Abstract
A multilocus sequence analysis (MLSA) scheme was developed for characterization of strains and species from the genus Achromobacter, which are increasingly recovered from patients with cystic fibrosis (CF). Five conserved housekeeping genes were selected for the MLSA, which was applied to a diverse collection of 77 strains originating from Europe, Asia, and South America and including type strains of the seven recognized Achromobacter species, six environmental strains, eight non-CF clinical strains, and 56 CF clinical strains. The discriminatory power of MLSA, based on 2,098 nucleotides (nt), was much superior to a 16S rRNA gene comparison based on 1,309 nt. Congruence was observed between single-gene trees and a concatenated gene tree. MLSA differentiated all seven current Achromobacter species and also demonstrated the presence of at least four novel potential species within the genus. CF isolates were predominantly Achromobacter xylosoxidans (64%), an undescribed Achromobacter species (18%), and Achromobacter ruhlandii (7%). A clone of Achromobacter, which has spread among patients from Danish CF centers in Aarhus and Copenhagen, was identified as Achromobacter ruhlandii. MLSA facilitates the specific identification of isolates of Achromobacter necessary for describing their role in clinical infections.
INTRODUCTION
Achromobacter xylosoxidans is an aerobic, Gram-negative bacillus found in a variety of aquatic environments such as moist soil (3), well water (29), and swimming pools (24) and also in chlorhexidine (38) and dialysis solutions (24). The bacterium has proved able to survive on inanimate surfaces in hospital settings (8), which is connected to its role as a nosocomial colonizer. It has been associated with a wide range of clinical infections such as catheter-related bacteremia (33, 35), mesh infection (11), meningitis (18), necrotizing pancreatitis (6), urinary tract infections (2, 34), endocarditis (1, 37), and pneumonia (2). It is generally considered an opportunistic pathogen and has attracted attention as an emerging pathogen in cystic fibrosis (CF) (5, 23, 30). Reported prevalence rates of A. xylosoxidans have increased in recent years although this may in part result from growing attention or improved microbiologic techniques. During a period of unaltered sample processing at our clinic, the proportion of CF patients with at least one airway sample positive for A. xylosoxidans increased from 6% in 2005 to 10% in 2009 (25).
The clinical impact of A. xylosoxidans infection in CF patients is unclear. However, recent data indicate that chronic infection with A. xylosoxidans may result in accelerated decline in lung function and an inflammatory response comparable to that observed for Pseudomonas aeruginosa (12, 17) although other studies have failed to document similar observations (32). Infection caused by A. xylosoxidans is of significant concern to CF patients because of its inherent antibiotic resistance and its ability to develop resistance to virtually all available antibiotics (26). In addition, patient-to-patient transmission of A. xylosoxidans is increasingly reported (15, 19, 21, 22, 36) and has also been observed in Danish study populations (25, 27).
Seven species are currently described within the genus Achromobacter, namely, A. xylosoxidans (type species of the genus), Achromobacter denitrificans, Achromobacter insolitus, Achromobacter marplatensis, Achromobacter piechaudii, Achromobacter ruhlandii, and Achromobacter spanius (7; List of Prokaryotic Names with Standing in Nomenclature [http://www.bacterio.cict.fr/]). Species identification of Achromobacter isolates is difficult, and clinical isolates of Achromobacter are generally referred to as A. xylosoxidans.
The aim of this study was to characterize a diverse collection of Achromobacter strains deriving mainly from CF patients using multilocus sequence analysis (MLSA) and 16S rRNA gene sequencing.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The 77 Achromobacter strains used in this study are listed in Table 1. They include the seven Achromobacter type strains (A. xylosoxidans [LMG 1863], A. denitrificans [CCUG 407], A. insolitus [CCUG 47057], A. marplatensis [CCUG 56371], A. piechaudii [CCUG 724], A. ruhlandii [CCUG 38886], and A. spanius [CCUG 47062]), six environmental strains, eight non-CF clinical strains, and 56 clinical strains from CF patients. Clinical strains were collected from Europe, Asia, and South America in order to achieve a geographically diverse collection of strains. The original method of species identification was not stated, but all clinical strains were received as A. xylosoxidans. Isolates were cultured on 5% blood agar at 35°C.
Table 1.
