Abstract
Corynebacterium tuberculostearicum is a lipophilic corynebacterium validly characterized in 2004. We provide clinical information on 18 patients from whom this organism was isolated. The majority of the patients were hospitalized and had a history of prolonged treatment with broad-spectrum antimicrobials. In 7 (38.9%) of the 18 cases, the isolates were found to be clinically relevant. The present report also includes detailed data on the biochemical and molecular identification of C. tuberculostearicum, as well as its identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Our data demonstrate that routine biochemical tests do not provide reliable identification of C. tuberculostearicum. MALDI-TOF MS represents a helpful tool for the identification of this species, since all of the strains matched C. tuberculostearicum as the first choice and 58.3% (7/12) of the strains processed with the full extraction protocol generated scores of >2.000. Nevertheless, partial 16S rRNA gene sequencing still represents the gold standard for the identification of this species. Due to the challenging identification of C. tuberculostearicum, we presume that this organism is often misidentified and its clinical relevance is underestimated. The antimicrobial susceptibility profile of C. tuberculostearicum presented here reveals that 14 (87.5%) of the 16 strains analyzed exhibited multidrug resistance.
INTRODUCTION
Gram-positive, aerobically growing rods belonging to the genus Corynebacterium colonize the skin and mucosal surfaces of humans. They are frequently isolated from clinical specimens. Interpretation of their clinical relevance is often difficult. Lipophilic corynebacteria are a particularly relevant subgroup of corynebacteria since they might be involved in infections of hospitalized patients and often show multiresistance to antimicrobials (9).
The taxonomic characterization of C. tuberculostearicum is intricate and is described here briefly. In 1984, Brown et al. studied 16 so-called leprosy-derived coryneform (LDC) strains and named the isolates “C. tuberculostearicum” because their fatty acid profile comprised tuberculostearic acid (3). Riegel et al. then showed that “C. tuberculostearicum” strain LDC8, as well as three strains of “C. pseudogenitalium,” another not validly named lipophilic Corynebacterium species, and the reference strain of Centers for Disease Control and Prevention (CDC) coryneform group G-2 (CDC G5840) clustered within one group (genomospecies I) based on DNA-DNA-hybridization analysis (15). In 2004, the species C. tuberculostearicum was characterized validly. The authors demonstrated that “C. pseudogenitalium” strain ATCC 33035 also represents C. tuberculostearicum and showed that this species forms a distinct phylogenetic lineage together with C. accolens and C. macginleyi (8). Unfortunately, strains of CDC group G-2 were not included in this taxonomic study, which would clarify the phylogenetic relationship to C. tuberculostearicum (10). In the 1990s, several clinically relevant isolates of CDC group G-2 showing partial multiple antibiotic resistance were described (16, 23, 24). Up to now, there has been only one publication in the recent literature describing the isolation of C. tuberculostearicum from human infections. In 2002, Paviour and colleagues reported on the isolation of corynebacterial species, including C. tuberculostearicum, from pus from women with mastitis, implicating their possible involvement in cases of inflammatory breast disease (14).
In this report, we provide clinical and microbiological information on 18 patients from whom C. tuberculostearicum was isolated and elaborate on its clinical significance and involvement in the infectious process. Furthermore, we present detailed data on its microbiological identification, including biochemistry, 16S rRNA gene sequencing, and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis, as well as the antimicrobial susceptibility profiles of 16 strains in our collection.
MATERIALS AND METHODS
Patients, clinical isolates, and culture conditions.
Fifteen isolates of C. tuberculostearicum were recovered from inpatients at three hospitals in Basel, Switzerland, from 2007 to 2011, i.e., the University Hospital (13 patients), the Bruderholz Hospital (2 patients), and the University Children's Hospital (1 patient). Two isolates originated from ambulatory patients at a private general practice in Zürich. C. tuberculostearicum was isolated from 8 biopsy specimens, 3 aspirates, 3 deep-wound swabs, 2 superficial-wound swabs, and 2 urine samples (Table 1). The antibiotic resistance testing and MALDI-TOF MS analysis of 16 strains in our collection were done retrospectively. Clinical specimens were cultured on Columbia agar supplemented with 5% sheep blood (BD Diagnostic Systems, Allschwil, Switzerland), while bone biopsy specimens were cultivated by direct placement into fluid thioglycolate medium (BD Diagnostic Systems). Plates were incubated aerobically at 37°C and examined for growth on the first and second days. Bone biopsy specimens in enrichment broth were cultivated aerobically at 37°C for 6 days with daily visual monitoring of growth.
Table 1.
