Abstract
Avian oncogenic viruses include Marek's disease virus (MDV), a highly contagious herpesvirus, as well as retroviruses such as avian leukosis virus (ALV) subgroups A to J and reticuloendotheliosis virus (REV). In this study, we examined the incidence of these viruses in suspected samples collected from poultry layer farms of South India, mainly in the Namakkal district of Tamil Nadu, a highly dense poultry-growing area in India. The histopathology-positive tissue sections were identified and further confirmed by immunohistochemistry using virus-specific antibodies. The viruses belonging to all 3 groups (MDV, ALV, and REV) were isolated in a cell culture system and confirmed by immunofluorescence using virus-specific antibodies. PCR appeared to be the method of choice for rapid and accurate diagnosis of these viruses. The multiplex PCR primers specific to MDV, ALV, REV, and chicken DNA were designed for rapid differential diagnosis. The specificity of the primers was checked by amplification of DNA from virus-infected cell culture in comparison with uninfected samples, and sensitivity was evaluated by calculating the minimum copy number at which amplification occurs in the cloned PCR products. The sequences of the amplicons were compared by BLAST analysis. PCR tests demonstrated the presence of single, dual, or triple viruses in some of the samples. Of 169 samples screened by multiplex PCR, 9 samples were positive for MDV, 17 samples were positive for ALV, 12 samples were positive for REV, and 17 samples were positive for both ALV and REV. Three samples were positive for all three viruses. ALV-positive samples were further subjected to subgroup-specific PCR, which gave positive results for subgroups B and D but not for subgroup J. Multiplex PCR appeared to be a useful technique for rapid differential diagnosis of avian oncogenic viruses and detection of multiple infections of avian oncogenic viruses under field conditions.
INTRODUCTION
Avian oncogenic viruses are considered significant pathogens of poultry, with huge economic significance to the poultry industry. These viruses include Marek's disease virus (MDV) (35), avian leukosis virus (ALV), containing subgroups A to J, and the reticuloendotheliosis virus (REV) (34). MDV is classified in the Alphaherpesvirus genus and transforms T lymphocytes, not only resulting in the formation of skin and visceral tumors but also causing immunosuppression, neurological symptoms, and ocular lesions until tumors become visible (25). ALV subgroups belong to the Alpharetrovirus genus and are generally associated with lymphoid leukosis, with tumors primarily in the bursa of Fabricius and visceral organs (14), but ALV subgroup J (ALV-J) targets cells of the myeloid lineage, inducing late-onset myelocytomatosis (32). REV is in the Gammaretrovirus genus and causes a group of disease syndromes that are unrelated to those caused by the leukosis/sarcoma (L/S) group of viruses; it transforms pre-B and pre-T lymphocytes, causing bursal and T-cell lymphomas in chicken and turkeys (7). Reports on multiple oncogenic virus infections are available, and commercial poultry flocks surveyed in Israel between 1993 and 2004 showed multiple oncogenic virus infections in 25% of commercial chicken and turkey flocks (7). At times ALV and REV have been detected as contaminants in Marek's disease vaccines (18, 23, 37).
The classical differential diagnosis of avian oncogenic viruses is based on virus isolation and histopathological examination of tumor tissues. Diagnosis based on virus isolation is laborious and time-consuming and can be further complicated by multiple virus infections in which adaptation of each virus to cell culture is difficult and involves different systems. Although histopathological diagnosis may be able to distinguish between MDV and ALV-J tumors, the lesions of lymphoid tumors induced by different viruses are often difficult to distinguish. Immunohistochemistry can be used for differential diagnosis of avian oncogenic viruses, but virus-specific antibodies are needed (9). PCR appears to be the method of choice for the diagnosis of avian oncogenic viruses because it overcomes many of the challenges encountered in the differential diagnosis and enables the detection of multiple viral infections (7). Hence, a rapid and precise multiplex PCR was developed to differentiate avian oncogenic viruses circulating in south Indian states, an area of India with a very high poultry population.
MATERIALS AND METHODS
Sample collection.