Strains used in this study
| Strain identificationa | Strain no. | Geographic origin | Source | MLSA cluster assignment (organism) |
|---|---|---|---|---|
| A. xylosoxidans LMG 1863T | Osaka, Japan | Ear | I (A. xylosoxidans) | |
| A. denitrificans CCUG 407T | Soil | Unclustered | ||
| A. piechaudii CCUG 724T | Pharynx | Unclustered | ||
| A. ruhlandii CCUG 38886T | Soil | II (A. ruhlandii) | ||
| A. insolitus CCUG 47057T | Wound | IV (A. insolitus) | ||
| A. spanius CCUG 47062T | Blood | Unclustered | ||
| A. marplatensis CCUG 56371T | Buenos Aires, Argentina | soil | Unclustered | |
| CCUG 723 | A7 | France | Antiseptic solution | I (A. xylosoxidans) |
| CCUG 3353 | A8 | Soil or water | Unclustered | |
| CCUG 14603 | A9 | Lyon, France | Water | I (A. xylosoxidans) |
| CCUG 27767 | A10 | Stockholm, Sweden | Unclustered | |
| CCUG 47056 | A11 | Laboratory sink | IV (A. insolitus) | |
| CCUG 52128 | A12 | Sweden | Environmental control | Unclustered |
| V8-27-5-17 90225-58779 | A13 | Nijmegen, the Netherlands | CF clinical isolate | I (A. xylosoxidans) |
| V9-31-7-87 400110 | A15 | Nijmegen, the Netherlands | CF clinical isolate | I (A. xylosoxidans) |
| V9-31-7-78 405530 X | A16 | Nijmegen, the Netherlands | CF clinical isolate | I (A. xylosoxidans) |
| E2916 | A18 | Edinburgh, Scotland, United Kingdom | CF clinical isolate | I (A. xylosoxidans) |
| E3809 | A19 | Edinburgh, Scotland, United Kingdom | CF clinical isolate | I (A. xylosoxidans) |
| E4539 | A22 | Edinburgh, Scotland, United Kingdom | CF clinical isolate | I (A. xylosoxidans) |
| E4712 | A23 | Edinburgh, Scotland, United Kingdom | CF clinical isolate | III |
| E4828 | A25 | Edinburgh, Scotland, United Kingdom | CF clinical isolate | Unclustered |
| E4857 | A26 | Edinburgh, Scotland, United Kingdom | CF clinical isolate | I (A. xylosoxidans) |
| CF-387-II | A27 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| CF-421-VIII | A28 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| CF-421-IX | A29 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| CF-429 | A30 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| CF-464 | A33 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| CF-470 | A34 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| S-636-III | A35 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| S-714-II | A36 | Münster, Germany | CF clinical isolate | I (A. xylosoxidans) |
| 1 G. D. | A37 | Genoa, Italy | CF clinical isolate | I (A. xylosoxidans) |
| 2. G. G. | A38 | Genoa, Italy | CF clinical isolate | I (A. xylosoxidans) |
| 4 G. D. | A40 | Genoa, Italy | CF clinical isolate | IV (A. insolitus) |
| 5. D. S. | A41 | Genoa, Italy | CF clinical isolate | I (A. xylosoxidans) |
| 6 S. T. | A42 | Genoa, Italy | CF clinical isolate | I (A. xylosoxidans) |
| Karolinska 09-2709 | A44 | Stockholm, Sweden | CF clinical isolate | I (A. xylosoxidans) |
| A0 9126 1295 | A47 | Toulouse, France | CF clinical isolate | I (A. xylosoxidans) |
| A0 9145 3458 | A48 | Toulouse, France | CF clinical isolate | I (A. xylosoxidans) |
| A0 9112 2770 | A49 | Toulouse, France | CF clinical isolate | I (A. xylosoxidans) |
| A0 9223 1515 | A50 | Toulouse, France | CF clinical isolate | IV (A. insolitus) |
| HM 26/5052/08 | A52 | Ljubljana, Slovenia | CF clinical isolate | V |
| MA 269/12551/08 | A54 | Ljubljana, Slovenia | CF clinical isolate | III |
| VN 524/6957/09 | A55 | Ljubljana, Slovenia | CF clinical isolate | I (A. xylosoxidans) |
| KD 570/8308/09 | A56 | Ljubljana, Slovenia | CF clinical isolate | III |
| S 7448 | A57 | Dublin, Ireland | CF clinical isolate | I (A. xylosoxidans) |
| S 7802 Q | A58 | Dublin, Ireland | CF clinical isolate | III |
| S 7900 R | A59 | Dublin, Ireland | CF clinical isolate | I (A. xylosoxidans) |
| S 7937 N | A60 | Dublin, Ireland | CF clinical isolate | I (A. xylosoxidans) |
| S 7951 M | A61 | Dublin, Ireland | CF clinical isolate | I (A. xylosoxidans) |
| S 7969 V | A62 | Dublin, Ireland | CF clinical isolate | I (A. xylosoxidans) |
| S 8076 L | A63 | Dublin, Ireland | CF clinical isolate | I (A. xylosoxidans) |
| 1609233396 | A64 | Hamad, Qatar | non-CF clinical isolate, blood | I (A. xylosoxidans) |