Microbiological and clinical findings on patients from whom C. tuberculostearicum was isolated
| Patient no. | Age/sexa | Specimen type | Gram stain (amt)b | Culture (amt)b | Other materials (amt)b | Clinical presentation and underlying disease | Pre-existing antimicrobial therapy | Clinical relevance of C. tuberculostearicume |
|---|---|---|---|---|---|---|---|---|
| 1 | 52/M | Biopsy specimen (chest wall) | Cell debris (++), Gram-positive diplococci (+), Gram-negative rods (+) | C. tuberculostearicum (++), CNSc (+) | C. tuberculostearicum in two other biopsy specimens (++) and one aspirate (++) | Osteomyelitis of chest wall and ribs following polytrauma with multiple rib fractures and empyema with Enterobacter cloacae | Cefepime | Relevant/relevant |
| 2 | 81/M | Bone biopsy specimen (sternum) | Not done | C. tuberculostearicum detected | C. tuberculostearicum detected in VACd sponge and in one bone biopsy specimen from the sternum | Deep sternal wound infection with osteomyelitis 3 weeks after aortocoronary bypass operation | None | Relevant/relevant |
| 3 | 63/M | Urine from extended-dwell catheter | Not done | C. tuberculostearicum 105 CFU/ml | No other materials analyzed | Persistent fever without clinical focus 3 weeks after meningoencephalitis with Streptococcus pneumoniae | Meropenem, ceftriaxone | Not considered relevant/relevant |
| 4 | 64/M | Biopsy specimen (wound) | Erythrocytes (++), cell debris (+), no bacteria | C. tuberculostearicum (++), CNSc (++) | C. tuberculostearicum in two other biopsy specimens (+ and ++) together with CNSc (+ and ++, respectively) | Superinfection of sternum and sternoclavicular joint after Staphylococcus aureus sepsis with infective endocarditis of mitral valve and septic sternoclavicular arthritis | Flucloxacillin, penicillin | Relevant/relevant |
| 5 | 43/F | Biopsy specimen (pericostal) | Erythrocytes (+++), cell debris (++), no bacteria | C. tuberculostearicum (+++) | C. tuberculostearicum in two other biopsy specimens (+), pleura aspirate (++), and bone biopsy specimen (detected) | Superinfection of empyema of chest wall after pneumococcal pneumonia and recurrent empyema with detection of Aspergillus fumigatus | Piperacillin-tazobactam, voriconazole | Relevant/relevant |
| 6 | 75/M | Deep-wound swab (sternum) | Leukocytes (+), cell debris (+), Gram-positive cocci (+), Gram-positive coryneform rods (++) | C. tuberculostearicum (+++) | No other materials analyzed | Persistent secretion of sternal wound in patient treated until recently for deep sternal wound infection with coagulase-negative staphylococci after aortocoronary bypass and mitral valve replacement | None | Not considered relevant/relevant |
| 7 | 61/F | Aspirate (abdomen) | Not known | C. tuberculostearicum (from enrichment broth), Candida albicans (quantity not known) | Not known | Perforated gastric ulcer with polymicrobial peritonitis and sepsis with Candida albicans | Not known | Not considered relevant/not considered relevant |
| 8 | 72/M | Hematoma aspirate (knee) | Not known | C. tuberculostearicum from enrichment broth, Staphylococcus epidermidis (quantity not known) | Not known | Infected hematoma and wound healing disorder following total knee prosthesis | Not known | Not considered relevant/not considered relevant |
| 9 | 18/F | Biopsy specimen (toe tissue) | Cell debris (+), epithelial cells (+), Gram-positive cocci (+) | C. tuberculostearicum (+) | No other materials with C. tuberculostearicum | Not known | Not known | Not known |
| 10 | 50/F | Biopsy specimen (phalanx) | Cell debris (+), no bacteria | C. tuberculostearicum (+), CNSc (++) | No other materials with C. tuberculostearicum | Pseudoarthrosis 7 months after a fracture of proximal phalanx IV of left foot with posttraumatic plegia of the foot | None | Possibly relevant/possibly relevant |
| 11 | 47/F | Deep-wound swab (abdomen) | Leukocytes (++), erythrocytes (++), no bacteria | C. tuberculostearicum (+), Klebsiella pneumoniae (from enrichment broth) | No other materials analyzed | Tertiary peritonitis after septic shock due to abdominal perforation and peritonitis with Clostridium perfringens, Klebsiella pneumoniae, and Escherichia coli following resection of sigma cancer | Imipenem, amoxicillin-clavulanic acid | Not considered relevant/possibly relevant |
| 12 | 32/M | Wound swab (genital ulcus) | Epithelial cells (+), cell debris (+), Gram-positive cocci (+) | C. tuberculostearicum (++), Staphylococcus epidermidis (++) | No other materials analyzed | Recurrent genital ulcerations | Not known | Not known |
| 13 | 24/M | Urethral swab | Leukocytes (+), cell debris (+), Gram-positive cocci (+) | C. tuberculostearicum (++), Corynebacterium glucuronolyticum (++), Ureaplasma urealyticum (+) | No other materials analyzed | Urethritis caused by Chlamydia trachomatis | Not known | Not considered relevant/not considered relevant |