A total of 169 suspected tissue samples, i.e., liver, spleen, bursa, and kidney from commercial layer chickens and liver, spleen, and intestine from turkeys, were collected during necropsy in and around the Indian states of Kerala, Karnataka, and Tamil Nadu. One portion of the tissue was stored at −80°C, and the second portion was collected in formalin for histopathological studies. Gross pathological lesions were recorded at the time of necropsy.
Histopathological examination and immunohistochemistry.
Forty-seven samples were initially screened for histopathological examination by using paraffin-fixed sections. Thin sections (4 to 6 mm) were cut by microtome and stained with hematoxylin and eosin using a standard procedure (2). Based on histopathology results, the samples were further analyzed by immunohistochemistry using mouse monoclonal antibodies against MDV (Santa Cruz Biotechnology) (1:500), chicken polyclonal antibodies against ALV (US Biologicals) (1:100), and chicken polyclonal antibodies against REV (US Biologicals) (1:100) (27). Peroxidase-conjugated antispecies secondary antibodies were used in this study. Positive signals were developed with urea H2O2 (5-mg tab; Sigma) as the substrate (2.0 mg/ml) and DAB-3 (3′-diaminobenzidine tetrahydrochloride) (3.5-mg tab; Sigma) as chromogen (0.7 mg/ml).
Isolation of viruses and DNA.
Samples that were found positive for MDV, ALV, and REV by histopathology and immunohistochemistry (two samples for each virus) were used for virus isolation. Primary isolation of MDV was done in chicken kidney cultures (CKC) and further maintained in chicken embryo fibroblast culture (CEF). Isolation of ALV and REV was done in CEF. Virus-adapted cells were grown in a coverslip culture system, and immunofluorescence assays were carried out for detection of viruses using virus-specific primary antibodies: mouse monoclonal antibodies against MDV (Santa Cruz Biotechnology) (1:1,000), chicken polyclonal antibodies against ALV (US Biologicals) (1:100), and chicken polyclonal antibodies against REV (US Biologicals) (1:100). Fluorescence-conjugated antispecies secondary antibodies were used for development of positive fluorescence after washing with phosphate-buffered saline (PBS). DNA was isolated from cell culture-adapted viruses as previously described (24). DNA was also isolated from all 169 tissue samples as previously described (21) for screening by multiplex PCR.
Primers for multiplex PCR.
Primers for the three avian oncogenic viruses and chicken DNA were designed using PRIMER3.0 software (frodo.wi.mit.edu/primer3/input.htm) and are presented in Table 1.
Table 1.
Targets of primers used in this study
| Virus or control | Target gene | Location(s) of the target (bp) (virus strain GenBank accession no.) | Size (bp) |
|---|---|---|---|
| MDV | meq | 5687–4825, 136663–137518 (EF523390.1) | 856 |
| ALV | p27 | 1369–1982 (AY013303.1) | 613 |
| REV | LTR | 301–504, 8023–8226 (FJ496333.1) | 204 |
| Chicken DNA (control) | β-actin | 3618-4013 (X00182.1) | 396 |
Optimization of multiplex PCR.
DNAs isolated from cell culture-passaged MDV, ALV, and REV were used as templates for standardizing individual PCRs with sequence-specific primers (13).
Specificity and sensitivity.
Cross-reactivities of multiplex PCR primers were tested individually using DNA isolated from cell culture-passaged viruses as the template. Marek's disease meq gene primers were tested against ALV and REV. Avian leukosis virus p27 gene-specific primers were tested against MDV and REV. Similarly, LTR sequence-specific primers of REV were tested against MDV and ALV.
The purified PCR products of MDV, ALV, and REV were inserted into the TA cloning vector (GeNei Instant TM cloning kit; GeNei, India) according to the manufacturer's instructions. Briefly, ligation was accomplished using T4 DNA ligase at 25°C for 1 h followed by 4°C overnight at a 1:3 molar ratio of vector to insert. The mixture (3 μl) was used to transform Escherichia coli DH5α competent bacterial cells, which were then plated on LB agar plates supplemented with ampicillin (100 μg/ml). Ampicillin-resistant colonies were isolated using the HiYield plasmid minikit (Real Biotech Corporation, Taiwan), and plasmid minipreparations were screened for the PCR insert using NcoI digestion. The sensitivity was assessed based on the minimum plasmid copy number at which amplification occurred. Plasmid copy number was calculated as the DNA concentration in grams per μl times 6 × 1023 copies per mol/molecular weight of cloned plasmid in gram per mol (11).
