Abstract
We describe a rare case of Francisella novicida bacteremia following a near-drowning event in seawater. We highlight the challenges associated with laboratory identification of F. novicida and differences in the epidemiology of F. novicida and Francisella tularensis infections.
CASE REPORT
A healthy 69-year-old male from Pennsylvania suffered severe neck trauma and a near drowning while body surfing along the coast of South Carolina. On admission to the local hospital, he was intubated and was quadriplegic with C1 and C3 vertebral fractures and spinal cord contusion at C3 to C4. Chest computed tomography demonstrated moderate bilateral pulmonary opacities suggesting aspiration. He was treated with ampicillin-sulbactam and dexamethasone. After respiratory cultures yielded Enterobacter aerogenes and methicillin-sensitive Staphylococcus aureus, antibiotics were changed to ceftriaxone. On hospital day 7, the patient developed a fever of 38.7°C. Blood cultures grew Staphylococcus epidermidis on 3 consecutive days, and respiratory culture grew Serratia marcescens. Antibiotics were switched to vancomycin and imipenem-cilastatin.
On hospital day 10, the patient was transferred to a tertiary care hospital in Pennsylvania for additional neurosurgical evaluation. Two sets of peripheral blood cultures were obtained the day after transfer. The anaerobic bottle of one set yielded a coagulase-negative Staphylococcus after 24 h, and the aerobic bottle from the same set yielded a pleomorphic Gram-negative bacillus after 3 days of incubation. Chest radiographs demonstrated left lower lobe air space opacity on hospital days 10 to 12; bronchoscopy cultures grew E. aerogenes and anaerobes. Cefepime, linezolid, and metronidazole were prescribed, and indwelling lines were changed but not cultured; his fevers gradually improved. Unfortunately, the patient remained quadriplegic, ventilator dependent, and unresponsive. Following neurosurgical evaluation and review of advanced directives, the family withdrew life support. The patient died of respiratory failure 13 days after the initial injury; a postmortem exam was not conducted.
The pleomorphic Gram-negative organism recovered from the patient's blood grew on blood and chocolate agar within 1 day of incubation but not on MacConkey agar. The isolate was oxidase negative. A Haemophilus sp. was suspected based on Gram stain findings and slow and fastidious growth; however, X and V factors were not necessary for growth. Fatty acid methyl ester analysis by gas chromatography (GC-FAME) (MIDI, Inc., Newark, DE) identified the organism as Francisella tularensis. The Pennsylvania State Department of Health was notified, and the isolate was sent to the Pennsylvania State Public Health Laboratory (PASPHL) on the same day. Due to concern for laboratory transmission of F. tularensis, antimicrobial prophylaxis was offered to 16 potentially exposed laboratory staff; 14 opted to take doxycycline, 1 chose ciprofloxacin, and 1 declined prophylaxis.
At PASPHL, the Laboratory Response Network real-time PCR assay was positive for three of three targets suggesting identification as F. tularensis. However, direct fluorescent antibody (DFA) testing using fluorescein isothiocyanate (FITC)-labeled anti-whole cell F. tularensis was indeterminate, and a commercial slide agglutination test for F. tularensis (Becton, Dickinson, Franklin Lakes, NJ) was negative. The isolate was forwarded to the Centers for Disease Control and Prevention (CDC), Division of Vector-Borne Diseases, where a separate real-time PCR assay (24) for F. tularensis (previously shown to also detect Francisella novicida) was positive for three of three targets, whereas real-time PCR assays for the two subspecies of F. tularensis responsible for causing human tularemia, F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B) (11), were both negative. A repeated DFA for F. tularensis was also negative. DNA sequencing of the 16S rRNA, pgm, and pdpD genes was performed. Primers and PCR conditions for the amplification and sequencing of the 16S rRNA were as described previously (10). Primers for pdpD amplification and sequencing were the same as used for real-time PCR (11). Primers for pgm amplification and sequencing were 5′ GADGCTTTWGGTGGBATYRTATTWTC 3′ (forward) and 5′ AAYTTCCAWCCTGTWGGAGT 3′ (reverse). PCR annealing temperatures for pdpD and pgm were 55°C and 50°C, respectively. All nucleotide positions included in analyses were sequenced at least twice. Neighbor-joining trees were constructed using the Jukes-Cantor algorithm in MEGA (version 5.0) with 1,000 bootstrap replicates.