| 1609226775 | A65 | Hamad, Qatar | non-CF clinical isolate, blood | I (A. xylosoxidans) |
| 1605503152 | A66 | Hamad, Qatar | non-CF clinical isolate, ear swab | I (A. xylosoxidans) |
| 1609282189 | A67 | Hamad, Qatar | non-CF clinical isolate, central line/blood | I (A. xylosoxidans) |
| 1608228057 | A68 | Hamad, Qatar | non-CF clinical isolate, blood | II (A. ruhlandii) |
| 1608207854 | A69 | Hamad, Qatar | non-CF clinical isolate, blood | II (A. ruhlandii) |
| 1607230268 | A70 | Hamad, Qatar | non-CF clinical isolate, blood | I (A. xylosoxidans) |
| 1609217638 | A71 | Hamad, Qatar | non-CF clinical isolate, blood | I (A. xylosoxidans) |
| C04 | A72 | Brazil | CF clinical isolate | II (A. ruhlandii) |
| 246 | A73 | Brazil | CF clinical isolate | II (A. ruhlandii) |
| L47 | A74 | Brazil | CF clinical isolate | I (A. xylosoxidans) |
| 610 | A75 | Brazil | CF clinical isolate | III |
| 481 | A76 | Brazil | CF clinical isolate | I (A. xylosoxidans) |
| 423 | A77 | Brazil | CF clinical isolate | I (A. xylosoxidans) |
| H13 | A79 | Brazil | CF clinical isolate | I (A. xylosoxidans) |
| J15059 | A80 | Aarhus, Denmark | CF clinical isolate | III |
| J20454 | A81 | Aarhus, Denmark | CF clinical isolate | I (A. xylosoxidans) |
| J21615 | A82 | Aarhus, Denmark | CF clinical isolate | II (A. ruhlandii) |
| J14493 | A83 | Aarhus, Denmark | CF clinical isolate | II (A. ruhlandii) |
| J16935 | A85 | Aarhus, Denmark | CF clinical isolate | I (A. xylosoxidans) |
| J17668 | A88 | Aarhus, Denmark | CF clinical isolate | III |
| J22077 | A89 | Aarhus, Denmark | CF clinical isolate | III |
| J10478 | A90 | Aarhus, Denmark | CF clinical isolate | III |
| J19995 | A91 | Aarhus, Denmark | CF clinical isolate | III |
| J15976 | A92 | Aarhus, Denmark | CF clinical isolate | I (A. xylosoxidans) |
| J15977 | A93 | Aarhus, Denmark | CF clinical isolate | V |
T, type strain; CCUG, Culture Collection, University of Göteborg, Sweden; LMG, Belgian Co-ordinated Collections of Micro-organisms; other designations as specified by the donating laboratory.
Primers for amplification and sequencing.
Five housekeeping loci were selected for MLSA: atpD (ATP synthase, β-subunit), icd (isocitrate dehydrogenase), recA (recombinase A), rpoB (RNA polymerase, β-subunit), and tyrB (aromatic amino acid transferase). Primers for the five loci were designed using corresponding sequences derived from the genome sequences of A. xylosoxidans A8 (GenBank accession number CP002287), A. piechaudii ATCC 43553 (GenBank accession number ADMS01000000), Bordetella bronchiseptica strain RB50 (GenBank accession number BX470250), Bordetella parapertussis 12822 (GenBank accession number BX470249), and Bordetella petrii CCUG 43448T (GenBank accession number AM902716). Primers were designed using the software CLC Main Workbench (CLC Bio) and Primer3 (28). Primers for icd were adapted from the Bordetella multilocus sequence typing (MLST) scheme (14), introducing one degenerate position in the forward primer and three in the reverse primer. Primers for 16S rRNA PCR and sequencing were as described by Gomila et al. (9). Primers are listed in Table 2.
Table 2.
Primers used for amplification and sequencing, PCR annealing temperatures, and the region used for phylogenetic analysis
| Gene | Primer | Nucleotide sequence (5′–3′) | Amplicon size (bp) | Annealing temp (°C) | Region used for MLSA (bp)a |
|---|---|---|---|---|---|
| atpD | atpD-35F | CCGTGGTGGATATTCAGT | 491 | 50 | 85–480 |
| atpD-522R | CTTGGCGATGTTGTTGAT | ||||
| icd | icd-685F | CTGGTSCACAAGGGCAACAT | 532 | 55 | 832–1167 |
| icd-1216R | ACACCTGVGTSGCVCCTTC | ||||
| recA | recA-64F | TCGCAGATCGAAAAGCAGTT | 549 | 50 | 133–591 |
| recA-615R | CATGCGGATCTGGTTGATGAAG | ||||
| rpoB | rpoB-903F | GCTGGCCAAGAACATCGT | 586 | 50 | 958–1470 |
| rpoB-1488R | GTGCGGCATYAGGTTTTC | ||||
| tyrB | tyrB-391F | CCSAGCTGGGAAAACCAYCG | 477 | 58 | 454–843 |
| tyrB-867R | CGGGTTGSAGTAGWTGGYG | ||||
| 16S rRNA | 16f27 | AGAGTTTGATCMTGGCTCAG | 1495 | 55 | 89–1397 |
| 16r1492 | TACGGYTACCTTGTTACGACTT | ||||
| 16f357 | ACTCCTACGGGAGGCAGCAG |
Position relative to A. xylosoxidans A8 genomic sequence.
Amplification and sequencing.
