Abstract
There was a high percentage of macrolide resistance in Mycoplasma pneumoniae clinical isolates in China. The genetic relatedness of macrolide-resistant M. pneunomiae strains was investigated using the multilocus variable-number tandem-repeat assay (MLVA). Among 152 M. pneunomiae isolates, the 137 macrolide-resistant strains were clustered into 15 MLVA types, indicating that the high macrolide resistance rate in M. pneumoniae is a result of the dissemination of the multiple resistant clones.
TEXT
Mycoplasma pneumoniae is one of the most common etiological agents of community-acquired respiratory tract infections, and macrolides are the first-choice antibiotics for treatment of M. pneumoniae infections (14). Prior to the year 2000, macrolide resistance in M. pneunomiae was rare around the world. However, several studies from China have described high percentages of macrolide-resistant isolates of M. pneumoniae, ranging from 69% to 92%, obtained from both children and adults between 2003 and 2010 (1, 7, 8, 15). While it is apparent that macrolide-resistant M. pneumoniae isolates are spreading rapidly in certain parts of Asia, the epidemiological mechanism is still unknown. There was no clear association between the macrolide-resistant M. pneumoniae isolates and the P1 restriction fragment length polymorphism (RFLP) subtypes or pulsed-field gel electrophoresis (PFGE) subtypes in previous studies (1).
Different typing methods such as conventional PCR (6), restriction fragment length polymorphism (RFLP) (3), and pulsed-field gel electrophoresis (3) were developed to differentiate subtypes of M. pneumoniae. However, M. pneumoniae is a highly homogeneous organism, and all of these methods have a limited power of discrimination. Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA), which determines the number of tandem repeat (TR) sequences at different loci in a bacterial genome (13), is highly discriminative and has been successfully applied for typing of M. pneumoniae clinical isolates, including the macrolide-resistant strains, in Europe (4, 5). In this study, MLVA was used to determine the genetic relationships of the 137 macrolide-resistant M. pneumoniae strains from Shanghai, China, over a 5-year period.
M. pneumoniae strains.
One hundred fifty-two unique M. pneumoniae clinical isolates were obtained from bronchial aspirates of children with lower respiratory infections in Shanghai, China, from December 2005 to July 2009. The distributions of the isolates over the period of these 5 years was as follows: a total of 3 to 10 strains were collected in March, June, July, and August, 11 to 16 strains were collected in February, October, December, and September, and 20 to 21 strains were collected in January, April, and May. M. pneumoniae culture, PCR amplification of the P1 gene for species identification, and antimicrobial susceptibility testing were carried out as described previously (7). The CLSI breakpoint (erythromycin resistance, ≥1 μg/ml) was used to define an isolate as resistant (2). A total of 137 macrolide-resistant isolates (90.1%) were identified from our collection.
PCR-RFLP analysis.
P1 gene PCR-RFLP typing of all 152 clinical strains of M. pneumoniae was performed as previously described (7). M. pneumoniae clinical isolates could only be classified into two types by the PCR-RFLP typing method. Of 152 clinical strains, 138 strains belonged to type I, 12 were type II, and 2 could not be identified.
VNTR locus selection and analysis.
A total of 5 variable-number tandem-repeat (VNTR) loci with core sequences of >9 bp were selected according to the website (http://minisatellites.u-psud.fr/ASPSamp/base_ms/bact.php; see Table S1 in the supplemental material), and the corresponding PCR primers that were the same as those previously described were used for MLVA typing (4). PCRs were carried out in a 50-μl volume containing 25 μl of 2× GC PCR buffer (TaKaRa Biotechnology, Dalian, China), 200 μM each of the four deoxynucleoside triphosphates (dNTPs), 10 μM each primer set, 0.5 U TaKaRa La Taq polymerase (TaKaRa Biotechnology, Dalian, China), 5 μl of template DNA, and 9.5 μl of water. The PCR conditions were as follows: initial denaturation at 95°C for 10 min and then 40 cycles of 95°C for 1 min, 53°C for 1 min, and 72°C for 1 min, followed by a final polymerase extension step at 72°C for 10 min. All PCR products were sequenced, and copy numbers were calculated according to the number of repeats for each locus. Two reference strains of M. pneumoniae, M129 (ATCC 29342) and FH (ATCC 15531), were also included as the positive control for the MLVA.