Sequencing of PCR products.
To further confirm the multiplex PCR results, MDV-, ALV-, and REV-specific positive PCR products were gel purified and sequenced in an ABI Prism 3130 genetic analyzer (Applied Biosystems), using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). The sequences obtained were analyzed by BLASTN to check for the homology. The aligned sequences were submitted to the NCBI database.
Multiplex PCR.
DNAs from 169 tissue samples were subjected to multiplex PCR along with internal control β-actin primers. The reaction conditions were as follows: initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, and extension at 72°C for 45 s, and final extension at 72°C for 7 min. PCR products were checked in 1.5% agarose gels (13).
Avian leukosis virus subgroup-specific PCR.
ALV-positive samples were further subjected to H5, H7, and AD1 amplification by touchdown PCR (33). H5- and AD1-amplified PCR products were sequenced and analyzed by constructing a phylogenetic tree using MEGA 5.0 software (www.megasoftware.net) (30) with similar sequences available in GenBank. ALV-positive samples were subjected to ALV subgroup-specific PCR for identification of ALV subgroups. PCR primers for all ALV subgroups (A, B, C, D, E, and J) were used in this study as previously described (28). REV-positive samples were further analyzed with an envelope glycoprotein gene-specific PCR as previously described (22). The amplicons were purified and sequenced, and the aligned sequences were submitted to the NCBI database.
Nucleotide sequence accession numbers.
The 538-bp sequence of envelope glycoprotein gene from REV (JN900392), the 849-bp sequence of envelope glycoprotein gene from ALV (JF913207), the 783-bp sequence of meq gene from MDV (JF921127), the 204-bp LTR sequences of REV (JF913197, JF913198, JF913199, JF913200, JF913201, and JF913202), the 537-bp sequence of p27 gene from ALV (JF913208), the 960-, 983-, and 964-bp sequences of the pol-flanked sequence from ALV (JF913205, JF913204, and JF913206), and the 355-bp sequence of the β-actin gene from Gallus gallus (JF921128) are available in GenBank.
RESULTS
Gross pathology and histopathology.
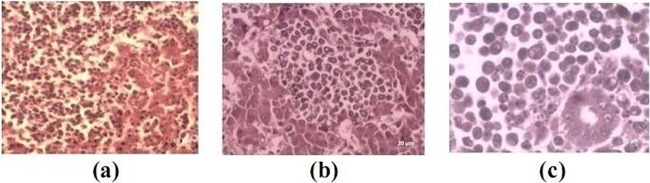
Grossly, in MD-suspected birds, the liver, spleen, proventriculus, and ovaries were affected. Discrete grayish white nodules of various sizes were seen in the liver. In birds suspected for lymphoid leukosis, liver was soft, friable, and grayish white and showed diffuse enlargement. The bursa of Fabricius was six times larger than the normal size with multiple grayish white nodules. In the cases of REV infection, nodular lesions were noticed in the visceral organs, especially in the liver and spleen, and tumors in intestine were also noticed in some positive cases. Of 47 samples subjected to histopathological examination (liver, spleen, kidney, oviduct, and intestine), 4 showed infiltration of pleomorphic lymphoid cells (Fig. 1a), giving a diagnosis of Marek's disease, and monomorphic lymphoblastic cell infiltration was observed in 6 samples (Fig. 1b and c).
Fig 1.
Histopathological lesions caused by avian oncogenic viruses. Pleomorphic infiltration of lymphoid cells in MDV-infected liver (a). Homogenous monomorphic infiltration of lymphoid cells in liver and kidney (b and c).
Immunohistochemistry.
