Abstract
Matrix-associated laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a rapid and simple microbial identification method. Previous reports using the Biotyper system suggested that this technique requires a preliminary extraction step to identify Gram-positive rods (GPRs), a technical issue that may limit the routine use of this technique to identify pathogenic GPRs in the clinical setting. We tested the accuracy of the MALDI-TOF MS Andromas strategy to identify a set of 659 GPR isolates representing 16 bacterial genera and 72 species by the direct colony method. This bacterial collection included 40 C. diphtheriae, 13 C. pseudotuberculosis, 19 C. ulcerans, and 270 other Corynebacterium isolates, 32 L. monocytogenes and 24 other Listeria isolates, 46 Nocardia, 75 Actinomyces, 18 Actinobaculum, 11 Propionibacterium acnes, 18 Propionibacterium avidum, 30 Lactobacillus, 21 Bacillus, 2 Rhodococcus equi, 2 Erysipelothrix rhusiopathiae, and 38 other GPR isolates, all identified by reference techniques. Totals of 98.5% and 1.2% of non-Listeria GPR isolates were identified to the species or genus level, respectively. Except for L. grayi isolates that were identified to the species level, all other Listeria isolates were identified to the genus level because of highly similar spectra. These data demonstrate that rapid identification of pathogenic GPRs can be obtained without an extraction step by MALDI-TOF mass spectrometry.
INTRODUCTION
Agerobically growing Gram-positive rods (GPRs) form a very heterogeneous and extensive group of bacterial species (23). Some of them, such as Corynebacterium diphtheriae, Listeria monocytogenes, and Bacillus anthracis are highly pathogenic. These bacterial species are associated with severe community-acquired infections and may be associated with outbreaks, thus requiring rapid identification for both therapeutic and infection control measures. Other clinically important pathogenic GPR species include Nocardia spp. and Rhodococcus equi, which can cause severe opportunistic infections. Corynebacterium urealyticum and Corynebacterium jeikeium are, respectively, associated with urinary tract infections and vascular catheter infections, prosthetic endocarditis, and septicemia and can be multidrug resistant (31, 49). Finally, during the last two decades, a number of species of low pathogenicity, such as Propionibacterium acnes and new actinomycetes (Actinobaculum schaalii, Actinomyces neuii, Actinomyces turicensis, and Actinomyces radingae), have been associated with various infectious conditions (5, 25, 37). Rapid identification of GPRs is therefore increasingly required for therapeutic and/or epidemiological concerns.
Systematic identification of GPRs is a challenge. Phenotypic identification methods are accurate for identification of pathogenic GPRs. However, it isn't easy to identify most commensal GPR isolates that are frequently cultured in microbiology laboratories by these techniques, due to weak or variable reactivity among single species or because additional tests are required in up to 50% of cases to obtain a 90% correct identification rate (22). In addition, as most recently described GPR species have been characterized using molecular techniques such as 16S rRNA gene sequencing, these species cannot be identified using phenotypic methods in the absence of appropriate identification systems or updated databases (1, 10, 21, 22, 45, 54). In some cases, 16S ribosomal gene sequencing poorly discriminates closely related species, and alternative molecular methods have been developed to solve these issues (28, 36, 38). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a rapid bacterial identification technique that is increasingly used in microbiology laboratories (13, 30, 48). MALDI-TOF MS identification systems are based on the comparison of the tested isolate mass spectrum with reference databases. Several databases and identification strategies have been developed, including the Biotyper (Bruker Daltonics, Bremen, Germany), the Saramis (bioMérieux, Marcy l'Etoile, France), and the Andromas (Paris, France) systems. Previous studies reported that the Bruker MALDI-TOF MS system accurately identified GPR species such as Listeria spp., Nocardia spp., and Corynebacterium species (3, 6, 29, 51). In these studies, a preliminary extraction step was mandatory to obtain satisfactory results, a procedure that is more cumbersome and time-consuming than the direct colony method that can be used to identify Gram-negative bacilli (43). Available MALDI-TOF MS databases have also been evaluated in the routine use of clinical microbiology laboratories, but few GPRs were included, and in some cases, poor identification results were reported (7, 47).