Template DNA was prepared by suspending a single bacterial colony in 1 ml of sterile water. PCR amplification was carried out in a reaction mixture containing 25 μl of AmpliTaq Gold 360 Master Mix (Applied Biosystems), 20 pmol of each primer, and 5 μl of DNA template. Cycling parameters were as follows: 95°C for 10 min, followed by 30 cycles of 95°C for 30 s, the appropriate annealing temperature for 30 s, and 72°C for 60 s, with a final extension at 72°C for 7 min. Annealing temperatures are given in Table 2. Amplification of the 16S rRNA gene was performed as described by Gomila et al. (9). Sequencing of housekeeping genes was performed with PCR forward primers, and sequencing of the 16S rRNA gene was performed with primers 16f27, 16f357, and 16r1492 (Table 2).
Sequence and phylogenetic analyses.
Sequence chromatograms were edited using ChromasPro (Technelysium Pty. Ltd.). Satisfactory sequence quality was ensured by visual examination of electropherograms and by translation into amino acid sequences, and ambiguities were resolved by sequencing both strands. Phred values were evaluated using CLC Main Workbench (CLC Bio). Nucleotide sequence alignments were made with MEGA5 (31). Sequences were concatenated using the CLC Main Workbench (CLC Bio). Cluster analysis of individual gene sequences as well as concatenated sequences was done employing a neighbor-joining (NJ) algorithm with 1,000 bootstrap replications. All positions containing gaps and missing data were eliminated. The genome sequence of B. petrii CCUG 43448T was used as the outgroup.
Splits decomposition analysis and the pairwise homoplasy index (Phi) were calculated by SplitsTree, version 4 (13). The option genetic algorithms for recombination detection (GARD) was also employed (www.datamonkey.org). The method single likelihood ancestor counting (SLAC) (www.datamonkey.org) was used to estimate the ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS). Congruence of dendrograms was assessed by calculating Pearson product-moment correlations with the unweighted-pair group method using average linkages (UPGMA) algorithm implemented in BioNumerics, version 6.6 (Applied Maths).
Nucleotide sequence and strain accession numbers.
DNA sequences generated in this study have been deposited in the GenBank under accession numbers JQ746037 to JQ746113 (atpD), JQ746114 to JQ746190 (icd), JQ746191 to JQ746267 (recA), JQ746268 to JQ746344 (rpoB), JQ746345 to JQ746421 (tyrB), and JQ746422 to JQ746498 (16S rRNA). A. ruhlandii A83 has been deposited in the German Collection of Microorganisms and Cell Cultures (DSMZ) under accession number DSM 25711.
RESULTS
MLSA.
We developed a multilocus sequence analysis (MLSA) scheme based on five gene fragments: atpD (399 nucleotides [nt]), icd (336 nt), recA (459 nt), rpoB (513 nt), and tyrB (390 nt), resulting in a concatenated sequence of 2,098 nt. The MLSA scheme was applied on the 77 reference and clinical strains listed in Table 1. To test for the role of recombination in generating allelic variation, splits decomposition analysis, GARD, and Phi were calculated. Splits decomposition analysis and GARD found no evidence of recombination in any of the five loci, in contrast to Phi analysis, which detected recombination in atpD (P value of 0.026) and recA (P value of 0.022). To assess the influence of natural selection on generating allelic variability, the ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS) was estimated by SLAC. Low dN/dS values (<0.1) were estimated for the five genes, indicating that they have evolved in the absence of strong positive selection. A high degree of congruence was found between single-gene trees and the concatenated gene tree, as shown by Pearson product-moment correlation analysis (Table 3). Combined, these tests indicate that the five selected housekeeping loci are suitable for phylogenetic analysis of the genus Achromobacter.
Table 3.
Congruence of tree topologies estimated by Pearson product-moment correlation coefficients
| Gene | % Congruence |
|||||
|---|---|---|---|---|---|---|
| Concatemer | tyrB | recA | rpoB | atpD | icd | |
| Concatemer | 100 | |||||
| tyrB | 97.62 | 100 | ||||
| recA | 96.75 | 95.79 | 100 | |||
| rpoB | 93.41 | 91.06 | 89.7 | 100 | ||
| atpD | 91.29 | 85.03 | 83.29 | 87.95 | 100 | |
| icd | 86.42 | 79.41 | 79.99 | 71.59 | 68.81 | 100 |
Phred scores were evaluated for all sequences, categorizing Phred scores above 20 as high-quality base calls. For each sequence, the number of high-quality base calls was counted, and the average frequency of high-quality base calls for each gene was determined as follows: atpD, 95%; icd, 94%; recA, 95%; rpoB, 96%; tyrB, 92%; and 16S rRNA gene, 92%.
Sequence analyses revealed the presence of 88 polymorphic sites in atpD, 96 sites in icd, 123 sites in recA, 102 sites in rpoB, and 135 sites in tyrB. Furthermore, a 3-bp insertion was found in atpD in four strains (in type strains of A. marplatensis, A. piechaudii, and A. spanius and in A. xylosoxidans A8). The number of alleles found in each gene was as follows: atpD, 35 alleles; icd, 47 alleles; recA, 49 alleles; rpoB, 55 alleles; tyrB, 39 alleles.