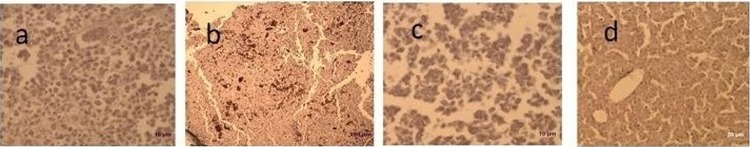
Marek's disease-affected liver tissue sections showed positive signals in the nuclei of the lymphoid cells (Fig. 2a). Avian leukosis virus-positive signals were seen extensively in the cytoplasm and nuclei of spleen tissue sections (Fig. 2b). For REV, the positive signals were seen in the cytoplasm and nuclei of affected liver tissue sections (Fig. 2c). The normal, uninfected liver sample serving as control did not show any viral particles in cytoplasm and nuclei (Fig. 2d).
Fig 2.
Immunohistochemistry of avian oncogenic viruses stained by DAB (bar = 10 μm). (a) MDV-affected liver showing positive signals in the nuclei. (b) ALV-affected spleen showing positive signals in the cytoplasm and nuclei. (c) REV-affected liver showing positive signals in the cytoplasm and nuclei. (d) Normal liver (control).
Isolation and identification of virus.
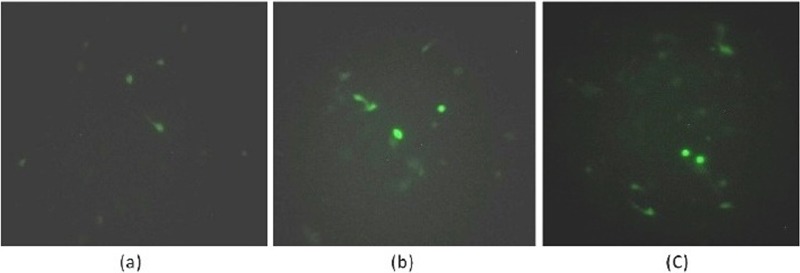
Two histopathology- and immunohistochemistry-positive samples for each virus were serially passaged five times in cell cultures. After five passages in CEF, MDV-positive samples showed clusters of cell plaques on the 5th day of infection. However, CEF infected with ALV and REV produced no cytopathic effects even after 5 passages. Nevertheless, the presence of virus in the infected cells was confirmed by virus-specific immunofluorescent signals using virus-specific antibodies (Fig. 3).
Fig 3.
Immunofluorescence of avian oncogenic viruses in cell culture systems stained by fluorescent secondary antibodies. (a) MDV-adapted CEF showing positive fluorescence; (b) ALV-adapted CEF showing fluorescence; (c) REV-adapted CEF showing positive fluorescence.
Optimization of multiplex PCR.
Optimization of PCR using positive DNA samples with individual multiplex PCR primers resulted in the amplification of the meq gene sequence (856 bp) for MDV, the p27 gene sequence (613 bp) for ALV, the LTR sequence (204 bp) for REV, and the β-actin sequence (396 bp) of chicken DNA (internal control) at an optimized annealing temperature of 55°C (Fig. 4).
Fig 4.