The Andromas identification strategy is based on a limited number of species-specific profiles for each entry (11, 16). A particularity of the Andromas database is that it was built without any extraction step (7, 12). Previous studies have shown that this identification strategy provides good identification of bacteria, mycobacteria, yeasts, and Aspergillus spp. by the direct colony method (2, 7, 11, 16, 18, 19, 32). We evaluated the accuracy of this method to identify a large collection of GPR isolates that included both pathogenic and commensal isolates.
MATERIALS AND METHODS
Bacterial isolates.
A collection of 659 GPR isolates was set up for this study, originating from French clinical microbiology laboratories and reference centers and from the clinical microbiology laboratory of the University of Buenos Aires, Argentina. Altogether, this collection comprised 16 bacterial genera and 73 bacterial species that are listed in Table 1. Except for Listeria, Nocardia, Corynebacterium diphtheriae, Corynebacterium pseudotuberculosis, and Corynebacterium ulcerans isolates, we used 16S rRNA gene sequencing as a reference technique to identify clinical isolates. 16S rRNA gene sequences were compared with those of bacterial strains obtained from the GenBank or Bioinformatic Bacterial Identification database (17). A ≥99.0% similarity cutoff was required for definite identification.
Table 1.
Identification of 659 Gram-positive rods using the MALDI-TOF MS Andromas systema
| Genus or species | No. of isolates tested | Identification result (no. of isolates) |
|||
|---|---|---|---|---|---|
| Species level | Genus level | Not identified | Error | ||
| Actinobaculum spp.b | 18 | 18 | 0 | 0 | 0 |
| A. schaalii | 14 | 14 | 0 | 0 | 0 |
| A. massiliense | 4 | 4 | 0 | 0 | 0 |
| Actinomyces spp.b | 76 | 75 | 1 | 0 | 0 |
| A. europaeus | 1 | 1 | 0 | 0 | 0 |
| A. funkei | 4 | 4 | 0 | 0 | 0 |
| A. naeslundii | 1 | 1 | 0 | 0 | 0 |
| A. neuii | 33 | 33 | 0 | 0 | |
| A. odontolyticus | 5 | 5 | 0 | 0 | 0 |
| A. radingae | 6 | 6 | 0 | 0 | 0 |
| A. turicensis | 23 | 23 | 0 | 0 | 0 |
| A. urogenitalis | 2 | 2 | 0 | 0 | 0 |
| A. viscosus | 1 | 0 | 1 | 0 | 0 |
| Bacillus spp.b | 21 | 21 | 0 | 0 | 0 |
| B. cereus/B. thuringiensis | 10 | 10 | 0 | 0 | 0 |
| B. licheniformis | 2 | 2 | 0 | 0 | 0 |
| B. pumilus/B. safensis | 4 | 4 | 0 | 0 | 0 |
| B. subtilis | 3 | 3 | 0 | 0 | 0 |
| B. simplex | 2 | 2 | 0 | 0 | 0 |
| Corynebacterium spp.b | 342 | 338 | 2 | 1 | 1 |
| C. amycolatum | 97 | 97 | 0 | 0 | 0 |
| C. aurimucosum | 30 | 27 | 2 | 0 | 1 |
| C. coyleae | 3 | 3 | 0 | 0 | 0 |
| C. diphtheriae | 40 | 39 | 0 | 1 | 0 |
| C. glucuronolyticum | 10 | 10 | 0 | 0 | 0 |
| C. imitans | 2 | 2 | 0 | 0 | 0 |
| C. jeikeium | 16 | 16 | 0 | 0 | 0 |
| C. massiliensis | 1 | 1 | 0 | 0 | 0 |
| C. pseudodiphtheriticum | 10 | 10 | 0 | 0 | 0 |
| C. pseudotuberculosis | 13 | 13 | 0 | 0 | 0 |
| C. simulans | 25 | 25 | 0 | 0 | 0 |
| C. singulare | 1 | 1 | 0 | 0 | 0 |
| C. striatum | 44 | 44 | 0 | 0 | 0 |
| C. tuberculostearicum | 18 | 18 | 0 | 0 | 0 |
| C. ulcerans | 19 | 19 | 0 | 0 | 0 |
| C. urealyticum | 10 | 10 | 0 | 0 | 0 |
| Lactobacillus spp.b | 30 | 30 | 0 | 0 | 0 |
| L. casei | 1 | 1 | 0 | 0 | |
| L. crispatus | 8 | 8 | 0 | 0 | 0 |
| L. delbrueckii | 1 | 1 | 0 | 0 | 0 |
| L. fermentum | 2 | 2 | 0 | 0 | 0 |