| 14 | 72/M | Bone biopsy specimen (fibula) | Not done | C. tuberculostearicum detected, Micrococcus group detected | No other materials with C. tuberculostearicum | Diabetes mellitus with Charcot foot deformation and chronic polymicrobial osteomyelitis with Staphylococcus aureus, Enterobacter cloacae, Pseudomonas aeruginosa, Streptococcus agalactiae, Enterococcus faecalis, Achromobacter xylosoxidans | Ciprofloxacin, trimethoprim-sulfamethoxazole, amoxicillin | Not considered relevant/not considered relevant |
| 15 | 67/F | Midstream urine | Not done | C. tuberculostearicum at 104 CFU/ml | C. tuberculostearicum at 104 CFU/ml in urine sample together with yeast at 103 CFU/ml | Febrile episode without clinical focus in patient treated with chemotherapy and antibiotics for poststenotic pneumonia due to small cell lung cancer | Piperacillin-tazobactam, amoxicillin-clavulanic acid | Not considered relevant/relevant |
| 16 | 54/F | Aspirate (pleura) | Leukocytes (+), erythrocytes (+++), no bacteria | C. tuberculostearicum (++) | C. tuberculostearicum in one aspirate from pleura (+) and one pleura biopsy specimen (+) | Superinfection of empyema with Streptococcus pneumoniae 2 weeks after initial operation | Piperacillin-tazobactam, vancomycin | Relevant/relevant |
| 17 | 75/M | Deep-wound swab (pericardium) | Leukocytes (++), erythrocytes (++), no bacteria | C. tuberculostearicum (+) | C. tuberculostearicum in two other biopsy specimens from pleura and sternum (+ and ++) and deep-wound swab from sternum (+) | Complicated course after aortic valve replacement with prolonged duration of heart-lung machine, multiple operative revisions, extracorporeal membrane oxygenation | Piperacillin-tazobactam | Relevant/relevant |
| 18 | 51/M | Biopsy specimen (rib cartilage) | Leukocytes (+), erythrocytes (+), no bacteria | C. tuberculostearicum (++) | C. tuberculostearicum in five other biopsy specimens from thorax (+) | Osteomyelitis of chest wall and ribs following empyema with unknown pathogen 1 month after pneumonectomy for non-small-cell lung cancer | Piperacillin-tazobactam | Relevant/relevant |
M, male; F, female.
Amounts: +, detached; ++, moderate; +++, numerous. CFU, colony-forming units.
CNS, coagulase-negative staphylococci.
VAC, vacuum-assisted closure.
CDC criteria/criteria of Funke and Bernard.
Criteria for estimation of the clinical significance of coryneform bacteria.
The C. tuberculostearicum infections were defined according to the criteria of the CDC (11). The clinical significance of C. tuberculostearicum isolates was also estimated on the basis of the criteria defined by Funke and Bernard (10).
Phenotypic identification.
Primary identification of coryneform bacteria was performed by colony morphology, Gram staining, and catalase reaction. Additionally, a CAMP test with Staphylococcus aureus ATCC 25923 was done. Lipophilia was tested for by subculturing the isolates on blood plates with and without 1% Tween 80. Routine biochemical identification of corynebacterial strains was performed with the API Coryne System (bioMérieux, Geneva, Switzerland) according to the manufacturer's instructions.
16S rRNA gene sequencing and sequence comparison.
16S rRNA gene amplification and sequencing were performed using the Fast MicroSeq 500 16S rRNA gene Bacterial Identification Sequencing Kit (Applied Biosystems, Rotkreuz, Switzerland). The resulting DNA sequences were compared with the reference sequences of the MicroSeqID 500 software (version 2.1.), as well as sequences deposited in the GenBank database at the National Center for Biotechnology Information using the BLAST algorithm. Comparative analysis of the partial 16S rRNA gene sequences of 18 C. tuberculostearicum strains was performed with the MegAlign 6.1 sequence analysis software (Lasergene; DNAStar, Madison, WI) by ClustalW analysis. The phylogenetic tree was constructed using default parameters with the help of the online software tool ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) from the European Bioinformatics Institute of the European Molecular Biology Laboratory.
MALDI-TOF MS analysis.
Colonies of 12 C. tuberculostearicum strains grown overnight on blood agar plates with 1% Tween 80 were smeared onto a steel MSP 96 target plate (Bruker Daltonics GmbH, Bremen, Germany) and overlaid with 1 μl of a matrix consisting of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile–2.5% trifluoroacetic acid (normal protocol). The short extraction protocol was performed by the addition of 1 μl of 70% formic acid to the bacterial smears before application of the matrix. Full extraction was done by the ethanol-formic acid protocol as recommended by the manufacturer. Measurements were performed with a Microflex LT MALDI-TOF MS system and analyzed with the BioTyper 3.0 SR software. The degrees of spectral concordance between the isolates tested and strains deposited in the database were expressed as logarithmic identification scores and interpreted according to the manufacturer's instructions. Scores of ≥2.300 indicated highly probable identification to the species level, scores of 2.000 to 2.299 indicated probable identification to the species level, scores of 1.700 to 1.999 indicated probable identification to the genus level, and scores of <1.700 indicated no reliable identification.
Antimicrobial agent susceptibility testing.
