine gDNA previously positive for E. canis, A. platys, and D. repens by PCR or when a feline gDNA previously positive for B. henselae by PCR was used as a template. Using a real-time PCR assay, 32/307 samples were positive, and the observed prevalence was 10.4% (7.2 to 14.4% [95% CI]). Concordant results were found in 301/307 (98.0%) cases (271 negative and 30 positive). Four samples which were positive when using nested PCR were negative when using real-time PCR and, vice versa, two samples which were negative when using nested PCR were positive when using real-time PCR.
Fig 5.
Calibration curve (upper panel) and the exponential PCR curves (lower panel) of the real-time PCR Wolbachia assay.
Features of the sample.
The analysis of medical records allowed the classification of the clinical problem in 258/307 cases (84.0%). The sample was divided into two subgroups based upon the Wolbachia status obtained using real-time PCR (227 Wolbachia PCR negative; 31 Wolbachia PCR positive); the involvement of the respiratory organ system occurred more frequently in the Wolbachia-positive group (odds ratio = 4.1 [95% CI = 1.7 to 10.0%; P = 0.002]). The complete findings of the analysis of the medical records are reported in Table 2.
Table 2.
Organ system involved at presentation according to the medical records of the convenience sample
| Organ system | No. of samples (%) |
||
|---|---|---|---|
| Total (n = 258) | PCR Wolbachia-negative group (n = 227) | PCR Wolbachia-positive group (n = 31) | |
| Respiratory system | 31 (12.0) | 22 (9.7) | 9 (29.0) |
| Digestive system | 83 (32.2) | 73 (32.2) | 10 (32.2) |
| Urogenital system | 40 (15.5) | 37 (16.3) | 3 (9.7) |
| Nervous system | 28 (10.9) | 24 (10.6) | 4 (12.9) |
| Others | 76 (29.5) | 71 (31.3) | 5 (16.1) |
DISCUSSION
In this study, setups for both the conventional nested PCR for D. immitis and Wolbachia and the real-time quantitative PCR for Wolbachia only were carried out. Both the conventional and the real-time assays gave satisfactory results in terms of sensitivity and specificity, even though the nested PCR could not differentiate between the endosymbiont of D. immitis and D. repens.
Whenever real-time thermal cyclers are available, the real-time technique is preferred due to the high sensitivity and even higher specificity achieved through the use of TaqMan hydrolysis probes. Furthermore, the reaction occurs in closed reaction tubes circumventing the risk of carryover of the amplicons. Conversely, in nested PCR, the risk of contamination due to PCR products is elevated due to the opening of the first-round tube. To limit this drawback of endpoint nested PCR, an additional improvement was represented by the introduction of internal controls known as mimics (1). The technique was intended to limit the possibility of carryover and false-positive results in nested reactions. In the present study, a slightly modified mimic technique was utilized. The modified mimics performed adequately without any detrimental effect on the sensitivity of the assay. Thereafter, although the real-time technique is very fast and effective, both endpoint and real-time PCRs were shown to readily and effectively detect their target cloned in plasmids.
The PCR assays were used for retrospectively assessing the presence of both filarial and Wolbachia DNA in feline gDNA purified from a convenience sample of cats examined at the Veterinary Teaching Hospital of the University of Bologna. Although the limit of detection of the PCR assays was almost identical, the calculated percentage of the positive samples varied markedly. Indeed, only 1 positive case of D. immitis was present as opposed to 34 and 32 positive cases for Wolbachia using conventional nested or real-time PCRs, respectively.
In the area of endemicity in northern Italy, the prevalence of feline filariosis estimated by the presence of antibodies against D. immitis was ca. 24%, whereas it was about 50 to 84% in untreated dogs (10). In the same area of endemicity, it was shown that 6.7% of cats harbored filarial nematodes (11). In this regard, the negligible prevalence (0.3%; 0.0 to 1.8% [95% CI]) assessed by nested PCR unquestionably represents an underestimated value. Likely explanations are related to feline filarial pathobiology; cats harbor fewer (fewer than 6) parasites with a shorter lifespan than dogs. Cats are frequently parasitized by one or only a few adult male nematodes; there are also very few microfilariae, and their presence is transient due to the strong immune response of cats (21, 27). Conversely, the remarkably higher observed prevalence of Wolbachia (11.1% [95% CI = 7.8 to 15.1%] using nested PCR or 10.4% [95% CI = 7.2 to 14.4%] using real-time PCR) is more consistent with the actual prevalence of filariosis.