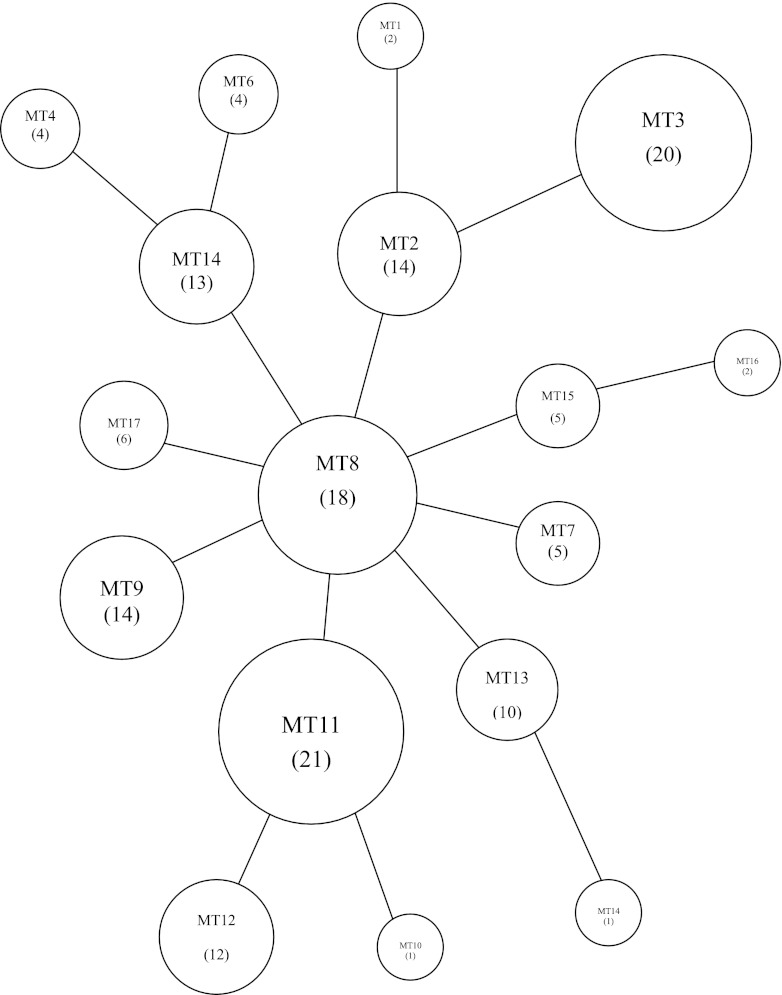
All data were input into BioNumerics version 6.5 software (Applied Maths, Belgium), and then cluster analysis and minimal spanning tree (MST) analysis were performed. The 152 M. pneumoniae clinical isolates were divided into 17 MLVA types (Table 1). Of these 152 isolates, 137 macrolide-resistant strains formed 15 MLVA types. There were 8 most common MLVA types in the resistant isolates, each containing more than 10 resistant strains and accounting for 84% of the total 137 isolates (Table 1). There was no evidence of resistant clonal outbreaks according to diversified MLVA patterns during the 5-year study period. The diversified pattern was also observed in isolates collected in the same time period: e.g., 11 resistant isolates from September 2008, 15 resistant strains from April 2009, and 21 resistant strains from May 2009 were clustered into 7, 8, and 9 MLVA types, respectively. MST population modeling also indicated the diversity among the tested isolates, and no dominant MLVA type could be determined (Fig. 1).
Table 1.
MLVA type distribution of 152 M. pneumoniae clinical isolates
| MLVA typea | No. of isolates: |
Distribution by yr (no. of resistant strains) |
|||||
|---|---|---|---|---|---|---|---|
| Macrolide susceptibleb | Macrolide resistantc | 2005 | 2006 | 2007 | 2008 | 2009 | |
| 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 14 | 0 | 0 | 0 | 3 | 11 |
| 3 | 3 | 17 | 0 | 2 | 6 | 0 | 9 |
| 4 | 1 | 3 | 3 | 0 | 0 | 0 | 0 |
| 5 | 0 | 13 | 0 | 1 | 1 | 8 | 3 |
| 6 | 0 | 4 | 0 | 0 | 1 | 1 | 2 |
| 7 | 4 | 1 | 0 | 0 | 0 | 0 | 1 |
| 8 | 1 | 17 | 0 | 3 | 3 | 2 | 9 |
| 9 | 1 | 13 | 0 | 2 | 3 | 5 | 3 |
| 10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 1 | 20 | 0 | 3 | 5 | 8 | 4 |
| 12 | 1 | 11 | 0 | 1 | 2 | 5 | 3 |
| 13 | 0 | 10 | 0 | 0 | 2 | 4 | 4 |
| 14 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| 15 | 0 | 5 | 0 | 1 | 1 | 2 | 1 |
| 16 | 0 | 2 | 0 | 0 | 0 | 1 | 1 |
| 17 | 0 | 6 | 0 | 0 | 0 | 3 | 3 |
| Total | 15 | 137 | 3 | 13 | 24 | 43 | 54 |
The MLVA types of two reference strains were not included in this table. M. pneumoniae reference strain M129 belonged to MLVA type 13, and M. pneumoniae reference strain FH alone showed a separate MLVA type.
Susceptible, erythromycin MIC of ≤0.5μg/ml.
Resistant, erythromycin MIC of ≥1μg/ml. All erythromycin-resistant strains had a MIC of ≥64μg/ml in the present study.
Fig 1.
Minimal spanning tree of the MLVA profiles of 152 M. pneumoniae isolates. Each circle denotes a particular MLVA type (MT) indicated by a number corresponding to the genotype in parentheses. The size of the circle is proportional to the number of isolates belonging to the indicated MLVA genotype. Neighboring genotypes have the same distance.