MICs were determined with Etest strips (bioMérieux, Geneva, Switzerland) on Mueller-Hinton agar with blood (Heipha, Heidelberg, Germany) using inocula of 1 McFarland unit incubated in an atmosphere of 5% CO2 at 37°C for 48 h. Clinical and Laboratory Standards Institute (4) breakpoints were used for the interpretation of MICs of penicillin, ceftriaxone, cefepime, imipenem, erythromycin, clindamycin, tetracycline, gentamicin, ciprofloxacin, vancomycin, rifampin, linezolid, and daptomycin. Additionally, amoxicillin-clavulanic acid, piperacillin-tazobactam, cefuroxime, moxifloxacin, and tigecycline were interpreted by using the European Committee on Antimicrobial Susceptibility Testing (6) non-species-related breakpoints.
RESULTS
Clinical and microbiological findings.
Microbiological and clinical features of 18 patients from whom C. tuberculostearicum was isolated are presented in Table 1. C. tuberculostearicum was isolated from 8 biopsy specimens, 3 aspirates, 3 deep-wound swabs, 2 superficial-wound swabs, and 2 urine samples. The median age of the patients was 56 (range, 24 to 81) years. Fifteen of the 18 patients had a history of extensive surgery with long hospital stays, and a majority had undergone prolonged therapy with broad-spectrum antimicrobials (Table 1). Only 1 of the 12 direct Gram stains revealed Gram-positive coryneform rods. No Gram stain was made from bone biopsy specimens and urine samples. In 9 of 18 samples, C. tuberculostearicum was isolated in pure culture (including 2 urine samples), whereas in the remaining 9 samples, coagulase-negative staphylococci, Candida albicans, other corynebacteria, or Micrococcus group bacteria were isolated as concomitant organisms. In 8 of 18 cases, C. tuberculostearicum was recovered from multiple specimens. In the 10 remaining cases, C. tuberculostearicum was not isolated from other materials, no additional samples were examined, or no data were available. One urethral swab originating from a patient with urethritis was positive for Chlamydia trachomatis by PCR. Based on clinical criteria, C. tuberculostearicum was found to be an etiological agent of the surgical site infections of 7 patients (patients 1, 2, 4, 5, 16, 17, and 18 in Table 1), while the clinical relevance of the isolate from patient 10 could not be excluded (possibly relevant). In 8 cases, isolation of C. tuberculostearicum was found not to be clinically relevant, while in 2 remaining cases, no conclusion about clinical relevance could be reached due to the insufficiency of the clinical and microbiological data. Relevance estimated on the basis of the criteria of Funke and Bernard (10) was found to be in agreement with that estimated on the basis of the clinical CDC criteria (11) in all cases, except for patients 3, 6, and 15 (Table 1).
Phenotypic characteristics.
All isolated strains showed identical morphological characteristics. After 24 to 48 h of incubation, very small, convex, smooth, grayish colonies appeared on blood agar. Gram staining revealed coryneform Gram-positive rods. The catalase reaction was positive, but the CAMP test was negative. The lipophilia test was positive since the growth of the organism on 1% Tween 80-supplemented blood agar was better than its growth on an ordinary blood plate. The results of the biochemical identification of the strains with the API Coryne System are presented in Table 2. Identification probabilities and T values corresponded to good identification as C. macginleyi for 2 strains and as CDC group G for 1 strain, while 13 isolates matched C. macginleyi (6 isolates), C. jeikeium (2 isolates), and CDC group G (5 isolates) with scores for acceptable, good, very good, or excellent identification on the genus level. One isolate displayed a doubtful profile for identification as C. accolens, and one displayed low selectivity as C. jeikeium. The variable reactions in different strains leading to the score differences were nitrate reduction, pyrazinamidase, pyrrolidonyl arylamidase, alkaline phosphatase, and saccharose fermentation.
Table 2.
Identification of 18 C. tuberculostearicum strains by 16S rRNA gene sequencing, 18 strains by the API Coryne System,a and 12 strains by MALDI-TOF MS
| No. of strains | 16S rRNA gene sequencing | API Coryne |
Avg MALDI-TOF MS scoreb ± SD |
||||
|---|---|---|---|---|---|---|---|
| Numerical code | First choice listed (% IDd probability, T value) | ID interpretation | Normal protocol | Short extraction protocol | Full extraction protocol | ||
| 5 | C. tuberculostearicum | 4100305 | C. macginleyi (85.6, 0.79) | Good ID on genus level | 1.644 ± 0.31 | 1.917 ± 0.08 | 2.045 ± 0.07 |
| 2 | C. tuberculostearicum | 4100304 | C. jeikeium (59.2, 0.64) | Acceptable ID on genus level | NDc | ND | ND |
| 2 | C. tuberculostearicum | 5100304 | C. macginleyi (97.5, 0.79) | Good ID | 1.486 ± 0.05 | 1.863 ± 0.14 | 2.076 ± 0.10 |
| 2 | C. tuberculostearicum | 7100305 | CDC group G (83, 0.86) | Very good ID on genus level | 1.715 ± 0.37 | 1.82 ± 0.24 | 2.007 ± 0.11 |
| 2 | C. tuberculostearicum | 7100304 | CDC group G (43.5, 0.69) | Good ID on genus level | 1.633 ± 0.12 | 1.894 ± 0.04 | 1.955 ± 0.02 |
| 1 | C. tuberculostearicum | 6100305 | CDC group G (96.7, 0.98) | Good ID | 1.893 | 1.802 | 1.85 |
| 1 | C. tuberculostearicum | 2100305 | CDC group G (86.9, 1) | Excellent ID on genus level | ND | ND | ND |
| 1 | C. tuberculostearicum | 0100305 | C. macginleyi (84.3, 0.82) | Very good ID on genus level | ND | ND | ND |
| 1 | C. tuberculostearicum | 4000304 | C. accolens (90.5, 0.64) | Doubtful profile | ND | ND | ND |
| 1 | C. tuberculostearicum | 6100304 | C. jeikeium (60, 0.79) | Low selectivity | ND | ND | ND |
