Abstract
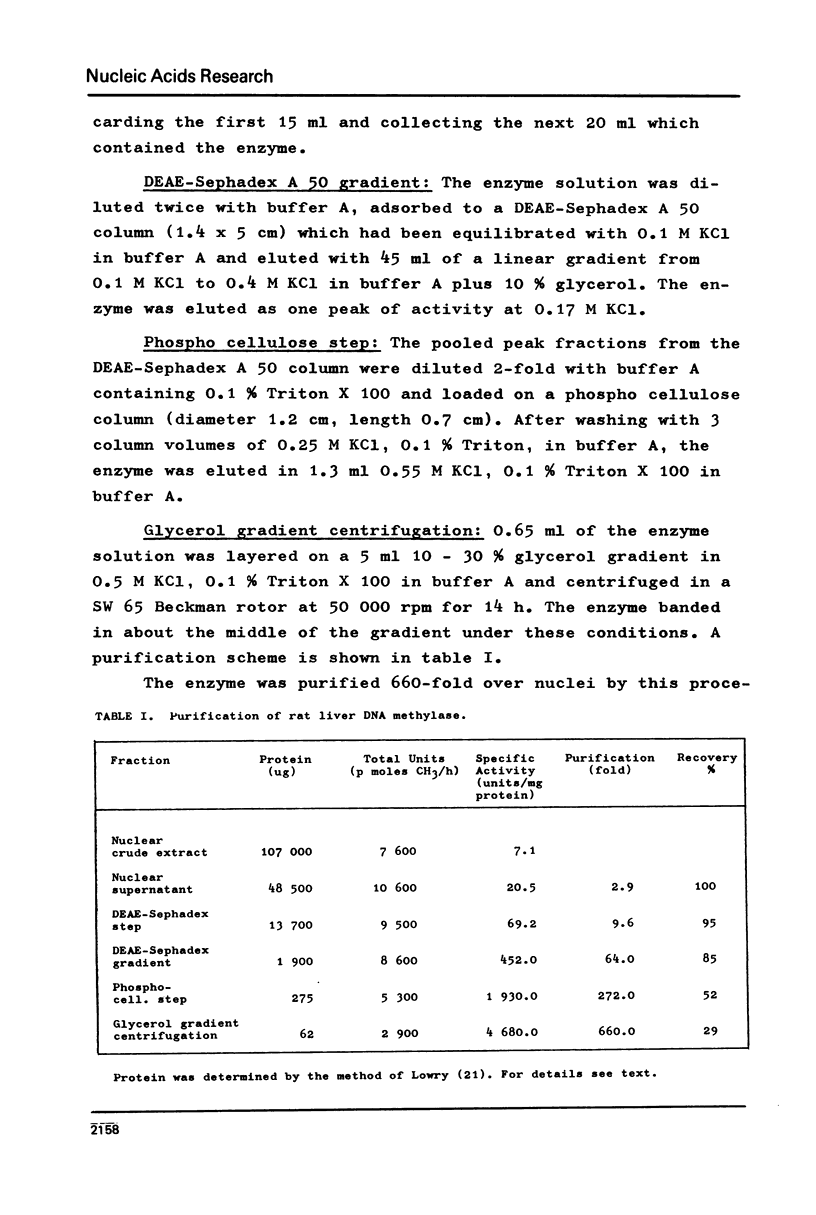
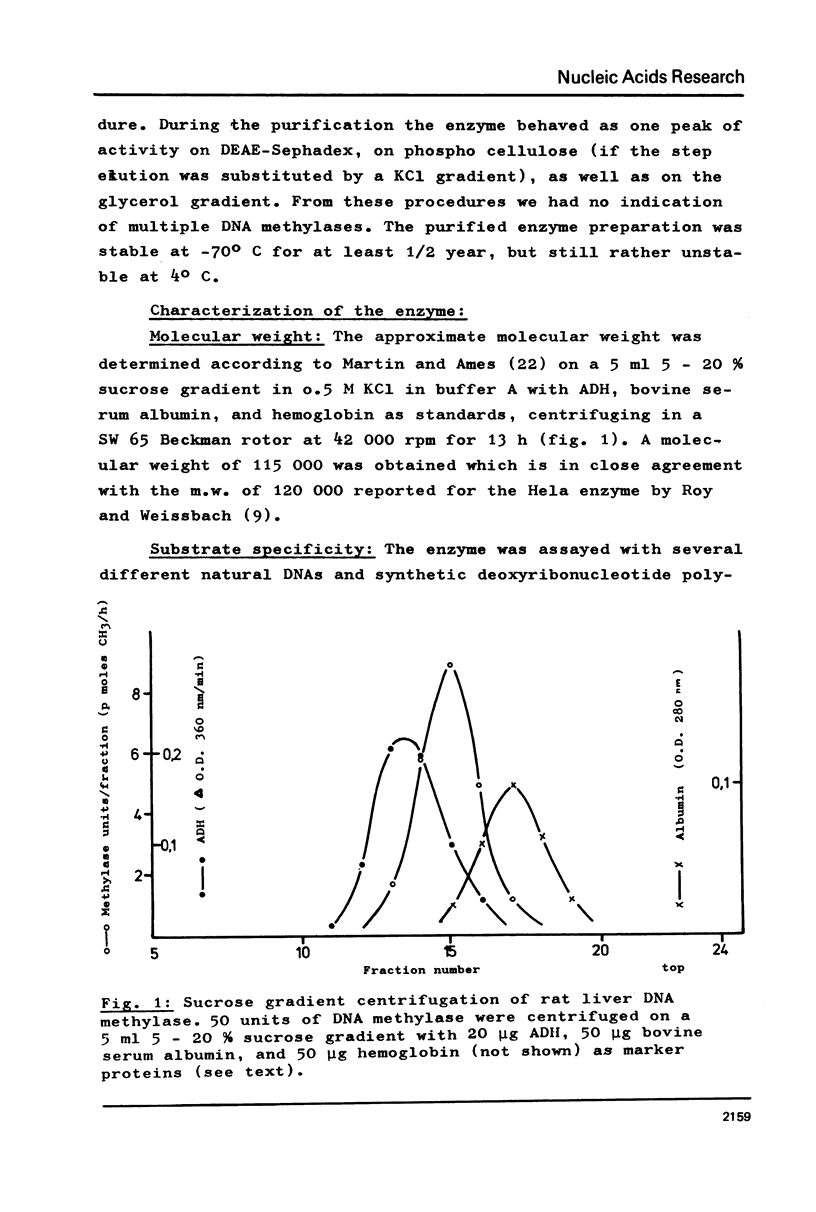
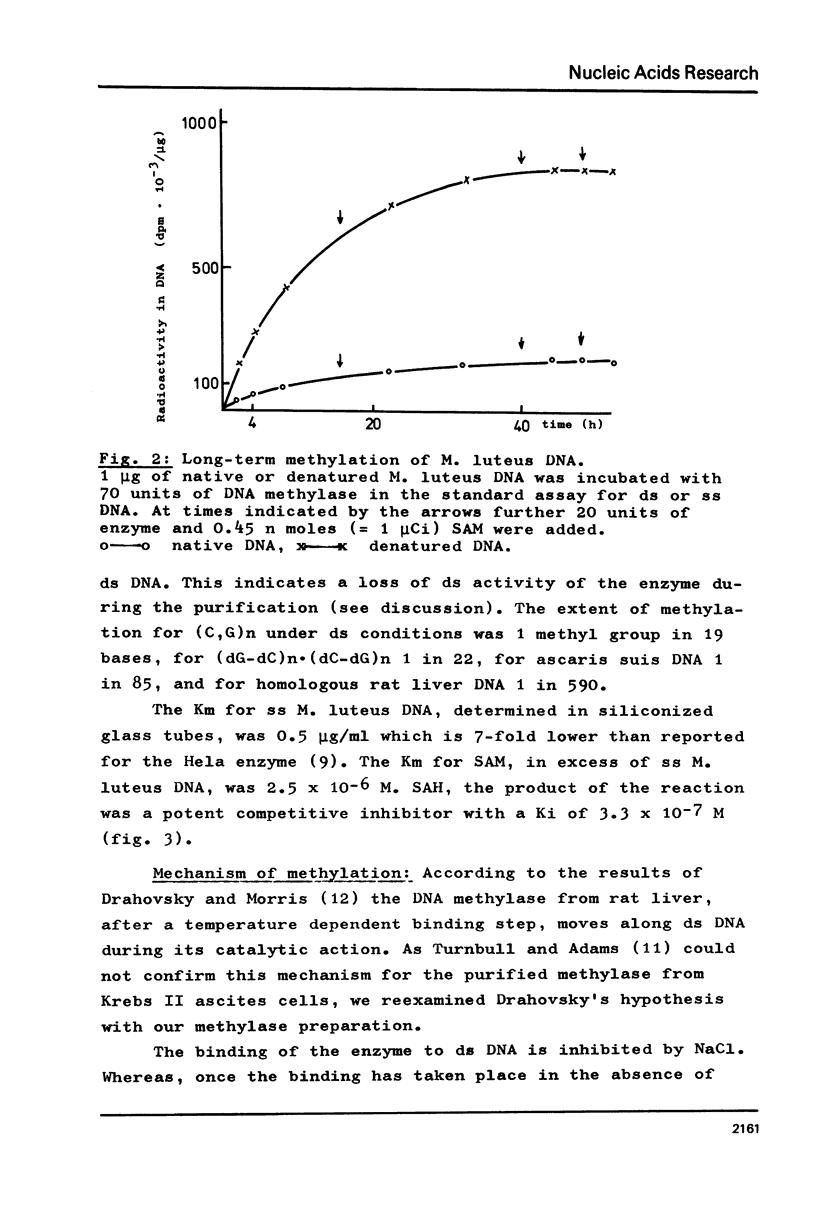
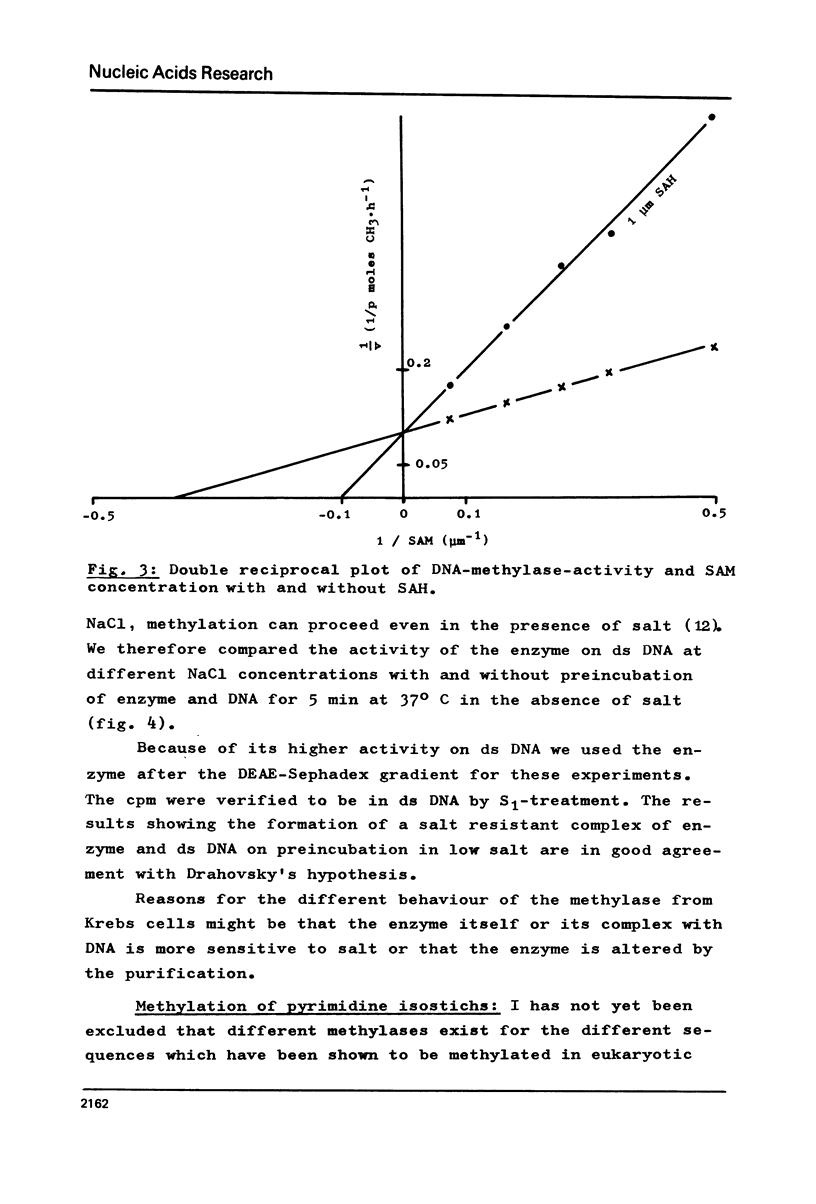
DNA methylase has been purified 660-fold from nuclei from regenerating rat liver. The enzyme is able to methylate single stranded (ss) and double stranded (ds) DNA, the only reaction product being 5-methylcytosine. Previously unmethylated double stranded DNA from prokaryotes (M.luteus) as well as from eukaryotes (Ascaris suis) can serve as substrates. The synthetic copolymers (dG-dC)n . (dC-dG)n and (dG,dC)n are also methylated. While SV40 DNA is almost not methylated, PM2 DNA is a good substrate even in the supercoiled form. The enzyme methylates 1 in 17 bases in heterologous M.luteus DNA, but only 1 in 590 in homologous rat liver DNA. The