Diversity of M. pneumoniae strains.
The Hunter-Gaston index (HGI) of the MLVA typing method was calculated. The formula was as follows: HGI = 1 − ∑[nj (nj − 1)]/[N (N − 1)], where N refers to the total number of experimental strains and nj is the number of strains belonging. Diversity indices of the 5 loci were between 0.000 and 0.812 (see Table S2 in the supplemental material). Locus VNTR1 had a higher HGI than other loci. The numbers of alleles of each of the 5 loci ranged between 1 and 7. Locus VNTR1 had the largest number of alleles (n = 7). MST modeling and cluster analysis showed the diversity among the tested isolates, and no dominant MLVA type could not be determined (Fig. 1).
Our earlier studies indicated that more than 90% of M. pneumoniae strains isolated in Shanghai, China, were resistant to macrolides (7, 8). It is known that most of the macrolide-resistant M. pneumoniae strains are caused by point mutations in 23S rRNA (1, 7, 8, 15). This study used MLVA as an individual identification method to type a large number of macrolide-resistant M. pneunomiae isolates to determine the origin of the resistant isolates, whether from a single colony or from multiple clones. In recent years, typing schemes based on MLVA have been designed and implemented for a number of microorganisms of public health importance (9, 10, 11, 12). Degrange et al. first applied this MLVA method in typing M. pneumoniae isolates and were able to group 265 strains (12 strains were macrolide resistant) into 26 MLVA types (4). The discrimination potential of MLVA was shown to be much higher than the typing approach based on the differences in the P1-encoding gene. However, MLVA analysis of the limited isolates from France and Japan did not reveal any link between a particular MLVA type and macrolide resistance (4). The present study represents the largest MLVA typing worldwide of the macrolide-resistant isolates so far. The 137 macrolide-resistant isolates were clustered into 15 MLVA types, and none of them could be determined as dominant, indicating both the absence of a particular emerging macrolide-resistant clone and that the high rate of macrolide resistance in China resulted from the dissemination of multiple resistant clones, instead of the spreading of a single colony.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 81000753).
We thank very much Ken B. Waites and Li Xiao for critical review of the manuscript.
Footnotes
Published ahead of print 30 May 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Cao B, et al. 2010. High prevalence of macrolide resistance in Mycoplasma pneumoniae isolates from adult and adolescent patients with respiratory tract infection in China. Clin. Infect. Dis. 51:189–194 [DOI] [PubMed] [Google Scholar]
- 2. Clinical and Laboratory Standards Institute 2011. Methods for antimicrobial susceptibility testing for human mycoplasmas; approved guideline M43-A. Clinical and Laboratory Standards Institute, Wayne, PA: [PubMed] [Google Scholar]
- 3. Cousin-Allery A, et al. 2000. Molecular typing of Mycoplasma pneumoniae strains by PCR-based methods and pulsed-field gel electrophoresis. Application to French and Danish isolates. Epidemiol. Infect. 124:103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Degrange S, et al. 2009. Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J. Clin. Microbiol. 47:914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dumke R, Jacobs E. 2011. Culture-independent multi-locus variable-number tandem-repeat analysis (MLVA) of Mycoplasma pneumoniae. J. Microbiol. Methods 86:393–396 [DOI] [PubMed] [Google Scholar]
- 6. Kong F, Gordon S, Gilbert GL. 2000. Rapid-cycle PCR for detection and typing of Mycoplasma pneumoniae in clinical specimens. J. Clin. Microbiol. 38:4256–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, et al. 2009. Antimicrobial susceptibility of Mycoplasma pneumoniae isolates and molecular analysis of macrolide-resistant strains from Shanghai, China. Antimicrob. Agents Chemother. 53:2160–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y, et al. 2010. Characterization of macrolide resistance in Mycoplasma pneumoniae isolated from children in Shanghai, China. Diagn. Microbiol. Infect. Dis. 67:355–358 [DOI] [PubMed] [Google Scholar]
- 9. Schouls LM, et al. 2005. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 43:2741–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schouls LM, et al. 2009. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One 4:e5082 doi:10.1371/journal.pone.0005082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schouls LM, van der Ende A, Damen M, van de Pol I. 2006. Multiple-locus variable-number tandem repeat analysis of Neisseria meningitidis yields groupings similar to those obtained by multilocus sequence typing. J. Clin. Microbiol. 44:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Top J, Schouls LM, Bonten MJ, Willems RJ. 2004. Multiple-locus variable number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 42:4503–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vergnaud G, Pourcel C. 2006. Multiple locus VNTR (variable number of tandem repeat) analysis (MLVA), p 83–104 In Stackebrandt E. (ed), Molecular identification, systematics and population structure of prokaryotes. Springer-Verlag, Berlin, Germany [Google Scholar]
- 14. Waites KB, Talkington DF. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xin DL, et al. 2009. Molecular mechanisms of macrolide resistance in clinical isolates of Mycoplasma pneumoniae from China. Antimicrob. Agents Chemother. 53:2158–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.