C. tuberculostearicum is not included in the API Coryne database.
Interpretation of score values: ≥2.300, highly probable identification to the species level; 2.000 to 2.299; probable identification to the species level, 1.700 to 1.999; probable identification to the genus level; <1.700, no reliable identification.
ND, not done.
ID, identification.
16S rRNA gene sequencing and sequence comparison.
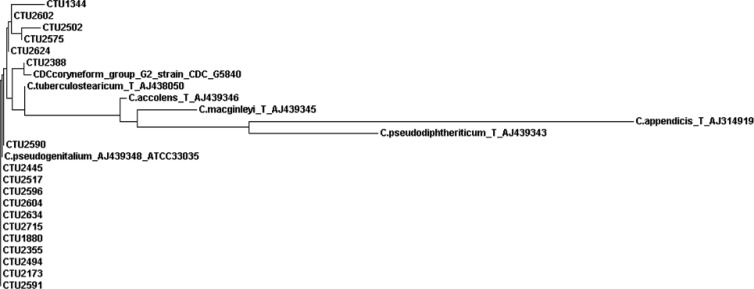
The 16S rRNA gene sequence comparison of 18 strains with the GenBank database revealed 100% identity with the C. tuberculostearicum type strain Medalle XT (= LDC-20T = CIP 107291T = CCUG 45418T = ATCC 35529T; GenBank accession no. NR_028975.1 or AJ438050), as well as 8 other deposited C. tuberculostearicum strains (accession no. AJ438042, AJ438043, AJ438044, AJ438045, AJ438046, AJ438047, AJ438049, and AJ438051) and C. pseudogenitalium ATCC 33035 (accession no. AJ439348) (8). Comparison with the MicroSeqID database matched C. tuberculostearicum with 100% identity. In addition, the sequence of CDC coryneform group G-2 strain ATCC 33035 (accession no. X84098) was 100% identical to our sequences. The second-best-matching Corynebacterium species in the GenBank and MicroSeqID databases was C. accolens, with 98% nucleotide identity. The alignment of the first third of the 16S rRNA gene sequence (positions 48 to 446) revealed 100% identity for all 18 strains, with the exception of 1 strain originating from patient number 7 (Table 1) showing a single nucleotide polymorphism at position 84 (G instead of A) compared to the type sequence NR_028975.1. In Fig. 1, our 18 partial sequences compared to the sequences of the C. tuberculostearicum type strain, “C. pseudogenitalium” strain ATCC 33035, CDC coryneform group G-2 strain CDC G5840, and other related taxa are presented in the form of a phylogenetic tree.
Fig 1.
Phylogenetic tree based on partial 16S rRNA gene sequences showing relationships between our 18 sequences (CTU) and related taxa.
MALDI-TOF MS analysis.
MALDI-TOF MS analysis of all 12 of the strains tested matched C. tuberculostearicum as the first choice. The highest scores were obtained by using the full extraction protocol. With 58.3% (7/12) of the strains, scores above 2.000 (range of probable identification to the species level) were achieved. The mean value of the scores obtained with the full extraction protocol was 2.013 ± 0.09. The short extraction protocol resulted in an average score of 1.878 ± 0.11, whereas 1 of the 12 strains was identified to the species level, 10 of the 12 strains were identified to the genus level, and for 1 of the 12 strains, the scores were consistent with no reliable identification. On the other hand, the mean value of the scores obtained with the normal protocol was 1.648 ± 0.25 and none of the 12 strains were identified to the species level. Six of the 12 strains were identified to the genus level, and for 6 of the 12 strains, the scores were consistent with no reliable identification (Table 2).
Susceptibility to antimicrobial agents.
The MICs for 16 of the strains investigated are presented in Table 3. All of the strains were sensitive to vancomycin, linezolid, and daptomycin. Eleven (68.7%) of the 16 strains were rifampin susceptible, and 13 (81.2%) of the 16 strains were tetracycline sensitive. For all of the strains tested, the tigecycline MICs were 0.5 mg/liter or higher, corresponding to intermediate and resistant phenotypes. Fourteen (87.5%) of the 16 strains were resistant to quinolones (ciprofloxacin and moxifloxacin). The MIC50s of all β-lactam antibiotics were >32 or >256 mg/liter, respectively. Ten (62.5%) of the 16 strains were cross resistant to all of the β-lactam antibiotics tested. On the other hand, 2 of the 16 strains tested were fully susceptible to β-lactams. Sensitivity to gentamicin was variable (6 strains resistant, 1 intermediate, and 9 sensitive).