high methylation level of M.luteus DNA, an analysis of the methylated pyrimidine isostichs and a preliminary dinucleotide analysis suggest that all the CpGs in a DNA can be methylated.
Full text
PDF





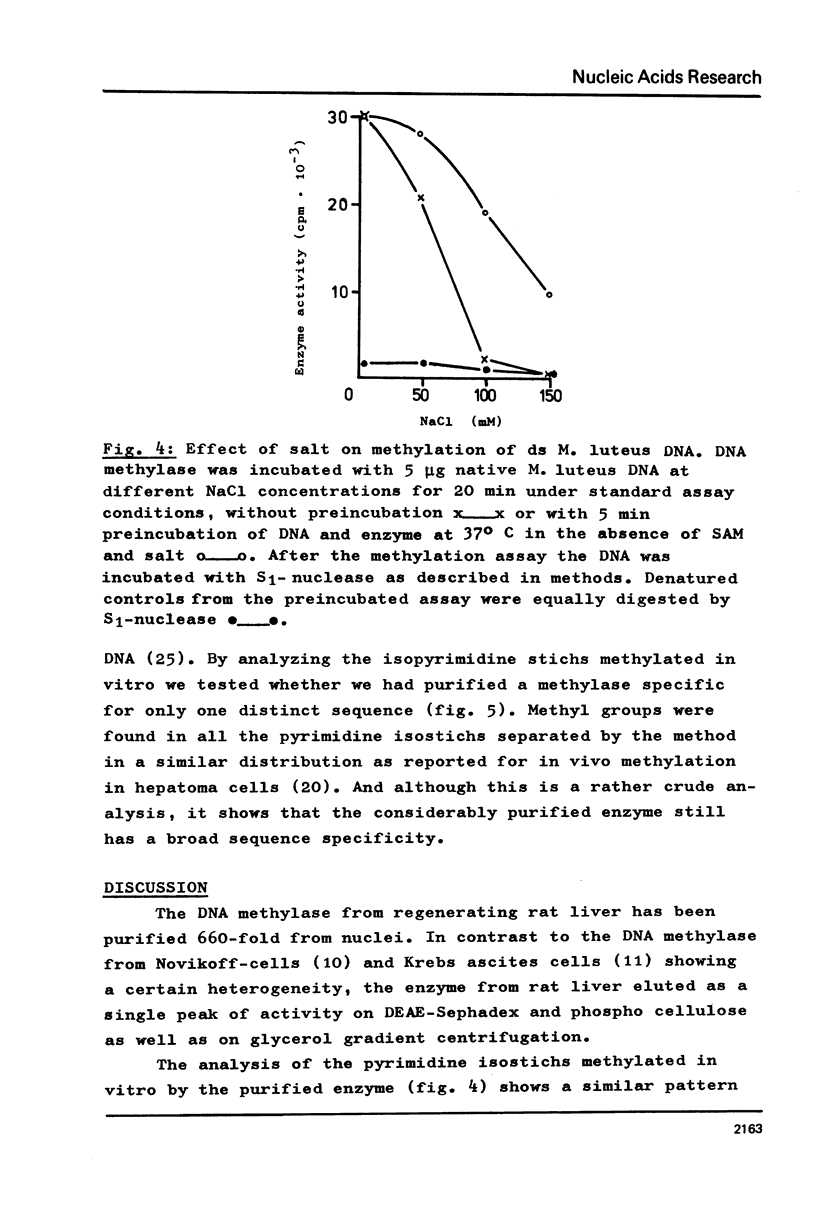
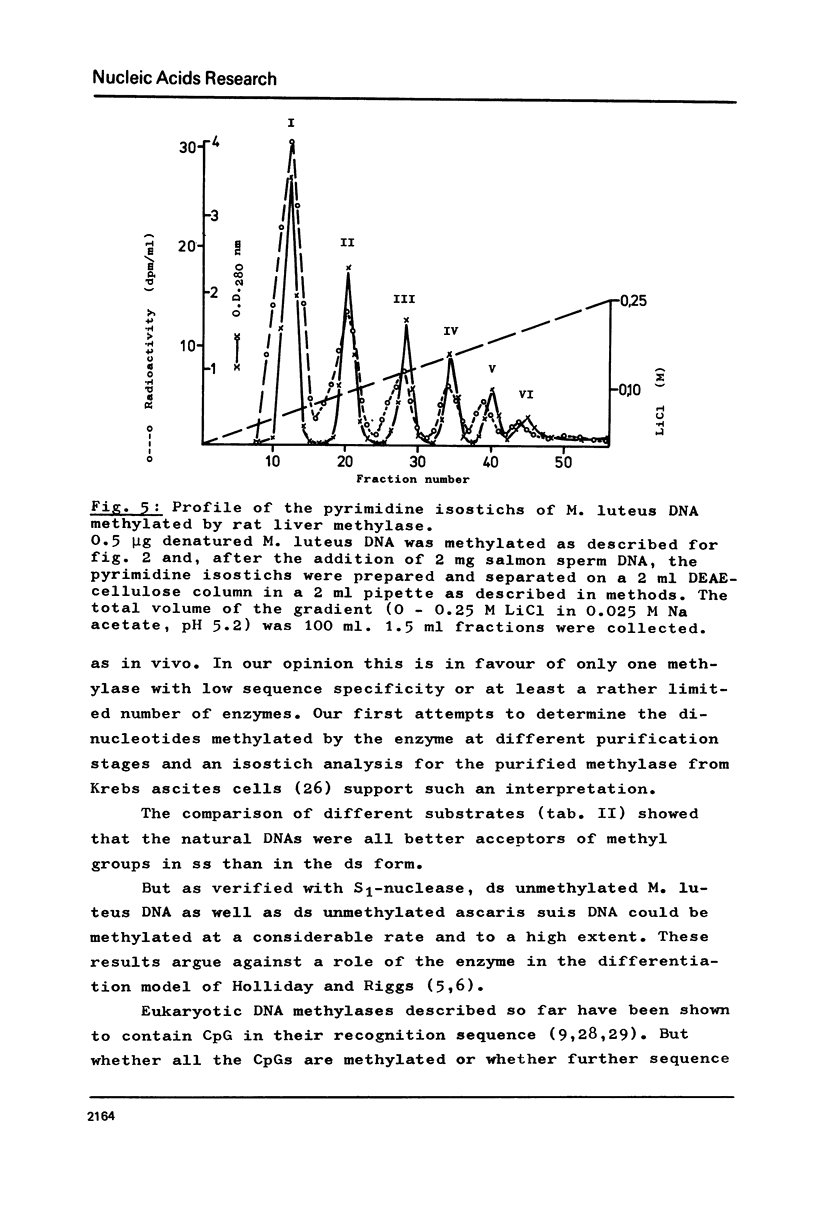
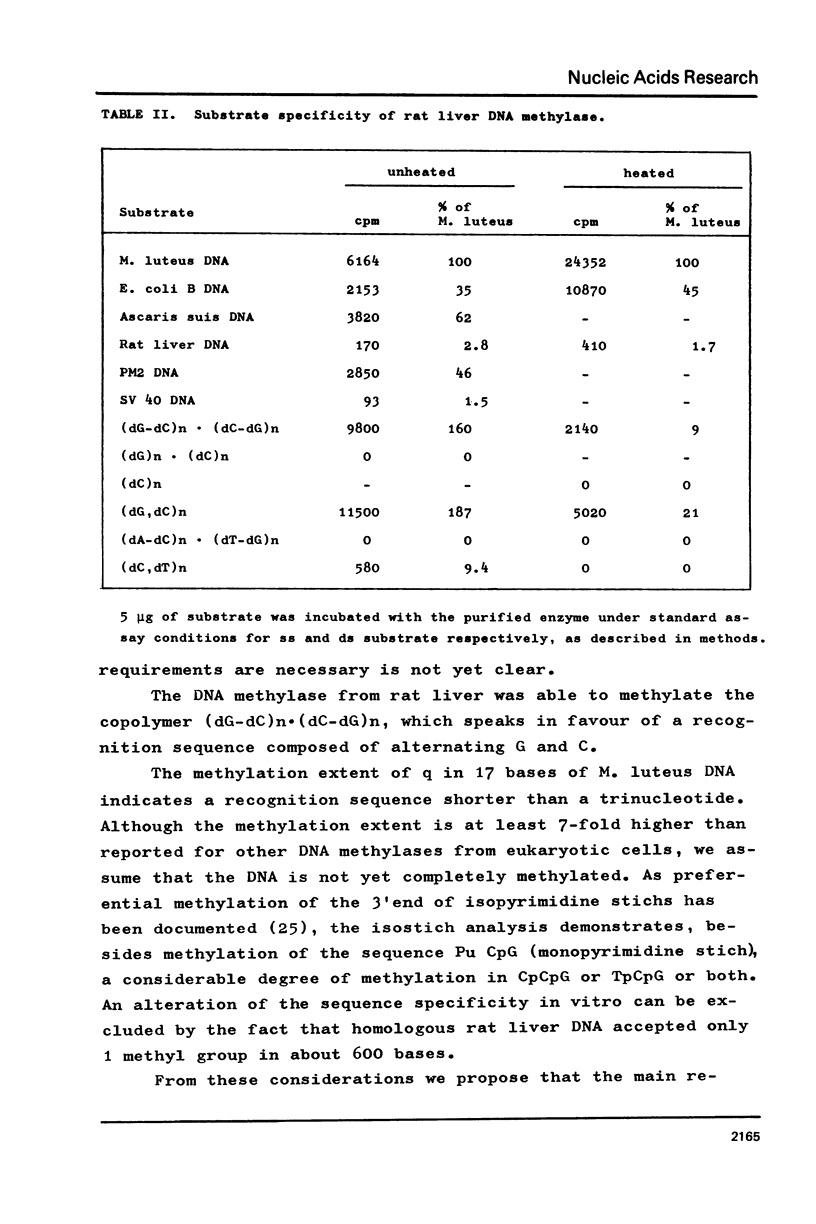


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W. DNA modification and restriction. Prog Nucleic Acid Res Mol Biol. 1974;14(0):1–37. doi: 10.1016/s0079-6603(08)60204-4. [DOI] [PubMed] [Google Scholar]
- Browne M. J., Turnbull J. F., McKay E. L., Adams R. L., Burdon R. H. The sequence specificity of a mammalian DNA methylase. Nucleic Acids Res. 1977 Apr;4(4):1039–1045. doi: 10.1093/nar/4.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Drahovsky D., Morris N. R. The mechanism of action of rat liver DNA methylase. 