MLSA segregated 77 strains of Achromobacter into five clusters, designated I to V, and nine evolutionary lineages represented by single strains (Fig. 1A). The five clusters included between 2 and 45 strains and were supported by bootstrap values of 99 to 100%. Three of the clusters encompassed a type strain and represent the species A. xylosoxidans (45 strains), A. ruhlandii (7 strains), and A. insolitus (4 strains). Two clusters did not include a type strain and probably represent undescribed species: cluster III (10 strains) and cluster V (2 strains). Nine strains branched separately, including the type strains of A. denitrificans, A. marplatensis, A. piechaudii, and A. spanius. Two of the separately branching strains, A12 and A8, probably represent undescribed species of Achromobacter. Two strains, A10 and A25, were only distantly related to other Achromobacter strains by MLSA although a 16S rRNA gene comparison suggested affiliation with the genus.
Fig 1.

Neighbor-joining dendrograms showing the relationship of 77 study strains using B. petrii DSM 12804 as an outgroup. (A) Comparison based on concatenated sequences of atpD, icd, recA, rpoB, and tyrB (2,098 nt). MLSA clusters I to V are indicated. (B) Comparison based on 16S rRNA gene sequences (1,309 nt). ▲, environmental isolate; ●, non-CF clinical isolate; ○, CF isolate. Type strains are highlighted in bold. Bootstrap support of clusters is indicated to the left of the node. Scale bar, 0.01 substitutions per site.
Comparison of nearly full-length 16S rRNA gene sequences (1,309 nt) revealed less variation and clustered strains differently (Fig. 1B). Bootstrap support of 16S rRNA gene clusters was, however, poor (bootstrap value range, 0 to 58%), and the overall topology of the tree appeared in conflict with current classification, as exemplified by the inability to clearly discriminate the type strains of A. spanius, A. piechaudii, A. marplatensis, and A. ruhlandii (Fig. 1B).
Specific identification of strains.
The 64 clinical strains of Achromobacter were represented in all five clusters. As expected, the majority of isolates (66%) clustered with the type strain of A. xylosoxidans, while six clinical strains clustered with the type strain of A. ruhlandii, and two clustered with the type strain of A. insolitus. Two strains in the A. ruhlandii cluster, strain A68 and A69, were non-CF isolates from blood. The 10 strains of cluster III all originated from CF patients; 5 strains were from our center at Aarhus University Hospital, Denmark, and 5 strains were from CF centers in Scotland, Ireland, Slovenia, and Brazil. The two strains of cluster V were recovered from CF patients in Slovenia and Denmark. The eight non-CF clinical strains belonged to either A. xylosoxidans or A. ruhlandii.
Strain A83 is a representative of the Achromobacter clone that has infected multiple patients at Danish CF centers in Aarhus and Copenhagen (25, 27). The clone was identified by MLSA as A. ruhlandii (Fig. 1A).
Of six environmental strains, two grouped with the type strain of A. xylosoxidans, and one grouped with the type strain of A. insolitus. The remaining three environmental isolates branched separately.
It is well known that A. xylosoxidans can cross-infect patients (15, 19, 21, 22, 25, 27, 36). In several cases we found strains of Achromobacter with identical sequences at all five MLSA loci. In four cases the patients were siblings (A37 and A38) or from the same center (A28 and A29, A59 and A63, and A66 and A71), which could indicate cross-infections. In the remaining five cases, strains with identical MLSA sequences originated from different geographical regions as diverse as the Netherlands and Qatar (A15 and A70) and Germany and Brazil (A28/A29 and A79).
Number of gene fragments needed for separation of MLSA clusters.
Neighbor-joining dendrograms were constructed from all combinations of one to four gene fragments (n = 30), and the bootstrap consensus trees were examined for the presence of the five clusters observed in the concatenated MLSA tree (Fig. 1A). Clusters IV and V were discriminated in all dendrograms, including the five single-gene trees, while separation of clusters I to III was more difficult as strains frequently did not cluster when fewer than three genes were used. The 68 strains were assigned to their MLSA clusters in 7 of 10 consensus trees based on the concatenated sequences of two genes, in 9 of 10 combinations based on three genes, and in 5 of 5 consensus trees based on four genes (Fig. 2). Bootstrap support was generally high for MLSA clusters when they were successfully discriminated in the consensus trees (Fig. 2).
Fig 2.