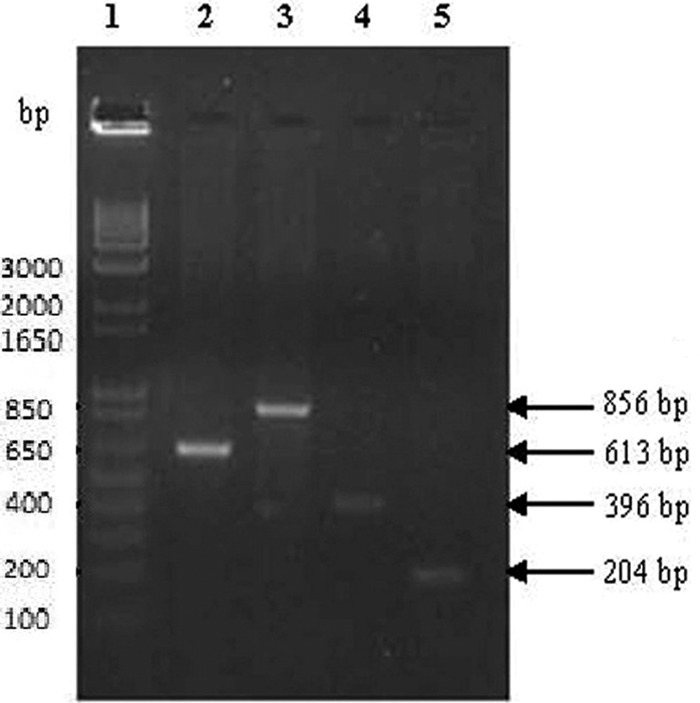
Optimization of PCR using individual tissue DNA with individual multiplex PCR primers. Lanes: 1, 1-kb DNA ladder; 2, ALV-positive cell culture DNA showing an amplicon size of 613 bp with p27 gene primers; 3, MDV-positive cell culture DNA showing an amplicon size of 856 bp with meq gene primers; 4, β-actin gene-specific primers used as internal control showing an amplicon size of 396 bp in CEF cell culture DNA; 5, REV-positive cell culture DNA showing an amplicon size of 204 bp with LTR sequence-specific primers.
Specificity of multiplex PCR primers.
The cross-reactivities of multiplex PCR primers were tested individually using DNA isolated from cell culture-passaged viruses. The results indicated that meq gene primers of MDV did not cross-react with ALV or REV (Fig. 5, left panel). p27 gene-specific primers of ALV amplified only ALV and did not amplify other avian oncogenic viruses (Fig. 5, middle panel). Similarly, REV LTR sequence-specific primers did not amplify MDV or ALV (Fig. 5, right panel).
Fig 5.
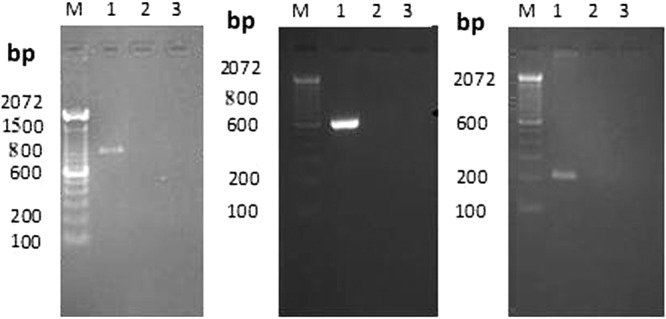
Specificity of multiplex PCR primers using cell culture-positive DNA. (Left) The meq gene primer amplicon (856 bp) was seen in MDV-positive cell culture DNA (lane 1) and not in ALV or REV (lanes 2 and 3). (Middle) The ALV p27 gene primer amplicon (613 bp) was seen in ALV-positive cell culture DNA (lane 1) and not in MDV or REV (lanes 2 and 3). (Right) The REV LTR primer amplicon (204 bp) was seen in REV-positive cell culture DNA (lane 1) and not in ALV or MDV (lanes 2 and 3).
Sensitivity of multiplex PCR primers.
Sensitivity was assessed based on the minimum plasmid copy number at which amplification occurred and was found to be 52 for the meq gene of MDV (Fig. 6a) and 53 for the p27 gene of ALV and the LTR sequence of REV (Fig. 6b and c).
Fig 6.
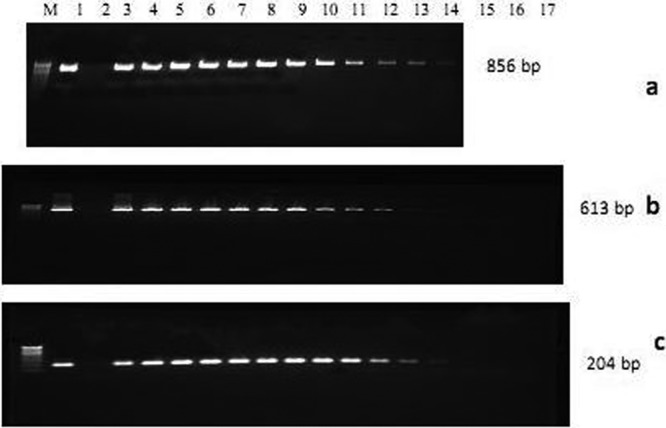
Sensitivity of multiplex PCR primers using PCR product cloned TA plasmid DNA. (a) The meq gene primer amplicon (856 bp) was seen in plasmid copy numbers of 514 (lane 1) and up to 52 in serial dilution (lanes 3 to 14). (b) The ALV p27 gene primer amplicon (613 bp) was seen in copy numbers of 515 (lane 1) and up to 53 in serial dilution (lanes 3 to 14). (c) The REV LTR sequence primer amplicon (204 bp) was seen in copy numbers of 515 (lane 1) and up to 53 in serial dilution (lanes 3 to 14). Lane 2, nontemplate control.