3. Nucleotide requirements for binding and methylation. Biochim Biophys Acta. 1972 Aug 25;277(2):245–250. doi: 10.1016/0005-2787(72)90404-2. [DOI] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. II. Interaction with single-stranded methyl-acceptor DNA. J Mol Biol. 1971 Oct 28;61(2):343–356. doi: 10.1016/0022-2836(71)90384-6. [DOI] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology. 1968 Apr;34(4):738–747. doi: 10.1016/0042-6822(68)90094-9. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Upholt W. B., Vinograd J. A buoyant method for the determination of the superhelix density of closed circular DNA. J Mol Biol. 1971 Nov 28;62(1):1–19. doi: 10.1016/0022-2836(71)90127-6. [DOI] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Hotani H. Interconversion between flagella and P-filament in vitro. J Mol Biol. 1971 May 14;57(3):575–587. doi: 10.1016/0022-2836(71)90110-0. [DOI] [PubMed] [Google Scholar]
- Johnson P. H., Grossman L. I. Electrophoresis of DNA in agarose gels. Optimizing separations of conformational isomers of double- and single-stranded DNAs. Biochemistry. 1977 Sep 20;16(19):4217–4225. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- Kalousek F., Morris N. R. The purification and properties of deoxyribonucleic acid methylase from rat spleen. J Biol Chem. 1969 Mar 10;244(5):1157–1163. [PubMed] [Google Scholar]
- Kappler J. W. The 5-methylcytosine content of DNA: tissue specificity. J Cell Physiol. 1971 Aug;78(1):33–36. doi: 10.1002/jcp.1040780106. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Razin A., Goren D., Friedman J. Studies on the biological role of DNA methylation: inhibition of methylation and maturation of the bacteriophage phichi174 by nicotinamide. Nucleic Acids Res. 1975 Oct;2(10):1967–1974. doi: 10.1093/nar/2.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Russell G. J., Walker P. M., Elton R. A., Subak-Sharpe J. H. Doublet frequency analysis of fractionated vertebrate nuclear DNA. J Mol Biol. 1976 Nov;108(1):1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- Sneider T. W. Methylation of mammalian deoxyribonucleic acid. II. The distribution of 5-methylcytosine in pyrimidine deoxyribonucleotide clusters in Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1971 Aug 10;246(15):4774–4783. [PubMed] [Google Scholar]
- Sneider T. W., Teague W. M., Rogachevsky L. M. S-adenosylmethionine: DNA-cytosine 5-methyltransferase from a Novikoff rat hepatoma cell line. Nucleic Acids Res. 1975 Oct;2(10):1685–1700. doi: 10.1093/nar/2.10.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. Methylation of mammalian deoxyribonucleic acid. 3. Terminal versus internal location of 5-methylcytosine in oligodeoxyribonucleotides from Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1972 May 10;247(9):2872–2875. [PubMed] [Google Scholar]
- Subak-Sharpe H., Bürk R. R., Crawford L. V., Morrison J. M., Hay J., Keir H. M. An approach to evolutionary relationships of mammalian DNA viruses through analysis of the pattern of nearest neighbor base sequences. Cold Spring Harb Symp Quant Biol. 1966;31:737–748. doi: 10.1101/sqb.1966.031.01.094. [DOI] [PubMed] [Google Scholar]
- Turnbull J. F., Adams R. L. DNA methylase: purification from ascites cells and the effect of various DNA substrates on its activity. Nucleic Acids Res. 1976 Mar;3(3):677–695. doi: 10.1093/nar/3.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Belozersky A. N., Kokurina N. A., Kadirova D. X. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature. 1968 Jun 15;218(5146):1066–1067. doi: 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Mazin A. L., Vasilyev V. K., Belozersky A. N. The content of 5-methylcytosine in animal DNA: the species and tissue specificity. Biochim Biophys Acta. 1973 Mar 28;299(3):397–403. doi: 10.1016/0005-2787(73)90264-5. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- WYATT G. R. The purine and pyrimidine composition of deoxypentose nucleic acids. Biochem J. 1951 May;48(5):584–590. doi: 10.1042/bj0480584. [DOI] [PMC free article] [PubMed] [Google Scholar]