| L. gasseri | 7 | 7 | 0 | 0 | 0 |
| L. jensenii | 6 | 6 | 0 | 0 | 0 |
| L. rhamnosus | 5 | 5 | 0 | 0 | 0 |
| Listeria spp.b | 56 | 4 | 52 | 0 | 0 |
| L. grayi | 4 | 4 | 0 | 0 | |
| L. innocua | 5 | 0 | 5 | 0 | 0 |
| L. ivanovii | 5 | 0 | 5 | 0 | 0 |
| L. monocytogenes | 32 | 0 | 32 | 0 | 0 |
| L. seeligeri | 5 | 0 | 5 | 0 | 0 |
| L. welshimeri | 5 | 0 | 5 | 0 | 0 |
| Nocardia spp.b | 46 | 42 | 4 | 0 | 0 |
| N. abscessus | 4 | 4 | 0 | 0 | 0 |
| N. asteroides | 2 | 2 | 0 | 0 | 0 |
| N. arthritidis | 1 | 0 | 1 | 0 | 0 |
| N. beijingensis | 3 | 3 | 0 | 0 | 0 |
| N. brasiliensis | 7 | 6 | 1 | 0 | 0 |
| N. cyriacigeorgica | 7 | 6 | 1 | 0 | 0 |
| N. farcinica | 11 | 10 | 1 | 0 | 0 |
| N. nova | 5 | 5 | 0 | 0 | |
| N. mexicana | 1 | 1 | 0 | 0 | 0 |
| N. otitidiscavium | 2 | 2 | 0 | 0 | 0 |
| N. paucivorans | 1 | 1 | 0 | 0 | 0 |
| N. veterana | 2 | 2 | 0 | 0 | 0 |
| Propionibacterium spp.b | 29 | 29 | 0 | 0 | 0 |
| P. acnes | 11 | 11 | 0 | 0 | 0 |
| P. avidum | 18 | 18 | 0 | 0 | 0 |
| Other GPR speciesc | 41 | 41 | 0 | 0 | 0 |
| Arthrobacter cumminsii | 1 | 1 | 0 | 0 | 0 |
| Brevibacterium casei | 3 | 3 | 0 | 0 | 0 |
| Brevibacterium paucivorans | 1 | 1 | 0 | 0 | 0 |
| Dermabacter hominis | 12 | 12 | 0 | 0 | 0 |
| Erysipelothrix rhusiopathiae | 2 | 2 | 0 | 0 | 0 |
| Gardnerella vaginalis | 3 | 3 | 0 | 0 | 0 |
| Paenibacillus motobuensis | 1 | 1 | 0 | 0 | 0 |
| Rhodococcus equi | 2 | 2 | 0 | 0 | 0 |
| Turicella otitidis | 16 | 16 | 0 | 0 | 0 |
The mass spectra of the isolates studied were compared with those of the whole Andromas database, which encompasses more than 700 bacterial species, including 33 genera and 156 species of aerobically growing Gram-positive rods (GPRs).
For these genera, the total numbers of taxa included in the Andromas database are 18 Actinomyces, 3 Actinobaculum, 13 Bacillus, 32 Corynebacterium, 24 Lactobacillus, 6 Listeria, 25 Nocardia, and 4 Propionibacterium species.
For these genera, the total numbers of taxa included in the Andromas database are 5 Arthrobacter, 6 Brevibacterium, 1 Dermabacter, 1 Erysipelothrix, 1 Gardnerella, and 3 Paenibacillus species, 1 Rhodococcus species, and 1 Turicella species.
Listeria isolates were identified by the French National Reference Centre for Listeria with the API Listeria identification system (bioMérieux, Marcy l'Etoile, France) (8, 20, 34) and additional phenotypic confirmation tests (41, 46). All Nocardia isolates were identified by the French Reference Center for Nocardia using a polyphasic approach, including morphological examination, biochemical profiling, antimicrobial susceptibility testing, and 16S rRNA gene sequencing using specific primers designed to identify Nocardia spp. (42). Partial sequencing of the hsp65 gene was done only for isolates identified as Nocardia abscessus, Nocardia arthritidis, and Nocardia beijingensis to confirm species identification (42). C. diphtheriae, C. ulcerans, and C. pseudotuberculosis isolates were identified by the French National Reference Centre of Corynebacteria of the Diphtheriae Complex using the API Coryne system (bioMérieux, Marcy l'Etoile, France), additional phenotypical assays, and specific molecular methods (a PCR targeting the dtxR gene for C. diphtheriae and a multiplex PCR targeting the 16S rRNA, rpoB, and pld genes for C. pseudotuberculosis and C. ulcerans) (36, 38).