These findings may have different explanations which, unfortunately, cannot be addressed here due to the inherent weakness of retrospective studies. Indeed, the discrepancy of the observed prevalence between D. immitis and Wolbachia could be because (i) both D. immitis and Wolbachia were present, although only endosymbiotic bacteria, released either by living adult worms lying in vessels or microfilariae, could readily be detected by PCR because they were more abundant; (ii) a massive release of Wolbachia in cats occurs within 90 days from infection during the migration of L5 immature adults before their arrival in the pulmonary vessels (it was shown that, unlike what happens in dogs, the majority of nematodes die prior to becoming adults due to the strong immune response of cats); (iii) Wolbachia bacteria entered the host as endosymbionts of the nematodes but remained, infecting host cells after the disappearance of the adult nematodes; or (iv) sources other than D. immitis could have released Wolbachia.
Verifying the hypothesis that there was a greater likelihood of detecting Wolbachia DNA in infected animals than detecting D. immitis DNA was within the aims of our study. Wolbachia could be found in all of the developmental stages of D. immitis; in adults, Wolbachia was present in the hypodermal cells of the lateral chords and in the reproductive organs of females (15, 16). Overall, one nematode harbors thousands of bacteria, and the bacteria are released en masse when the parasite is killed by adulticidal therapy or dies spontaneously (6). As a result of the above-mentioned evidence, a PCR targeting Wolbachia DNA would be more suitable than a PCR targeting D. immitis DNA, and the positive results of a Wolbachia PCR could be considered the surrogate of a positive filarial test and diagnosis thereof.
Nonetheless, indirect evidence supports alternative explanations. In the present study, the Wolbachia-positive samples showed a significantly higher prevalence of respiratory disorders, although positive cases showed a wide range of clinical syndromes. Typically, feline filariosis is characterized by physical signs that may vary greatly from asymptomatic to fatal cases. Many cats never present any signs during their lifetime; in other cases, the signs and symptoms may be acute or chronic and mild or severe (7, 11, 35). Typically, respiratory signs characterize the HARD syndromes. Even though arteritis and thromboembolic disease secondary to the presence of adult D. immitis in the pulmonary arteries could explain a few of the manifestations of feline filarial disease at postmortem examination, it was nevertheless demonstrated that severe pulmonary lesions occur in the absence of adult worms (5, 8). Macrophages containing Wolbachia have been found in the lungs, kidneys, and liver, of dogs infected with D. immitis (18). Evidence suggests that, in cats, even transient filarial infections cause enduring and even worsening lung lesions (4). Lung lesions are typically characterized by both parenchymal and vascular inflammation evolving into arteriolar occlusive hypertrophy. Typically, the WSP antigen was demonstrated in lung lesions (8, 17, 18). The WSP antigen of Wolbachia elicits a strong inflammatory response itself in the host, so a pivotal role in the HARD syndromes was hypothesized (24). However, a recent study failed to demonstrate a clear association between the presence of either Wolbachia DNA or WSPs in lung lesions and the severity of pulmonary lesions (8).
The possibility of other sources of Wolbachia organisms merits further investigation. The occurrence of cutaneous dirofilariosis sustained by D. repens has been reported in limited case series, and the presence of D. repens in cats was demonstrated in a central region of Italy (33). Both conventional and real-time PCR assays were designed on the nucleotide sequences of the Wolbachia endosymbiont of D. immitis since only partial FtsZ nucleotide sequences are available in public databases to date. Indeed, nested PCR, but not real-time PCR, could amplify the Wolbachia endosymbiont of D. repens. Although the possibility that the few discordant cases, in particular the nested PCR positives that were negative using the real-time PCR, could be ascribed to the presence of the D. repens endosymbiont; overall, the observed prevalences obtained using the two PCR methods overlap.
In addition to filaroid parasites, other sources, represented by arthropods, are possible and could not be ruled out. Indeed, if, on the one hand, common nematodes of cats were not reported to harbor Wolbachia (4), on the other hand, ticks and fleas harbor Wolbachia (29), and no studies have investigated the possibility that Wolbachia are transmitted during the feeding of hematophagous arthropods. In addition, the absence of FtsZ sequences of the Wolbachia endosymbiont in cat fleas and ticks impedes ruling out cross-amplification using in silico methods.
Although these uncertainties were not fully addressed here, a prospective study investigating a combination of filarial or even Wolbachia (13) antibody assays and PCR assays is warranted in order to clarify the issues related to the universality of Wolbachia primers and probes, and the role of D. repens, as well as the exact significance of PCR positivity. Such a study could also establish the accuracy of a combination of a D. immitis antibody assay and a Wolbachia PCR assay, as well as argue, for or against the use of antibiotic therapy targeting Wolbachia.