Table 3.
MICs for 16 C. tuberculostearicum strains
| Antimicrobial | MICa range | MIC50a | MIC90a | No. (%) of sensitive strains |
|---|---|---|---|---|
| Penicillin | 0.125–>32 | >32 | >32 | 3 (18.7) |
| Amoxicillin-clavulanic acid | 0.125–>256 | >256 | >256 | 6 (37.5) |
| Piperacillin-tazobactam | 0.125–>256 | >256 | >256 | 3 (18.7) |
| Cefuroxime | 0.25–>256 | >256 | >256 | 3 (18.7) |
| Ceftriaxone | 1–>256 | >256 | >256 | 2 (12.5) |
| Cefepime | 0.5–>256 | >256 | >256 | 3 (18.7) |
| Imipenem | 0.03–>32 | >32 | >32 | 6 (37.5) |
| Erythromycin | 0.125–>256 | >256 | >256 | 3 (18.7) |
| Clindamycin | 0.5–>256 | >256 | >256 | 1 (6.2) |
| Tetracycline | 0.05–32 | 1 | 16 | 13 (81.2) |
| Tigecycline | 0.5–2 | 0.5 | 2 | 0 (0) |
| Gentamicin | <0.06–64 | 2 | 32 | 9 (56.2) |
| Ciprofloxacin | 0.125–>32 | >32 | >32 | 2 (12.5) |
| Moxifloxacin | 0.06–>32 | 4 | >32 | 2 (12.5) |
| Vancomycin | 0.5–2 | 1 | 1 | 16 (100) |
| Rifampin | 0.06–>32 | 0.125 | 32 | 11 (68.7) |
| Linezolid | 0.5–1 | 1 | 1 | 16 (100) |
| Daptomycin | 0.06–1 | 0.25 | 0.5 | 16 (100) |
MICs are in milligrams per liter.
DISCUSSION
C. tuberculostearicum is a lipophilic species (8) about which very little information is available in the current literature. Therefore, we set out to provide clinical and microbiological information, including extended profiles of antimicrobial susceptibility presented here for the first time.
The biochemical analysis of C. tuberculostearicum strains with the API Coryne System produced scores consistent with the identification of other lipophilic corynebacteria with variable probabilities and T values (Table 2). This is due to the fact that C. tuberculostearicum is not included in the API Coryne database. Furthermore, it has already been reported that the identification of lipophilic corynebacteria could be quite challenging because most species show identical phenotypic characteristics when investigated with routine microbiological methods (15, 22). Since biochemical identification with the API Coryne System still represents the most widely used identification tool in clinical microbiological laboratories, we presume that C. tuberculostearicum is commonly misidentified as some other lipophilic Corynebacterium species. Six of the 18 strains tested in our study yielded numerical codes consistent with CDC group G, showing again the close relatedness to C. tuberculostearicum (10). Therefore, isolation of lipophilic, catalase-positive, and CAMP-negative corynebacterial species with API scores identical to those shown in Table 2 should raise suspicion of the presence of C. tuberculostearicum.
The best-matching organism for all of the strains analyzed with MALDI-TOF MS was C. tuberculostearicum, but the scores obtained showed great variation when different protocols were applied (Table 2). The difficulty in obtaining reliable scores for the identification of Gram-positive bacteria is due to the structural properties of their cell wall (1, 18). This problem can be partially solved by using the full extraction protocol. MALDI-TOF MS identification of 58.3% (7/12) of the strains tested in this study yielded scores of >2.000, consistent with identification to the species level by using the full extraction protocol in comparison to only one strain with short extraction and none with the normal protocol. Therefore, we recommend the use of the full extraction protocol for the testing of suspicious strains even though it is a rather lengthy and more complex protocol to perform. In addition, our MALDI-TOF MS results are in accordance with data very recently obtained by Alatoom et al. by analyzing among 92 isolates of Corynebacterium species 5 C. tuberculostearicum strains with the same extraction protocol (2). Another reason for the difficulties in the identification of this species could be the fact that only two strains of C. tuberculostearicum are present in the current database provided by Bruker. It is likely that the reliability of the MALDI-TOF MS identification could be improved if more strains were added to the Biotyper database in the future. Hence, identification by means of 16S rRNA gene sequencing remains the gold standard for the identification of C. tuberculostearicum since 17 of the 18 strains tested revealed 100% identity with 10 strains of C. tuberculostearicum (including the type strain) deposited in the MicroSeq and GenBank databases.