Instrumentation and data analysis.
Bacterial isolates were grown on 5% horse blood Columbia agar plates and incubated for 24 h (48 h for slow-growth bacteria) at 37°C in a 10% CO2 atmosphere. Whole-cell bacteria were deposited without an extraction step using a cotton swab on a MALDI-TOF MS target plate and allowed to dry at room temperature. Samples were then fixed by adding 1 μl of absolute ethanol to inactivate bacteria and allowed to dry at room temperature. Samples were then overlaid with 1 μl of saturated α-cyano-4-hydroxycinnamic acid and allowed to crystallize at room temperature. Samples were processed on the Microflex MALDI-TOF MS spectrometer with the FlexControl software (Bruker Daltonics, Bremen, Germany). Positive ions were extruded at an accelerating voltage of 20 kV in linear mode. Each spectrum was the sum of the ions obtained from 350 laser shots performed automatically on different regions of the same well. The spectra were analyzed in an m/z range of 3,500 to 20,000.
Evaluation of the Andromas MALDI-TOF MS system for identification of GPRs.
Bacterial isolates were identified using the Andromas software, while being unaware of the identification result of the reference method. This software compares the mass spectrum of the tested isolate with the spectra included in the Andromas database, taking into account a possible MS peak variation of ±10 m/z (7). The Andromas database was created using both reference and clinical isolates routinely used in our laboratory. To date, this database encompasses more than 700 bacterial species, including 33 genera and 156 species of aerobically growing Gram-positive rods. Using this software, species identification is considered to be valid if the percentage of common peaks is ≥68% of those of a species-specific profile of the database. A 10% difference between the first two species diagnostics having the best match in the database is also required to give species identification. If the latter condition is not fullfilled, the identification is considered to be correct at the genus or group level for closely related species if the first two matches belonged to the same genus or group of bacteria. In all other cases, results are considered to be inconclusive.
In this study, a misidentification was defined as a discrepancy between the identification result given by MALDI-TOF MS and the result obtained by 16S rRNA sequencing or other reference identification methods described above. Identifications to the genus level and absence of identification were not considered misidentifications.
Listeria species discrimination.
A similarity study of Listeria isolates' mass spectra was performed using the BioNumerics 5.1 software (Applied Maths, Belgium) for in-gel-view representation and calculation of dendrograms. Similarities between mass spectra were calculated using the Euclidean distance. Hierarchical clustering was performed using the unweighted-pair group method using average linkages (UPGMA) aggregation method.
RESULTS
Except for Listeria isolates, 98.5% (594/603) of strains were identified to the species level, and only 1.2% (7/603) of them could not be identified beyond the genus level (Table 1). Correct identifications were obtained for the closely related pathogenic species C. diphtheriae, C. ulcerans, and C. pseudotuberculosis, as well as for frequently encountered nonlipophilic commensal Corynebacterium species that are difficult to identify using biochemical tests: C. striatum or C. amycolatum, which have been misidentified as C. xerosis (21); C. simulans, which fits the biochemical profile of C. striatum or C. minutissimum (53), and C. aurimucosum, which is difficult to differentiate from C. minutissimum (55). MALDI-TOF MS also allowed a rapid identification of bacterial species that would have required molecular methods for definitive identification as Nocardia spp., Rhodococcus equi, Erysipelothrix rhusiopathiae, Actinomyces spp., and Bacillus and Lactobacillus spp. (9). In most cases, MALDI-TOF MS performed as well as extended phenotypic and specific molecular methods to identify GPR isolates.