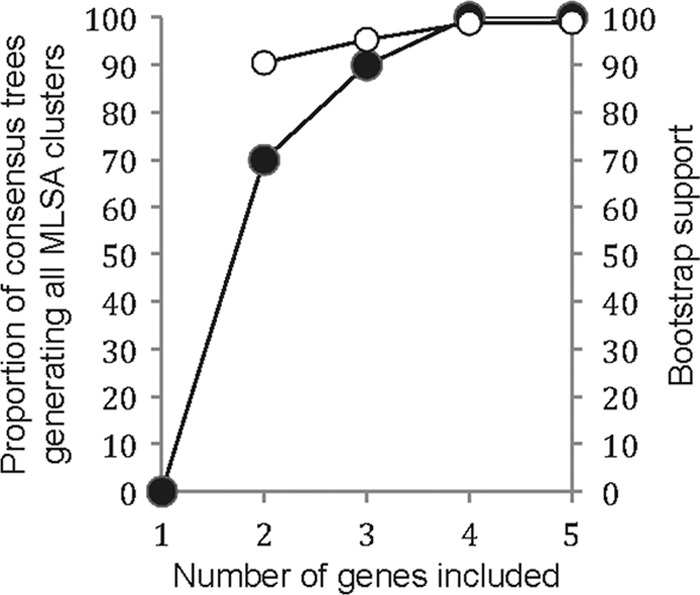
Generation of MLSA clusters in relation to the number of gene fragments included in analysis. Dendrograms were constructed from all combinations of the five housekeeping genes, and the proportion of bootstrap consensus trees assigning 68 strains to the MLSA clusters of Fig. 1 was calculated (●). The mean bootstrap support of successfully generated MLSA clusters is given (○).
DISCUSSION
Bacteria that were previously rare in clinical samples are now frequently recovered from respiratory secretions from CF patients, probably in consequence of the increased life span of the patients and the selective pressure imposed by the repeated use of antimicrobial agents; indeed, many of these “new” microorganisms are innately resistant to commonly prescribed antimicrobial agents. A. xylosoxidans is such a microorganism. It is generally considered an emerging CF pathogen although its pathogenic role in CF disease progression remains to be elucidated.
In the present study, we have developed an MLSA scheme capable of differentiating Achromobacter isolates at the species level. The genetic loci included in the analysis were selected based on several criteria: the loci should be housekeeping genes, which are expected to be relatively conserved; they should be located separately in the genome; and they should be single-copy genes (39). We chose the five loci atpD, icd, recA, rpoB, and tyrB since they fulfilled these criteria and are widely used in established MLST schemes. As only one of three tests detected recombination and that was in just two of five genes (atpD and recA), our tests indicate no overall evidence of recombination, and the low dN/dS ratio points to the absence of strong positive selection. Collectively, this indicates that the five loci are suitable for phylogenetic analyses.
When applied to a collection of 77 diverse Achromobacter isolates, MLSA differentiated the seven currently described Achromobacter species and also demonstrated the presence of at least four novel potential species within the genus. Due to the restricted number of strains and the taxonomic uncertainties regarding novel species, it is not possible to predict whether the resolution of a five-gene MLSA will be sufficient for separation of all putative species in the genus Achromobacter. Multilocus sequence typing (MLST) of Propionibacterium acnes based on nine gene fragments was shown to be superior to a seven-gene MLST when a tree based on 78 P. acnes genomes was used as a reference (16). However, MLST is applied on clones and types within a species, which by definition are closely related, in contrast to MLSA, which can compare different species within a genus. Accordingly, we did not observe inferior resolution of the five Achromobacter clusters by restricting our MLSA to four genes while analysis based on two or three gene fragments could be fallacious (Fig. 2). Our results have shown that the current classification of species within the genus Achromobacter is inadequate, and until a satisfying classification has been achieved, the full resolution of an MLSA scheme cannot be determined.
The most prevalent species of Achromobacter isolated from CF patients was A. xylosoxidans (64%), while the second largest group of CF strains (18%) belonged to a putative novel species of Achromobacter (MLSA cluster III). Half of the strains in this cluster originated from the CF center at Aarhus University Hospital, Denmark, but the strains belonged to separate MLSA types (Fig. 1A) and were unrelated by pulsed-field gel electrophoresis (25), thereby excluding cross-infections. Other strains from cluster III were recovered from CF patients from Europe and South America. CF clinical strains of Achromobacter were also identified as A. ruhlandii (7%) and A. insolitus (4%).
A clone of Achromobacter, designated the Danish epidemic strain (DES), has chronically infected 13 patients from the two Danish centers in Copenhagen (27) and Aarhus (25). Despite implementation of cohort segregation policies for patients infected with Achromobacter and discouraging social contact between infected and uninfected patients, spread of DES has not ceased, and transmission of DES occurred during 2011 (unpublished observations). The DES is, or has become, an exceptionally resistant clone of Achromobacter, and infection with DES is suspected if a patient presents with a first-time isolate of Achromobacter showing pan-resistance. A single representative of DES (A83/DSM 25711) was included in the present study and identified as A. ruhlandii.
The availability of an MLSA scheme for Achromobacter allows specific identification of clinical isolates. Robust identification may also be achieved by sequencing fewer genes (Fig. 2). We found 16S rRNA gene sequencing of little value below the genus level (Fig. 1B), in conflict with the recommendations from the Clinical and Laboratory Standards Institute (CLSI) (4). The approved CLSI guideline MM18-A suggests species resolution for A. xylosoxidans to be achievable based on a 400- to 600-nt 16S rRNA gene sequence; however, the cited literature in MM18-A precedes the publication of the majority of currently described species in the genus Achromobacter. Only three polymorphic positions differentiate the 16S rRNA genes of the type strains of A. xylosoxidans and A. ruhlandii, and only a single nucleotide separates A. piechaudii from A. spanius (alignments not shown). Such subtle differences do not provide the resolution necessary for differentiating species (10).