The majority of the strains investigated (14/16) exhibited resistance to at least one antimicrobial agent in three or more antimicrobial categories, which makes them multiresistant organisms (12). Nevertheless, all of the strains investigated were susceptible to vancomycin (Table 3). These results are in concordance with previously published data on the multiple antimicrobial resistance of investigated CDC group G-2 isolates and, conversely, their 100% susceptibility to vancomycin (23, 24). Vancomycin may therefore represent the empirical therapeutic option for serious infection while awaiting results of susceptibility testing since vancomycin is the treatment of choice for other multiresistant corynebacteria, including C. jeikeium (13). Furthermore, all of the strains investigated were susceptible to daptomycin and linezolid. Ten (62.5%) of the 16 strains displayed cross resistance to all β-lactams, 4 of them (25%) showed variable sensitivity to β-lactams, and 2 of them (12.5%), originating from ambulatory patients, were completely sensitive to all of the β-lactams tested. Although the mechanisms of β-lactam resistance in corynebacteria has not been widely studied, it is generally thought that resistance is due to decreased outer membrane permeability to or affinity for these antibiotics (17). To date, β-lactamases have been found only in coryneform bacteria of the genus Brevibacterium (17). For all 16 of the strains tested, the tigecycline MICs ranged from 0.5 to 2 mg/liter, consistent with intermediate or resistant phenotypes. This is in contrast to the data presented by Fernandez-Roblas and colleagues, who recommended tigecycline as a good alternative for infections caused by non-C. diphtheriae corynebacteria (7). On the basis of the results presented in this study, we are of the opinion that tigecycline does not represent a good therapeutic option for the treatment of C. tuberculostearicum infections. Taking these variable susceptibility patterns into consideration, antimicrobial susceptibility testing of every strain isolated should be performed.
It has already been reported in the literature that a prolonged hospital stay, treatment with broad-spectrum antibiotics, and impaired skin integrity are risk factors for the development of infection with the multiresistant, lipophilic species C. jeikeium (5, 13). Likewise, the multiresistant strains investigated in our study originated from patients with extended hospital stays, undergoing invasive surgical interventions, or with serious underlying comorbidities. The majority of these patients were treated with different broad-spectrum antimicrobials (Table 1). According to the clinical criteria defined by the CDC, our findings show a clear association of C. tuberculostearicum with the infectious process in 7 of the 18 clinical cases presented in this study (patients 1, 2, 4, 5, 16, 17, and 18 in Table 1), where this bacterium was involved in the infection of a surgical site with osteomyelitis. C. tuberculostearicum isolated in pure culture from the urine of patients 3 and 15 could be considered relevant based on the criteria described by Funke and Bernard (10). Since no clinical manifestation was observed in these two patients, the bacteriological cultures positive for C. tuberculostearicum could therefore be interpreted as asymptomatic bacteriuria. The isolate from patient 6 could be interpreted as significant on the basis of the Funke and Bernard criteria due to the great quantity of C. tuberculostearicum bacteria that grew in culture and their presence in the direct Gram stain. The clinical significance of C. tuberculostearicum isolates originating from patients 9 and 12 is questionable because limited patient data are available and none of the clinical criteria or the criteria described by Funke and Bernard (10) were met. Therefore, it is probable that C. tuberculostearicum may be found as a frequent colonizer on the skin of hospitalized patients, causing or not causing infection, which has already been reported for other lipophilic and potentially multiresistant corynebacteria like C. jeikeium and C. urealyticum (9, 19, 20, 21).
Interestingly, the two strains originating from patients 12 and 13, who presented at a private general practice because of urethritis and genital ulceration, were sensitive to all of the antimicrobials tested except macrolides. Therefore, it is possible that community-acquired strains probably exhibit susceptible phenotypes, in contrast to hospital-acquired strains, but this observation needs further confirmation.
In summary, we strongly suspect that the rate of isolation of C. tuberculostearicum from human clinical samples is underreported since this species is not included in the API Coryne database and is therefore systematically misidentified as other lipophilic corynebacteria such as C. macginleyi or C. jeikeium. The intricate taxonomy of this species, including the probable synonym CDC coryneform group G-2, may contribute to this situation. MALDI-TOF MS proved to be a helpful tool for the identification of C. tuberculostearicum using the full extraction protocol. It is likely that the reliability of identification by MALDI-TOF MS could be improved if more strains were added to the Biotyper database. Currently, the most reliable identification to the species level can be achieved by partial 16S rRNA gene sequencing. Multiple-antibiotic exposure and extended hospital stays are risk factors for colonization and infection with C. tuberculostearicum since most strains exhibit multiresistance.
ACKNOWLEDGMENTS
We thank Moritz Grubenmann and Biljana Savic of the Laborgemeinschaft 1 diagnostic laboratory in Zürich for kindly providing two C. tuberculostearicum strains. We are thankful to Michèle Ackermann for her help with MALDI-TOF MS analysis. We are also grateful to Susanne Graf and Sebastian Wirz for providing clinical information on two patients hospitalized at the Bruderholz Hospital in Basel.