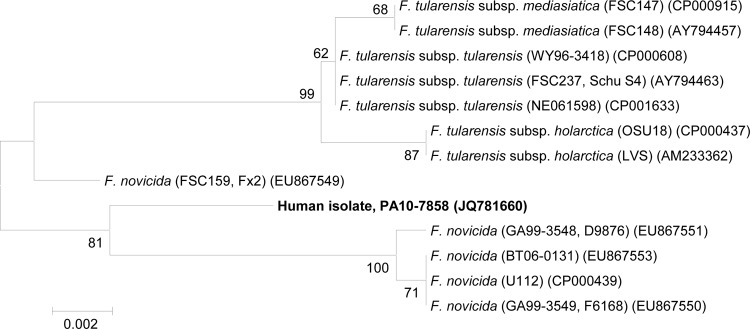
Analysis of the 16S rRNA gene sequences grouped the patient isolate with F. tularensis subspp. tularensis (type A), holarctica (type B), and mediasiatica and with F. novicida and showed 100% identity to the F. novicida strain Fx2 (Fig. 1). Sequencing of the pgm gene indicated that the clinical isolate clustered with F. novicida strains as opposed to F. tularensis strains (Fig. 2). Similarly, sequencing of a 224-bp region of the pdpD gene (GenBank accession no. JX070223) indicated 100% identity to other F. novicida strains for which pdpD sequences were available (U112, GA99-3549, and Fx1). F. novicida strains have a 144-bp insertion in the pdpD gene compared with F. tularensis type A strains, and the pdpD gene is entirely absent in F. tularensis type B strains (11, 16). Taken together, these data identified the isolate as F. novicida. Antibiotic susceptibility testing was performed using Clinical and Laboratory Standards Institute (CLSI) broth microdilution for F. tularensis (6); the isolate was susceptible to ciprofloxacin, doxycycline, gentamicin, levofloxacin, streptomycin, and tetracycline.
Fig 1.
Neighbor-joining tree showing the relationship of the clinical isolate, PA107858, to other members of the Francisellaceae based on an 835-bp region of the 16S rRNA gene. Bootstrap support values >60% are indicated. GenBank accession numbers are indicated following the strain designations.
Fig 2.
Neighbor-joining tree showing the relationship of the clinical isolate, PA107858, to other members of the Francisellaceae members based on a 507-bp region of the pgm gene. Bootstrap support values >60% are indicated. GenBank accession numbers are indicated following the strain designations.
The Francisella species most commonly associated with human infection is F. tularensis, the cause of tularemia. This is transmitted to humans via arthropod bites, contact with infected animals, inhalation of contaminated aerosols, or consumption of contaminated freshwater (18). Tularemia patients usually present with fever, cutaneous ulcer, and regional lymphadenopathy. Less common syndromes include pneumonia, oculoglandular tularemia, and typhoidal tularemia.
Human infection with F. novicida is exceedingly rare, with only six cases published in the English literature (2, 5, 8, 14). Clinical manifestations of reported cases include two otherwise healthy individuals with regional lymphadenopathy without fever and four immunocompromised patients with fever and nonlocalizing symptoms. Sources of human infection with F. novicida remain largely unknown; unlike F. tularensis, F. novicida has not been shown to be associated with arthropod vectors or animals in nature. F. novicida has been detected in brackish and saltwater sources (1, 12, 19); however, this appears to be the first reported case associated with a near-drowning event. In this instance, F. novicida was not detected until hospital day 14, 1 day after the patient's death. Potential explanations include faster growth in culture by other organisms, inhibition of growth by other organisms such as Staphylococcus spp. (20), intermittent or low levels of F. novicida bacteremia, and progressive immunosuppression of the patient due to dexamethasone administration. Although the patient received multiple antibiotics, the patient was treated predominantly with beta-lactams, which have limited to no activity against Francisella species (7, 15).
Given the rarity of human illness caused by F. novicida, clinical and laboratory identification of cases can be challenging. Despite marked differences in virulence (17), F. tularensis and F. novicida share an average nucleotide identity of 99.2% over 1.1 Mbp of genome sequence (13). Consequently, F. novicida has been considered a subspecies of F. tularensis, and controversy exists regarding the nomenclature of F. novicida (9). This high level of genetic relatedness limits the ability of many DNA-based assays to accurately differentiate F. novicida from F. tularensis. Additionally, many bacterial identification systems that use biochemical or fatty acid profiles (including MIDI) do not include F. novicida in their databases. These systems may misidentify F. novicida and other rare Francisella spp. as F. tularensis.
In reference laboratories, tests generally available for identifying F. tularensis include slide agglutination, DFA staining, PCR, and 16S rRNA sequencing. Polyclonal antibodies to whole killed F. tularensis, used in both the direct fluorescence antibody and slide agglutination tests (25), generally react poorly or not at all with F. novicida due to differences in the O antigens of the lipopolysaccharides of the two organisms (21). Consequently, F. novicida should be suspected when PCR assays, fatty acid analysis, or 16S rRNA gene sequencing is positive for F. tularensis but DFA or slide agglutination is equivocal or negative. F. tularensis type A- and type B-specific PCR assays can also be used to distinguish F. novicida from F. tularensis. Further resolution between F. novicida and F. tularensis can be completed by sequencing genes such as pdpD, sdhA, pdpD, uup, aroA, atpA, pgm, tpiA, trpE, and parC (1, 2). Development of PCR assays for F. tularensis that do not cross-react with F. novicida will be important in the future for eliminating misidentification of F. novicida as F. tularensis.