The present analysis of a relatively restricted number of strains has shown that there exists a broader diversity of species in the genus Achromobacter than previously described. However, only three of seven validated species were detected among our clinical isolates, while a putative novel species accounted for 18% of the CF isolates. Specific identification of potentially pathogenic bacteria is of importance, as has been shown for, among others, the Burkholderia cepacia complex. Species identification of Burkholderia cepacia complex isolates from CF patients has shown that chronic infection with Burkholderia cenocepacia is associated with an increased mortality risk after lung transplantation, compared with other species of the complex (20); in consequence, some centers are reluctant to perform lung transplantation on patients with chronic B. cenocepacia infection (20). Specific identification of clinical isolates of Achromobacter will enable elucidation of possible differences in tropism and pathogenicity between the different species of the genus.
ACKNOWLEDGMENTS
We are grateful to Sabine Meijers (Nijmegen, the Netherlands), Catherine Doherty (Edinburgh, Scotland, United Kingdom), Barbara Kahl (Münster, Germany), Graziana Manno (Genoa, Italy), Shah Jalal (Stockholm, Sweden), Christine Segonds (Toulouse, France), Katja Seme (Ljubljana, Slovenia), Lisa Whelan (Dublin, Ireland), Anand Desmukh (Doha, Qatar), and Carlos Levy (Campinas, Brazil) for donation of clinical strains.
The study was financially supported by the Torben and Alice Frimodt Fund and the Aase and Einar Danielsens Fund.
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Ahmed MS, Nistal C, Jayan R, Kuduvalli M, Anijeet HK. 2009. Achromobacter xylosoxidans, an emerging pathogen in catheter-related infection in dialysis population causing prosthetic valve endocarditis: a case report and review of literature. Clin. Nephrol. 71:350–354 [DOI] [PubMed] [Google Scholar]
- 2. Aisenberg G, Rolston KV, Safdar A. 2004. Bacteremia caused by Achromobacter and Alcaligenes species in 46 patients with cancer (1989–2003). Cancer 101:2134–2140 [DOI] [PubMed] [Google Scholar]
- 3. Busse H-J, Auling G. 2005. Genus II. Achromobacter Yabuuchi and Yano 1981, 477VP emend. Yabuuchi, Kawamura, Kosako and Ezaki 1998a, 1083, p 658–662 In Brenner DJ, et al. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer Verlag, New York, NY [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI document MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Davies JC, Rubin BK. 2007. Emerging and unusual gram-negative infections in cystic fibrosis. Semin. Respir. Crit. Care Med. 28:312–321 [DOI] [PubMed] [Google Scholar]
- 6. Eshwara VK, Mukhopadhyay C, Mohan S, Prakash R, Pai G. 2011. Two unique presentations of Achromobacter xylosoxidans infections in clinical settings. J. Infect. Dev. Ctries. 5:138–141 [DOI] [PubMed] [Google Scholar]
- 7. Euzéby JP. 1997. List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int. J. Syst. Bacteriol. 47:590–592 [DOI] [PubMed] [Google Scholar]
- 8. Festini F, et al. 2007. Patient risk of contact with respiratory pathogens from inanimate surfaces in a cystic fibrosis outpatient clinic. A prospective study over a four-year period. Pediatr. Pulmonol. 42:779–784 [DOI] [PubMed] [Google Scholar]
- 9. Gomila M, et al. 2005. Identification of culturable bacteria present in haemodialysis water and fluid. FEMS Microbiol. Ecol. 52:101–114 [DOI] [PubMed] [Google Scholar]
- 10. Gomila M, et al. 2011. Achromobacter marplatensis sp. nov., isolated from a pentachlorophenol-contaminated soil. Int. J. Syst. Evol. Microbiol. 61:2231–2237 [DOI] [PubMed] [Google Scholar]
- 11. Gupta V, Nirkhiwale S, Gupta P, Phatak S. 2012. Achromobacter xylosoxidans mesh related infection: a case of delayed diagnosis and management. J. Infect. 64:e1–e5 doi:10.1016/j.jinf.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 12. Hansen CR, et al. 2010. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J. Cyst. Fibros. 9:51–58 [DOI] [PubMed] [Google Scholar]
- 13. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 14. Jolley KA, Chan MS, Maiden MC. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86 doi:10.1186/1471-2105-5-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanellopoulou M, et al. 2004. Persistent colonization of nine cystic fibrosis patients with an Achromobacter (Alcaligenes) xylosoxidans clone. Eur. J. Clin. Microbiol. Infect. Dis. 23:336–339 [DOI] [PubMed] [Google Scholar]