Sequencing of PCR products.
The PCR amplicons of MDV, ALV, REV, and β-actin were sequenced, and the sequences obtained were analyzed using BLASTN (www.ncbi.nlm.nih.gov/blast) to check for homology. The sequences of MDV, ALV, REV, and β-actin were found to have more than 90% homology with respective virus strains and chicken β-actin.
Multiplex PCR.
The results of screening of tissue DNA samples by multiplex PCR are presented in Table 2. The results of multiplex PCR indicated the presence of multiple infections in the samples tested (Fig. 7).
Table 2.
Number and types of samples screened by multiplex PCR primers for avian oncogenic viruses
| Species | Sample type | Total no. of samples screened | No. of samples positive for: |
||||
|---|---|---|---|---|---|---|---|
| MDV | ALV | REV | ALV + REV | MDV + ALV + REV | |||
| Chicken | Liver | 96 | 9 | 10 | 3 | 12 | 3 |
| Spleen | 48 | 0 | 6 | 5 | 5 | 0 | |
| Bursa | 7 | 0 | 0 | 0 | 0 | 0 | |
| Kidney | 5 | 0 | 1 | 0 | 0 | 0 | |
| Turkey | Liver | 5 | 0 | 0 | 2 | 0 | 0 |
| Spleen | 5 | 0 | 0 | 0 | 0 | 0 | |
| Intestine | 3 | 0 | 0 | 2 | 0 | 0 | |
| Total | 169 | 9 | 17 | 12 | 17 | 3 | |
Fig 7.
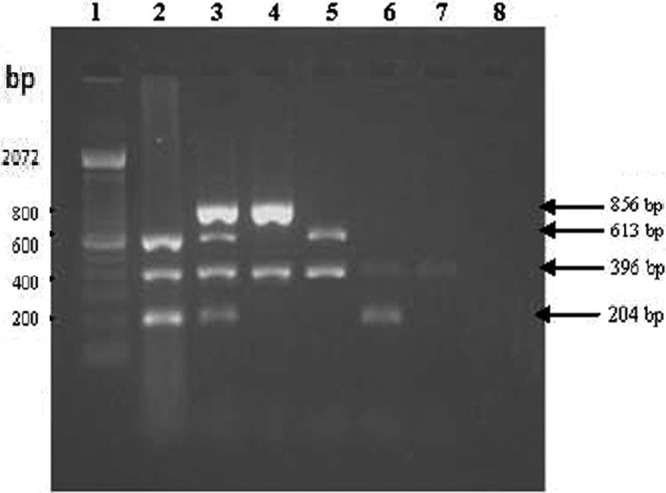
Multiplex PCR amplification of tissue DNA. All the tissue samples show an amplicon size of 396 bp for the β-actin internal control, except the nontemplate control (lane 8). Lane 1, 1-kbp DNA ladder. Lane 2, ALV- and REV-positive tissue DNA showing the amplicon sizes of 613 bp and 204 bp, respectively. Lane 3, MDV-, ALV-, and REV-positive tissue DNA showing the amplicon sizes of 856 bp, 613 bp, and 204 bp, respectively. Lane 4, MDV-positive tissue DNA showing the amplicon size of 856 bp. Lane 5, ALV-positive tissue DNA showing the amplicon size of 613 bp. Lane 6, REV-positive tissue DNA showing the amplicon size of 204 bp. Lane 7, normal chicken liver tissue DNA used as control.