In this complex scenario, the findings presented here add a small piece to the puzzle and further support the role of Wolbachia in the pathogenesis of filarial-associated syndromes and the HARD syndrome, in particular. In conclusion, our findings indicate that testing Wolbachia by means of PCR could be suitable for reaching a diagnosis of the filaria-associated syndrome in cats and could therefore be convincingly introduced into the clinical setting.
Footnotes
Published ahead of print 30 May 2012
REFERENCES
- 1. Ballagi-Pordány A, Belák S. 1996. The use of mimics as internal standards to avoid false negatives in diagnostic PCR. Mol. Cell. Probes 10:159–164 [DOI] [PubMed] [Google Scholar]
- 2. Berdoulay P, et al. 2004. Comparison of serological tests for the detection of natural heartworm infection in cats. J. Am. Anim. Hosp. Assoc. 40:376–384 [DOI] [PubMed] [Google Scholar]
- 3. Blagburn BL, Dillon AR. 2007. Feline heartworm disease: solving the puzzle. Vet. Med. 2007(Suppl):7–14 [Google Scholar]
- 4. Bordenstein SR, Fitch DH, Werren JH. 2003. Absence of Wolbachia in nonfilariid nematodes. J. Nematol. 35:266–270 [PMC free article] [PubMed] [Google Scholar]
- 5. Browne LE, Carter TD, Levy JK, Snyder PS, Johnson CM. 2005. Pulmonary arterial disease in cats seropositive for Dirofilaria immitis but lacking adult heartworms in the heart and lungs. Am. J. Vet. Res. 66:1544–1549 [DOI] [PubMed] [Google Scholar]
- 6. Cross HF, Haarbrink M, Egerton G, Yazdanbakhsh M, Taylor MJ. 2001. Severe reactions to filarial chemotherapy and release of Wolbachia endosymbionts into blood. Lancet 358:1873–1875 [DOI] [PubMed] [Google Scholar]
- 7. Dillon AR, Brawner AR, Jr, Robertson-Plouch CK, Guerrero J. 2000. Feline heartworm disease: correlations of clinical signs serology and other diagnostics: results of a multicenter study. Vet. Ther. 1:176–182 [PubMed] [Google Scholar]
- 8. Dingman P, et al. 2010. Association of Wolbachia with heartworm disease in cats and dogs. Vet. Parasitol. 170:50–60 [DOI] [PubMed] [Google Scholar]
- 9. Dunn KF, Levy JK, Colby KN, Michaud RI. 2011. Diagnostic treatment and prevention protocols for feline heartworm infection in animal sheltering agencies. Vet. Parasitol. 176:342–349 [DOI] [PubMed] [Google Scholar]
- 10. Genchi C, Guerrero J, McCall JW, Venco L. 2007. Epidemiology and prevention of Dirofilaria infections in dogs and cats, p 145–161 In Genchi C, Rinaldi L, Cringoli G. (ed), Dirofilaria immitis and D. repens in dog and cat and human infections. Mappe Parassitologiche 8. Mendeley, Naples, Italy [Google Scholar]
- 11. Genchi C, Venco L, Ferrari N, Mortarino M, Genchi M. 2008. Feline heartworm (Dirofilaria immitis) infection: a statistical elaboration of the duration of the infection and life expectancy in asymptomatic cats. Vet. Parasitol. 158:177–182 [DOI] [PubMed] [Google Scholar]
- 12. Gentilini F, et al. 2009. Use of combined conventional and real-time PCR to determine the epidemiology of feline haemoplasma infections in northern Italy. J. Feline Med. Surg. 11:277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grandi G, Morchon R, Kramer L, Kartashev V, Simon F. 2008. Wolbachia in Dirofilaria repens, an agent causing human subcutaneous dirofilariasis. J. Parasitol. 94:1421–1423 [DOI] [PubMed] [Google Scholar]
- 14. Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281:215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozek WJ. 2005. What is new in the Wolbachia-Dirofilaria interaction? Vet. Parasitol. 133:127–132 [DOI] [PubMed] [Google Scholar]
- 16. Kramer LH, Passeri B, Corona S, Simoncini L, Casiraghi M. 2003. Immunohistochemical/immunogold detection and distribution of the endosymbiont Wolbachia of Dirofilaria immitis and Brugia pahangi using a polyclonal antiserum raised against WSP (Wolbachia surface protein). Parasitol. Res. 89:381–386 [DOI] [PubMed] [Google Scholar]