- 16. Kilian M, Scholz C, Lomholt HB. 18 January 2012. Multilocus sequence typing (MLST) and phylogenetic analysis of Propionibacterium acnes. J. Clin. Microbiol. 50:1158–1165 [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lambiase A, et al. 2011. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 30:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manckoundia P, et al. 2011. A case of meningitis due to Achromobacter xylosoxidans denitrificans 60 years after a cranial trauma. Med. Sci. Monit. 17:CS63–CS65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moissenet D, et al. 1997. Colonization by Alcaligenes xylosoxidans in children with cystic fibrosis: a retrospective clinical study conducted by means of molecular epidemiological investigation. Clin. Infect. Dis. 24:274–275 [DOI] [PubMed] [Google Scholar]
- 20. Olland A, Falcoz PE, Kessler R, Massard G. 2011. Should cystic fibrosis patients infected with Burkholderia cepacia complex be listed for lung transplantation? Interact. Cardiovasc. Thorac. Surg. 13:631–634 [DOI] [PubMed] [Google Scholar]
- 21. Pereira RH, et al. 2011. Achromobacter xylosoxidans: characterization of strains in Brazilian cystic fibrosis patients. J. Clin. Microbiol. 49:3649–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raso T, Bianco O, Grosso B, Zucca M, Savoia D. 2008. Achromobacter xylosoxidans respiratory tract infections in cystic fibrosis patients. APMIS 116:837–841 [DOI] [PubMed] [Google Scholar]
- 23. Razvi S, et al. 2009. Respiratory microbiology of patients with cystic fibrosis in the United States, 1995 to 2005. Chest 136:1554–1560 [DOI] [PubMed] [Google Scholar]
- 24. Reverdy ME, et al. 1984. Nosocomial colonization and infection by Achromobacter xylosoxidans. J. Clin. Microbiol. 19:140–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ridderberg W, Bendstrup KE, Olesen HV, Jensen-Fangel S, Norskov-Lauritsen N. 2011. Marked increase in incidence of Achromobacter xylosoxidans infections caused by sporadic acquisition from the environment. J. Cyst. Fibros. 10:466–469 [DOI] [PubMed] [Google Scholar]
- 26. Rolston KV, Messer M. 1990. The in-vitro susceptibility of Alcaligenes denitrificans subsp. xylosoxidans to 40 antimicrobial agents. J. Antimicrob. Chemother. 26:857–860 [DOI] [PubMed] [Google Scholar]
- 27. Ronne Hansen C, Pressler T, Hoiby N, Gormsen M. 2006. Chronic infection with Achromobacter xylosoxidans in cystic fibrosis patients; a retrospective case control study. J. Cyst. Fibros. 5:245–251 [DOI] [PubMed] [Google Scholar]
- 28. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 29. Spear JB, Fuhrer J, Kirby BD. 1988. Achromobacter xylosoxidans (Alcaligenes xylosoxidans subsp. xylosoxidans) bacteremia associated with a well-water source: case report and review of the literature. J. Clin. Microbiol. 26:598–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steinkamp G, et al. 2005. Prospective evaluation of emerging bacteria in cystic fibrosis. J. Cyst. Fibros. 4:41–48 [DOI] [PubMed] [Google Scholar]
- 31. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tan K, Conway SP, Brownlee KG, Etherington C, Peckham DG. 2002. Alcaligenes infection in cystic fibrosis. Pediatr. Pulmonol. 34:101–104 [DOI] [PubMed] [Google Scholar]
- 33. Tena D, et al. 2005. Outbreak of long-term intravascular catheter-related bacteremia due to Achromobacter xylosoxidans subspecies xylosoxidans in a hemodialysis unit. Eur. J. Clin. Microbiol. Infect. Dis. 24:727–732 [DOI] [PubMed] [Google Scholar]
- 34. Tena D, Gonzalez-Praetorius A, Perez-Balsalobre M, Sancho O, Bisquert J. 2008. Urinary tract infection due to Achromobacter xylosoxidans: report of 9 cases. Scand. J. Infect. Dis. 40:84–87 [DOI] [PubMed] [Google Scholar]
- 35. Turgutalp K, Kiykim A, Ersoz G, Kaya A. 3 June 2011, posting date Fatal catheter-related bacteremia due to Alcaligenes (Achromobacter) xylosoxidans in a hemodialysis patient. Int. Urol. Nephrol. doi:10.1007/s11255-011-0003-1 [DOI] [PubMed] [Google Scholar]
- 36. Van Daele S, et al. 2005. Shared genotypes of Achromobacter xylosoxidans strains isolated from patients at a cystic fibrosis rehabilitation center. J. Clin. Microbiol. 43:2998–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Hal S, Stark D, Marriott D, Harkness J. 2008. Achromobacter xylosoxidans subsp. xylosoxidans prosthetic aortic valve infective endocarditis and aortic root abscesses. J. Med. Microbiol. 57:525–527 [DOI] [PubMed] [Google Scholar]
- 38. Vu-Thien H, et al. 1998. Investigation of an outbreak of wound infections due to Alcaligenes xylosoxidans transmitted by chlorhexidine in a burns unit. Eur. J. Clin. Microbiol. Infect. Dis. 17:724–726 [DOI] [PubMed] [Google Scholar]
- 39. Zeigler DR. 2003. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int. J. Syst. Evol. Microbiol. 53:1893–1900 [DOI] [PubMed] [Google Scholar]