Footnotes
Published ahead of print 16 May 2012
REFERENCES
- 1. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alatoom AA, Cazanave CJ, Cunningham SA, Ihde SM, Patel R. 2012. Identification of non-diphtheriae Corynebacterium by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:160–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown S, Lanéelle MA, Asselineau J, Barksdale L. 1984. Description of Corynebacterium tuberculostearicum sp. nov., a leprosy-derived Corynebacterium. Ann. Microbiol. (Paris) 135B:251–267 [DOI] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing, twenty-first informational supplement M100-S21, vol. 31, no. 1 Clinical and Laboratory Standards Institute; Wayne, PA [Google Scholar]
- 5. Coyle MB, Lipsky BA. 1990. Coryneform bacteria in infectious diseases: clinical and laboratory aspects. Clin. Microbiol. Rev. 3:227–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. European Committee on Antimicrobial Susceptibility Testing 2011. Breakpoint tables for interpretation of MICs and zone diameters. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden [Google Scholar]
- 7. Fernandez-Roblas R, et al. 2009. In vitro activity of tigecycline and 10 other antimicrobials against clinical isolates of the genus Corynebacterium. Int. J. Antimicrob. Agents 33:453–455 [DOI] [PubMed] [Google Scholar]
- 8. Feurer C, et al. 2004. Taxonomic characterization of nine strains isolated from clinical and environmental specimens, and proposal of Corynebacterium tuberculostearicum sp. nov. Int. J. Syst. Evol. Microbiol. 54:1055–1061 [DOI] [PubMed] [Google Scholar]
- 9. Funke G, von Graevenitz A, Clarridge JE, Bernard KA. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Funke G, Bernard KA. 2011. Coryneform gram-positive rods, p 413–442 In Versalovic J, et al. (ed), Manual of clinical microbiology 10th ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 11. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. 1988. CDC definitions for nosocomial infections. Am. J. Infect. Control 16:128–140 [DOI] [PubMed] [Google Scholar]
- 12. Magiorakos AP, et al. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281 [DOI] [PubMed] [Google Scholar]
- 13. Meyer DK, Reboli AC. 2005. Other coryneform bacteria and Rhodococcus, p 2465–2478 In Mandell GL, Bennett JE, Dolin R. (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 6th ed, vol 2 Churchill Livingstone; Philadelphia, PA [Google Scholar]
- 14. Paviour S, et al. 2002. Corynebacterium species isolated from patients with mastitis. Clin. Infect. Dis. 35:1434–1440 [DOI] [PubMed] [Google Scholar]
- 15. Riegel P, et al. 1995. Genomic diversity and phylogenetic relationships among lipid-requiring diphtheroids from humans and characterization of Corynebacterium macginleyi sp. nov. Int. J. Syst. Bacteriol. 45:128–133 [DOI] [PubMed] [Google Scholar]
- 16. Riegel P, Ruimy R, Christen R, Monteil H. 1996. Species identities and antimicrobial susceptibilities of corynebacteria isolated from various clinical sources. Eur. J. Clin. Microbiol. Infect. Dis. 15:657–662 [DOI] [PubMed] [Google Scholar]
- 17. Riegel P. 2010. Corynebacteria, p 379–388 In Courvalin P, Leclercq R, Rice LB. (ed), Antibiogram. ASM Press, Washington, DC [Google Scholar]
- 18. Smole SC, King LA, Leopold PE, Arbeit RD. 2002. Sample preparation of Gram-positive bacteria for identification by matrix assisted laser desorption/ionization time-of-flight. J. Microbiol. Methods 48:107–115 [DOI] [PubMed] [Google Scholar]
- 19. Soriano F, Rodriguez-Tudela JL, Fernández-Roblas R, Aguado JM, Santamaría M. 1988. Skin colonization by Corynebacterium groups D2 and JK in hospitalized patients. J. Clin. Microbiol. 26:1878–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soriano F, Aguado JM, Ponte C, Fernandez-Roblas R, Rodriguez-Tudela JL. 1990. Urinary tract infection caused by Corynebacterium group D2: report of 82 cases and review. Rev. Infect. Dis. 12:1019–1034 [DOI] [PubMed] [Google Scholar]
- 21. Telander B, Lerner R, Palmblad J, Ringertz O. 1988. Corynebacterium group JK in a hematological ward: infections, colonization and environmental contamination. Scand. J. Infect. Dis. 20:55–61 [DOI] [PubMed] [Google Scholar]
- 22. Watts JL, Lowery DE, Teel JF, Rossbach S. 2000. Identification of Corynebacterium bovis and other coryneforms isolated from bovine mammary glands. J. Dairy Sci. 83:2373–2379 [DOI] [PubMed] [Google Scholar]
- 23. Weiss K, Lavardière M, Rivest R. 1996. Comparison of antimicrobial susceptibilities of Corynebacterium species by broth microdilution and disk diffusion methods. Antimicrob. Agents Chemother. 40:930–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams DY, Selepak ST, Gill VJ. 1993. Identification of clinical isolates of nondiphtherial Corynebacterium species and their antibiotic susceptibility patterns. Diagn. Microbiol. Infect. Dis. 17:23–28 [DOI] [PubMed] [Google Scholar]