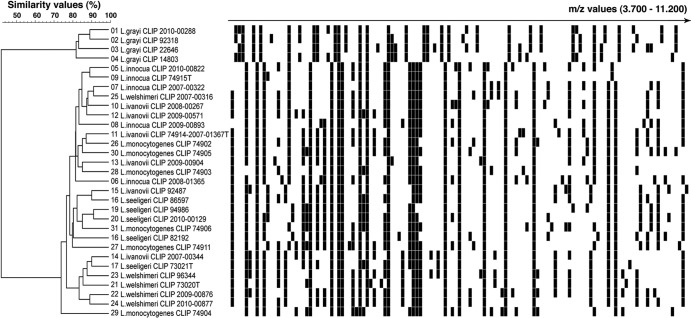
Conversely, except for Listeria grayi isolates that were identified to the species level, all other Listeria species isolates could not be identified beyond the genus level. Indeed, the mass spectrum of these isolates always matched more than one Listeria species-specific profile of the database with a high similarity value (≥68% of common peaks). Moreover, the 10% difference between the two best matchs that is required by the Andromas algorithm to provide species identification was not reached. Therefore, identification was only given at the genus level. The similarity analysis of the mass spectra of Listeria isolates accounts for these results, showing that Listeria grayi and non-grayi isolates form two distinct clusters with high intrinsic similarity values (≥70%), but these two clusters only share a 40% similarity value (Fig. 1). These findings corroborate molecular 16S and 23S rRNA phylogenetic studies that showed that two clusters can be identified within this genus: one for L. grayi and a second for other Listeria species (14, 44).
Fig 1.
MALDI-TOF MS discrimination of Listeria species. MALDI-TOF MS profiles of Listeria isolates, as obtained by the direct colony method, are visualized in a gel-view representation. Spectra were clustered using the UPGMA clustering algorithm. The scale represents the percentage of matching peak mass signals between individual spectra.
Only 7 other GPRs were identified to the genus level, including 4 Nocardia and 2 C. aurimucosum isolates and 1 A. viscosus isolate. The 4 Nocardia isolates, which belonged to 4 different species, were not identified to the species level because of poor mass spectra, a technical issue that has been previously reported for Nocardia spp. without an extraction step (51). The two C. aurimucosum isolates and the A. viscosus isolate had satisfactory mass spectra but matched another species-specific profile of the same genus with a less than 10% difference.
Finally, 1 C. aurimucosum isolate was misidentified as C. striatum, and 1 C. diphtheriae isolate could not be identified. Analysis of the mass spectrum of the C. aurimucosum isolate showed that it corresponded to a new specific C. aurimucosum profile. The C. diphtheriae isolate was not identified because its mass spectrum comprised only 5 peaks, whereas species-specific profiles of the closely related species C. diphtheriae, C. pseudotuberculosis, and C. ulcerans comprise a minimum of 23 peaks. In that case, a formic acid protein extraction step slightly improved the mass spectrum profile (13 peaks), but species identification could not be obtained because the two best matches in the database were too close (C. diphtheriae at 63% and C. pseudotuberculosis at 60%). This nontoxigenic isolate was recovered from a patient with bacteremia. It grew well on agar plates and was correctly identified by phenotypic and molecular assays.
All Bacillus cereus isolates were identified as Bacillus cereus/B. thurigiensis group since these two species display similar species-specific spectra in the Andromas database. Four additional Bacillus isolates were identified as B. pumilus/B. safensis for the same reason and because they cannot be identified to the species level using 16S rRNA gene sequencing.
DISCUSSION
This work is the first MALDI-TOF MS study that addressed the issue of the accuracy of this new bacterial identification method against a fairly extensive collection of Gram-positive rods, including both pathogenic and commensal species. The most important result of our study is that the vast majority of pathogenic GPRs could be identified to the species level. In addition, we also accurately identified nonpathogenic or low-pathogenicity species. As most of these strains were obtained from clinical microbiology laboratories and represent the GPRs that are most frequently recovered in this setting, our data suggest that this identification strategy can also be used to rule out pathogenic pathogens and to describe new GPR-associated infectious conditions.