- 17. Kramer L, Simón F, Tamarozzi F, Genchi M, Bazzocchi C. 2005. Is Wolbachia complicating the pathological effects of Dirofilaria immitis infections? Vet. Parasitol. 133:133–136 [DOI] [PubMed] [Google Scholar]
- 18. Kramer LH, et al. 2005. Immune response to and tissue localization of the Wolbachia surface protein (WSP) in dogs with natural heartworm (Dirofilaria immitis) infection. Vet. Immunol. Immunopathol. 106:303–308 [DOI] [PubMed] [Google Scholar]
- 19. Kramer L, et al. 2008. Wolbachia and its influence on the pathology and immunology of Dirofilaria immitis infection. Vet. Parasitol. 158:191–195 [DOI] [PubMed] [Google Scholar]
- 20. Lee SA, Lee SG, Choi EJ, Hyun C. 2008. Prevalence of the endosymbiont Wolbachia in heartworms (Dirofilaria immitis). Vet. Rec. 163:484–486 [DOI] [PubMed] [Google Scholar]
- 21. Litster AL, Atwell RB. 2008. Feline heartworm disease: a clinical review. J. Feline Med. Surg. 10:137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lorentzen L, Caola AE. 2008. Incidence of positive heartworm antibody and antigen tests at IDEXX Laboratories: trends and potential impact on feline heartworm awareness and prevention. Vet. Parasitol. 158:183–190 [DOI] [PubMed] [Google Scholar]
- 23. Martin C, Gavotte L. 2010. The bacteria Wolbachia in filariae a biological Russian dolls' system: new trends in antifilarial treatments. Parasite 17:79–89 [DOI] [PubMed] [Google Scholar]
- 24. McCall JW, Genchi C, Kramer LH, Guerrero J, Venco L. 2008. Heartworm disease in animals and humans. Adv. Parasitol. 66:193–285 [DOI] [PubMed] [Google Scholar]
- 25. Michalski ML, et al. 2010. Identification and phylogenetic analysis of Dirofilaria ursi (Nematoda: Filarioidea) from Wisconsin black bears (Ursus americanus) and its Wolbachia endosymbiont. J. Parasitol. 96:412–419 [DOI] [PubMed] [Google Scholar]
- 26. Morchón R, et al. 2004. Specific IgG antibody response against antigens of Dirofilaria immitis and its Wolbachia endosymbiont bacterium in cats with natural and experimental infections. Vet. Parasitol. 125:313–321 [DOI] [PubMed] [Google Scholar]
- 27. Nelson CT, et al. 2005. Executive Board of the American Heartworm Society 2005 guidelines for the diagnosis prevention and management of heartworm (Dirofilaria immitis) infection in cats. Vet. Parasitol. 133:267–275 [DOI] [PubMed] [Google Scholar]
- 28. Rishniw M, et al. 2006. Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet. Parasitol. 135:303–314 [DOI] [PubMed] [Google Scholar]
- 29. Rolain JM, Franc M, Davoust B, Raoult D. 2003. Molecular detection of Bartonella quintana, B. koehlerae, B. henselae, B. clarridgeiae, Rickettsia felis, and Wolbachia pipientis in cat fleas, France. Emerg. Infect. Dis. 9:338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rossi MI, et al. 2010. Detection of Wolbachia DNA in blood from dogs infected with Dirofilaria immitis. Exp. Parasitol. 126:270–272 [DOI] [PubMed] [Google Scholar]
- 31. Saint André A, et al. 2002. The role of endosymbiotic Wolbachia bacteria in the pathogenesis of river blindness. Science 295:1892–1895 [DOI] [PubMed] [Google Scholar]
- 32. Simoncini L, et al. 2001. Real-time PCR for quantification of the bacterial endosymbionts (Wolbachia) of filarial nematodes. Parasitologia 43:173–178 [PubMed] [Google Scholar]
- 33. Traversa D, et al. 2010. Autochthonous foci of canine and feline infections by Dirofilaria immitis and Dirofilaria repens in central Italy. Vet. Parasitol. 169:128–132 [DOI] [PubMed] [Google Scholar]
- 34. Unver A, Rikihisa Y, Kawahara M, Yamamoto S. 2003. Analysis of 16S rRNA gene sequences of Ehrlichia canis Anaplasma platys and Wolbachia species from canine blood in Japan. Ann. N. Y. Acad. Sci. 990:692–698 [DOI] [PubMed] [Google Scholar]
- 35. Venco L, Genchi C, Genchi M, Grandi G, Kramer LH. 2008. Clinical evolution and radiographic findings of feline heartworm infection in asymptomatic cats. Vet. Parasitol. 158:232–237 [DOI] [PubMed] [Google Scholar]
- 36. Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6:741–751 [DOI] [PubMed] [Google Scholar]