Avian leukosis virus subgroup-specific PCR.
The ALV pol-flanked sequences of H5 and AD1 amplicon were found to have 96 to 99% homology with reference strains by BLASTN analysis. The phylogenetic tree constructed using the pol-flanked (267-bp) sequence of the ALV field strain with reference strains present in the NCBI database (rooted by the midpoint method via the unweighted-pair group method using average linkages [UPGMA] and the maximum composite likelihood method) showed a group with SDO5O1 (China) and ev-1 (United States) sequences. A total of 37 ALV-positive DNA samples showed a positive amplicon size of 2.4 kb by using primers for all ALV subgroups. Of 37 samples, 1 sample showed amplification of a 1.1-kb product specific for ALV subgroups B and D and the other samples showed an amplicon with a size of 1.25 kb that was specific for ALV subgroup E. The REV-positive DNA samples also showed the expected amplicon size of 642 bp by using REV envelope gene-specific primers.
DISCUSSION
The gross lesions caused by avian oncogenic virus infections often overlap, and diagnosis based on gross lesions is difficult, necessitating specific laboratory diagnosis (9). Based on histopathology, the cytological picture of MDV could be typical, while histological differentiation between ALV and REV may not be possible (8). In such instances, histopathological results could be confirmed with immunological and molecular techniques. In our study, immunohistochemistry was used for identification of virus-specific proteins in suspected tissue sections, using virus-specific antibodies. By immunohistochemistry, Marek's disease virus-specific signals were observed in the nuclei of lymphoid cells in positive samples as previously described (4). Similarly, avian leukosis virus antigens were detected in the surface and also were concentrated in cytoplasmic discrete granules (12). Inclusion bodies were present in both the cytoplasm and the nuclei of cells infected with REV as previously described (27).
Primary isolation of low-passage-number (virulent) MDV isolates in CEF or embryonic CKC cultures has been considered less efficient than isolation in CKC or DEF (5). Considering this, CKC was used for the primary isolation of MDV, and subsequent passages in CEF showed characteristic cell plaques at the fifth passage in our study. CEF cultures were used for isolation of ALV (26) and REV (3). Most ALVs produce no visible morphological changes in culture, unlike sarcoma viruses (17). Cytopathic effects may not be seen on primary isolation of nondefective REV strains, in contrast to the defective REV-T strain (15). So, specific techniques are needed to confirm virus identification in cell culture. The presence of all three viruses in the in vitro-cultured cells was confirmed by virus-specific immunofluorescence (15).
About one-quarter of tumor-bearing commercial flocks in Israel carried mixed MDV and retrovirus infections (10). Hence, multiplex PCR was attempted for rapid differential diagnosis of avian oncogenic viruses in commercial layer flocks and turkeys. The meq gene and LTR sequences were used for designing primers for MDV and REV, respectively. Among the structural polypeptides (encoded by p27, p19, p15, p12, and p10) shared by all members of the L/S group of avian retroviruses, including endogenous and exogenous ALV, that encoded by p27 has been the most abundant protein (16). Hence, the p27 gene sequence was used for primer designing of ALV. Our poultry samples did not show ALV-J positivity, which is common in other Asian countries (31, 36). So, we concentrated on ALV subgroups A to D, as the sequence common to ALV-A to -D also amplifies endogenous ALV (ALV-E). So ALV-positive samples were further analyzed with sequence results and ALV subgroup-specific PCR for subgroup detection. In our study, ALV-negative samples were not ALV-E positive. The chicken β-actin gene was used as an internal control for chicken DNA to avoid false-positive results in multiplex PCR. The designed primers resulted in the expected amplicons of 856 bp, 613 bp, 204 bp, and 396 bp for the meq gene (MDV), p27 gene (ALV), LTR gene (REV), and β-actin gene (chicken DNA), respectively, at an annealing temperature of 55°C.