In this study, MALDI-TOF MS identification of bacterial isolates were obtained by the direct colony method—i.e., under workflow conditions that can be routinely used in all microbiology laboratories. Similar results were only obtained after a preliminary extraction step using the Bruker Biotyper system (3, 6, 29, 51). Each MALDI-TOF MS identification system has its own specificities, including the hardware itself (the mass spectrometer), the way the database was built, and the algorithms used to compare the mass spectrum of a sample with those of the database. Therefore, it is not an easy task to identify the reason why better results were obtained with the Andromas system by the direct colony method. It is well established that the quality of mass spectra varies according to bacterial genera. For instance, better MALDI-TOF MS acquisitions (i.e., higher number of peaks and/or higher relative intensity of peaks) are usually obtained for Gram-negative bacteria (except for mucoid colonies) compared to enterococci and Gram-positive rods (4). Consequently, using a MALDI-TOF database that was built with an extraction step, it is likely that identification errors will occur more frequently for Gram-positive bacteria than for Gram-negative bacteria. The Andromas database was built without any extraction step, a technical issue that probably had an important impact on the quality of results we obtained. To the best of our knowledge, the way the bioMérieux and Bruker databases were engineered has not been previously reported or published.
Our data suggest that species identification of Listeria spp. by the direct colony method can only be achieved at the genus level, except for L. grayi. It has been reported that species identification and typing of Listeria spp. can be obtained with the Biotyper system (6). Nevertheless, a complex extraction step and several centrifugations that may not be routinely implemented in clinical microbiology laboratories were required to obtain these results. As L. monocytogenes causes almost all cases of human listeriosis, rapid identification of a GPR isolate as Listeria sp. is significant progress. For a limited number of GPRs belonging to other genera, no identification was obtained at the species level, because the mass spectrum of the tested isolate matched two species-specific profiles of the same genus, because of an atypical mass spectrum, or due to poor mass spectrum acquisition. Altogether, these observations illustrate the limitations, albeit limited, of the Andromas MALDI-TOF MS strategy for identification of GPRs. By the direct colony method, we obtained species identification rates of Corynebacterium and Nocardia isolates similar to those reported by the extraction method using the Biotyper system, suggesting that a simpler approach can be used to identify GPRs.
Rapid identification of GPRs should have an important clinical impact on the diagnosis of GPR-associated infectious diseases. Rarely encountered diseases, such as granulomatous lymphadenitis due to C. pseudotuberculosis or cutaneous manifestations of C. ulcerans or C. diphtheriae, may be easier to diagnose (27, 33, 52). In that case, the main advantage of this direct colony method is to give rapid identification of colonies that cannot be distinguished from those of other nonlipophilic Corynebacterium species. MALDI-TOF MS should also improve the diagnosis of fastidious or recently described GPR species, like new Actinomyces species (26, 35, 39, 45). In addition, rapid identification of GPRs should improve the probabilistic antimicrobial treatment of patients. Indeed, some GPRs have specific and atypical susceptibility patterns, such as resistance to broad-spectrum cephalosporins for L. monocytogenes and resistance to glycopeptides for E. rhusiopathiae, some Nocardia spp., and some Lactobacillus spp. (15, 24, 50). Some GPRs, such as C. jeikeium, and, to a lesser extent, C. urealyticum and C. amycolatum, can be multidrug resistant (40).
To conclude, our data suggest that the Andromas strategy is an accurate method for identification and/or screening of pathogenic GPRs. As taxonomic changes and characterization of new species or genomospecies of GPRs are frequent, continuous enrichment of the MALDI-TOF MS databases will have to be performed as a dynamic process.
ACKNOWLEDGMENTS
We thank Gilles Quesnes for technical assistance.
This work was supported by grants from the Programme Hospitalier de Recherche Clinique (PHRC; grant no. BOS07001 and AOM08181).
Julie Leto and Brunhilde Dauphin are employees of Andromas. Xavier Nassif is a shareholder of Andromas.