Early in the identification process, laboratory manipulation of cultures of suspect Francisella species should be minimized and biosafety level 3 precautions should be used due to the risk of laboratory airborne transmission of F. tularensis (4, 22). Following potential laboratory exposure to F. tularensis, CDC's Select Agent Program should be notified (23) and exposed workers offered antimicrobial prophylaxis or a “fever watch” with immediate treatment if a fever develops (3). If Francisella species other than F. tularensis are suspected from preliminary testing of isolates, risks and benefits of antibiotic prophylaxis should be considered in the context of the patient history and laboratory data.
ACKNOWLEDGMENTS
We thank Andre Weltman with the Pennsylvania Department of Health for his assistance with the case investigation.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
There are no potential conflicts of interest.
Footnotes
Published ahead of print 12 June 2012
REFERENCES
- 1. Berrada ZL, Telford SR., III 2010. Diversity of Francisella species in environmental samples from Martha's Vineyard, Massachusetts. Microb. Ecol. 59:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Birdsell DN, et al. 2009. Francisella tularensis subsp. novicida isolated from a human in Arizona. BMC Res. Notes 2:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 22 March 2011, posting date Managing potential laboratory exposures to Francisella tularensis. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/tularemia/resources/lab/TularemiaLabExposureFactSheet.pdf [Google Scholar]
- 4. Centers for Disease Control and Prevention, American Society for Microbiology, and Association of Public Health Laboratories 2001. Sentinel level clinical microbiology laboratory guidelines for suspected agents of bioterrorism and emerging infectious diseases: Francisella tularensis. American Society for Microbiology, Washington, DC: http://www.asm.org/images/pdf/Clinical/Protocols/tularemia.pdf [Google Scholar]
- 5. Clarridge JE, III, et al. 1996. Characterization of two unusual clinically significant Francisella strains. J. Clin. Microbiol. 34:1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Cross JT, Jacobs RF. 1993. Tularemia: treatment failures with outpatient use of ceftriaxone. Clin. Infect. Dis. 17:976–980 [DOI] [PubMed] [Google Scholar]
- 8. Hollis DG, et al. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johansson A, et al. 2010. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int. J. Syst. Evol. Microbiol. 60:1717–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kugeler KJ, et al. 2008. Isolation and characterization of a novel Francisella sp. from human cerebrospinal fluid and blood. J. Clin. Microbiol. 46:2428–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kugeler KJ, Pappert R, Zhou Y, Petersen JM. 2006. Real-time PCR for Francisella tularensis types A and B. Emerg. Infect. Dis. 12:1799–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larson CL, Wicht W, Jellison WL. 1955. A new organism resembling P. tularensis isolated from water. Public Health Rep. 70:253–258 [PMC free article] [PubMed] [Google Scholar]
- 13. Larsson P, et al. 2009. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 5:e1000472 doi:10.1371/journal.ppat.1000472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leelaporn A, Yongyod S, Limsrivanichakorn S, Yungyuen T, Kiratisin P. 2008. Francisella novicida bacteremia, Thailand. Emerg. Infect. Dis. 14:1935–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maurin M, Mersali NF, Raoult D. 2000. Bactericidal activities of antibiotics against intracellular Francisella tularensis. Antimicrob. Agents Chemother. 44:3428–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nano FE, et al. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Owen CR, Buker EO, Jellison WL, Lackman DB, Bell JF. 1964. Comparative studies of Francisella tularensis and Francisella novicida. J. Bacteriol. 87:676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Penn RL. 2009. Francisella tularensis (tularemia), p 2927–2937 In Mandell GL, Bennett JE, Dolin R. (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 7th ed, vol 2 Churchill Livingstone, Philadelphia, PA [Google Scholar]
- 19. Petersen JM, et al. 2009. Direct isolation of Francisella spp. from environmental samples. Lett. Appl. Microbiol. 48:663–667 [DOI] [PubMed] [Google Scholar]
- 20. Petersen JM, et al. 2004. Methods for enhanced culture recovery of Francisella tularensis. Appl. Environ. Microbiol. 70:3733–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomas RM, et al. 2007. The immunologically distinct O antigens from Francisella tularensis subspecies tularensis and Francisella novicida are both virulence determinants and protective antigens. Infect. Immun. 75:371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, and National Institutes of Health 2009. Biosafety in microbiological and biomedical laboratories, 5th ed U.S. Government Printing Office, Washington, DC [Google Scholar]
- 23. U.S. Departments of Health and Human Services and Agriculture 22 December 2011, posting date National Select Agent Registry. U.S. Departments of Health and Human Services and Agriculture, Washington, DC [Google Scholar]
- 24. Versage JL, Severin DD, Chu MC, Petersen JM. 2003. Development of a multitarget real-time TaqMan PCR assay for enhanced detection of Francisella tularensis in complex specimens. J. Clin. Microbiol. 41:5492–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. World Health Organization 2007. WHO guidelines on tularemia, p 27–34 World Health Organization, Geneva, Switzerland [Google Scholar]