Footnotes
Published ahead of print 12 June 2012
REFERENCES
- 1. Adderson EE, et al. 2008. Identification of clinical coryneform bacterial isolates: comparison of biochemical methods and sequence analysis of 16S rRNA and rpoB genes. J. Clin. Microbiol. 46:921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alanio A, et al. 2011. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin. Microbiol. Infect. 17:750–755 [DOI] [PubMed] [Google Scholar]
- 3. Alatoom AA, Cazanave CJ, Cunningham SA, Ihde SM, Patel R. 2012. Identification of non-diphtheriae Corynebacterium by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:160–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of Gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bank S, Jensen A, Hansen TM, Soby KM, Prag J. 2010. Actinobaculum schaalii, a common uropathogen in elderly patients, Denmark. Emerg. Infect. Dis. 16:76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbuddhe SB, et al. 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bille E, et al. 27 September 2011. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin. Microbiol. Infect. [Epub ahead of print.] doi:10.1111/j.1469-0691.2011.03688.x [DOI] [PubMed] [Google Scholar]
- 8. Bille J, et al. 1992. API Listeria, a new and promising one-day system to identify Listeria isolates. Appl. Environ. Microbiol. 58:1857–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bizzini A, et al. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyd MA, Antonio MA, Hillier SL. 2005. Comparison of API 50 CH strips to whole-chromosomal DNA probes for identification of Lactobacillus species. J. Clin. Microbiol. 43:5309–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carbonnelle E, et al. 2007. Rapid identification of staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 45:2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carbonnelle E, et al. 2012. Robustness of two MALDI-TOF mass spectrometry systems for bacterial identification. J. Microbiol. Methods 89:133–136 [DOI] [PubMed] [Google Scholar]
- 13. Carbonnelle E, et al. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:104–109 [DOI] [PubMed] [Google Scholar]
- 14. Collins MD, et al. 1991. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:240–246 [DOI] [PubMed] [Google Scholar]
- 15. Danielsen M, Wind A. 2003. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 82:1–11 [DOI] [PubMed] [Google Scholar]
- 16. Degand N, et al. 2008. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting Gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Devulder G, Perriere G, Baty F, Flandrois JP. 2003. BIBI, a bioinformatics bacterial identification tool. J. Clin. Microbiol. 41:1785–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dupont C, et al. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 16:998–1004 [DOI] [PubMed] [Google Scholar]
- 19. Ferroni A, et al. 2010. Real-time identification of bacteria and Candida species in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujisawa T, Mori M. 1994. Evaluation of media for determining hemolytic activity and that of API Listeria system for identifying strains of Listeria monocytogenes. J. Clin. Microbiol. 32:1127–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Funke G, Lawson PA, Bernard KA, Collins MD. 1996. Most Corynebacterium xerosis strains identified in the routine clinical laboratory correspond to Corynebacterium amycolatum. J. Clin. Microbiol. 34:1124–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Funke G, Renaud FN, Freney J, Riegel P. 1997. Multicenter evaluation of the updated and extended API (RAPID) Coryne database 2.0. J. Clin. Microbiol. 35:3122–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Funke G, von Graevenitz A, Clarridge JE, III, Bernard KA. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gorby GL, Peacock JE., Jr 1988. Erysipelothrix rhusiopathiae endocarditis: microbiologic, epidemiologic, and clinical features of an occupational disease. Rev. Infect. Dis. 10:317–325 [DOI] [PubMed] [Google Scholar]
- 25. Hall V. 2008. Actinomyces—gathering evidence of human colonization and infection. Anaerobe 14:1–7 [DOI] [PubMed] [Google Scholar]
- 26. Hwang SS, et al. 2011. Actinomyces graevenitzii bacteremia in a patient with alcoholic liver cirrhosis. Anaerobe 17:87–89 [DOI] [PubMed] [Google Scholar]
- 27. Join-Lambert OF, et al. 2006. Corynebacterium pseudotuberculosis necrotizing lymphadenitis in a twelve-year-old patient. Pediatr. Infect. Dis. J. 25:848–851 [DOI] [PubMed] [Google Scholar]
- 28. Khamis A, Raoult D, La Scola B. 2005. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J. Clin. Microbiol. 43:1934–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Konrad R, et al. 2010. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry as a tool for rapid diagnosis of potentially toxigenic Corynebacterium species in the laboratory management of diphtheria-associated bacteria. Euro Surveill. 15:19699 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19699 [DOI] [PubMed] [Google Scholar]
- 30. La Scola B. 2011. Intact cell MALDI-TOF mass spectrometry-based approaches for the diagnosis of bloodstream infections. Expert Rev. Mol. Diagn. 11:287–298 [DOI] [PubMed] [Google Scholar]
- 31. Lavollay M, et al. 2009. The beta-lactam-sensitive D,D-carboxypeptidase activity of Pbp4 controls the L,D and D,D transpeptidation pathways in Corynebacterium jeikeium. Mol. Microbiol. 74:650–661 [DOI] [PubMed] [Google Scholar]
- 32. Lotz A, et al. 2010. Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:4481–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lowe CF, Bernard KA, Romney MG. 2011. Cutaneous diphtheria in the urban poor population of Vancouver, British Columbia, Canada: a 10-year review. J. Clin. Microbiol. 49:2664–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McLauchlin J. 1997. The identification of Listeria species. Int. J. Food Microbiol. 38:77–81 [DOI] [PubMed] [Google Scholar]
- 35. Nielsen HL, Soby KM, Christensen JJ, Prag J. 2010. Actinobaculum schaalii: a common cause of urinary tract infection in the elderly population. Bacteriological and clinical characteristics. Scand. J. Infect. Dis. 42:43–47 [DOI] [PubMed] [Google Scholar]
- 36. Pacheco LG, et al. 2007. Multiplex PCR assay for identification of Corynebacterium pseudotuberculosis from pure cultures and for rapid detection of this pathogen in clinical samples. J. Med. Microbiol. 56:480–486 [DOI] [PubMed] [Google Scholar]
- 37. Perry AL, Lambert PA. 2006. Propionibacterium acnes. Lett. Appl. Microbiol. 42:185–188 [DOI] [PubMed] [Google Scholar]
- 38. Pimenta FP, et al. 2008. A PCR for dtxR gene: application to diagnosis of non-toxigenic and toxigenic Corynebacterium diphtheriae. Mol. Cell. Probes 22:189–192 [DOI] [PubMed] [Google Scholar]
- 39. Reinhard M, et al. 2005. Ten cases of Actinobaculum schaalii infection: clinical relevance, bacterial identification, and antibiotic susceptibility. J. Clin. Microbiol. 43:5305–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Riegel P, Ruimy R, Christen R, Monteil H. 1996. Species identities and antimicrobial susceptibilities of corynebacteria isolated from various clinical sources. Eur. J. Clin. Microbiol. Infect. Dis. 15:657–662 [DOI] [PubMed] [Google Scholar]
- 41. Rocourt J, Buchrieser C. 2007. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification, p 1–20 In Ryser ET, Marth EH. (ed), Listeria, listeriosis, and food safety, 3rd ed, vol 1 CRC Press, Boca Raton, FL [Google Scholar]
- 42. Rodriguez-Nava V, et al. 2006. Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J. Clin. Microbiol. 44:536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saffert RT, et al. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of Gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sallen B, Rajoharison A, Desvarenne S, Quinn F, Mabilat C. 1996. Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int. J. Syst. Bacteriol. 46:669–674 [DOI] [PubMed] [Google Scholar]
- 45. Santala AM, et al. 2004. Evaluation of four commercial test systems for identification of Actinomyces and some closely related species. J. Clin. Microbiol. 42:418–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seeliger HPR, Jones D. 1986. Genus Listeria, p 1235–1245 In Holt JG, Krieg NR, Sneath PH. (ed), Bergey's manual of systematic bacteriology, vol 2 Williams & Wilkins, Baltimore, MD [Google Scholar]
- 47. Seng P, et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 48. Seng P, et al. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 5:1733–1754 [DOI] [PubMed] [Google Scholar]
- 49. Tauch A, et al. 2008. The lifestyle of Corynebacterium urealyticum derived from its complete genome sequence established by pyrosequencing. J. Biotechnol. 136:11–21 [DOI] [PubMed] [Google Scholar]
- 50. Troxler R, von Graevenitz A, Funke G, Wiedemann B, Stock I. 2000. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 6:525–535 [DOI] [PubMed] [Google Scholar]
- 51. Verroken A, et al. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Nocardia species. J. Clin. Microbiol. 48:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wagner J, et al. 2001. Infection of the skin caused by Corynebacterium ulcerans and mimicking classical cutaneous diphtheria. Clin. Infect. Dis. 33:1598–1600 [DOI] [PubMed] [Google Scholar]
- 53. Wattiau P, Janssens M, Wauters G. 2000. Corynebacterium simulans sp. nov., a non-lipophilic, fermentative Corynebacterium. Int. J. Syst. Evol. Microbiol. 50:347–353 [DOI] [PubMed] [Google Scholar]
- 54. Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]
- 55. Yassin AF, Steiner U, Ludwig W. 2002. Corynebacterium aurimucosum sp. nov. and emended description of Corynebacterium minutissimum Collins and Jones (1983). Int. J. Syst. Evol. Microbiol. 52:1001–1005 [DOI] [PubMed] [Google Scholar]