Preferential amplification often occurs in multiplex PCR as a result of various efficiencies of different primer pairs and because of the formation of primer dimers. This is more likely to happen as the number of primers increases (13), and the problem of amplification failure is compounded when the template DNA contains various copy numbers. This problem could be circumnavigated by optimization of the multiplex PCR, by changing primer sequences, concentrations, and cycling conditions (19). In this study, multiplex PCR primers were standardized at optimum concentrations that showed amplifications with MDV-, ALV-, and REV-infected tissue DNA at the annealing temperature of 55°C.
The specificity of the multiplex PCR primers was clearly indicated by the specific amplification of the viral genome in positive cell culture DNA without cross-reactivity with other oncogenic viruses. The sensitivity of these primers was checked using cloned PCR products. The copy number was calculated, and the minimum copy number at which amplification occurred was seen as sensitivity (1, 11). Virus-specific amplifications were also confirmed by sequencing the amplified PCR products. Results obtained in this study indicated that this assay could be useful for rapid differential diagnosis of avian oncogenic viruses and detection of multiple infections of avian oncogenic viruses under field conditions.
The phylogenetic tree construction by the midpoint method using UPGMA and the maximum composite likelihood method showed that the ALV sequence of the field strain was grouped with ALV endogenous virus strains SDO5O1 (China), ev-1 (United States), PDRC-3246 (United States), and ev-3 (United States). So, the ALV-positive samples were further grouped based on ALV subgroup-specific PCR as previously described (28). These results confirmed the specific subgroups, since of 37 samples, 36 were positive for ALV-E and 1 sample was positive for ALV-B and -D. In this study, endogenous ALV (ALV-E) was detected in most samples. Although endogenous viruses have little or no oncogenicity (29), they can affect induction of neoplasia and other production or performance traits by their interaction with exogenous ALV. Similarly, subgroup E recombinants of endogenous and exogenous viruses have been reported to be capable of inducing neoplasm (6). ALV-B and -D are exogenous viruses isolated less frequently than ALV-A from outbreaks of lymphoid leukosis (14). Nowadays, ALV-J is reported mostly in Asian countries (31, 36). However, in our study, the samples were screened for ALV-J, but no sample was found to be positive. For reticuloendotheliosis virus, envelope gene-specific primers were used to obtain an accurate diagnosis to differentiate virus strains that carried intact REV provirus from those that carried solo 5′ LTR sequences (20). Multiplex PCR appeared to be a technique for rapid differential diagnosis of avian oncogenic viruses and detection of multiple infections of avian oncogenic viruses under field conditions. These multiplex PCR primers can be useful in detecting the presence of ALV-J, with modification in the ALV primer sequences for detection of ALV-J's unique sequence.
ACKNOWLEDGMENTS
We thank the Indian Council of Agricultural Research, Government of India, for providing the necessary financial support to carry out this work through the ICAR Niche area of excellence in the Animal Biotechnology program.
We thank Venugopal Nair, Institute of Animal Health, Compton, United Kingdom, for his valuable suggestions through the BBSRC collaborative program.
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Abdul-Careem MF, et al. 2006. Development of a real-time PCR assay using SYBR Green chemistry for monitoring Marek's disease virus genome load in feather tips. J. Virol. Methods 133:34–40 [DOI] [PubMed] [Google Scholar]
- 2. Balachandran C, Pazhanivel N, Vairamuthu S, Murali Manohar B. 2009. Marek's disease and lymphoid leucosis in chicken—a histopathological survey. Tamilnadu J. Vet. Anim. Sci. 5:167–170 [Google Scholar]
- 3. Calvert JG, Nazerian K. 1994. An immunoperoxidase plaque assay for reticuloendotheliosis virus and its application to a sensitive serum neutralization assay. Avian Dis. 38:165–171 [PubMed] [Google Scholar]
- 4. Cho KO, Ohashi K, Onuma M. 1999. Electron microscopic and immunohistochemical localization of Marek's disease (MD) herpesvirus particles in MD skin lymphomas. Vet. Pathol. 36:314–320 [DOI] [PubMed] [Google Scholar]
- 5. Churchill AE, Biggs PM. 1967. Agent of Marek's disease in tissue culture. Nature 215:528–530 [DOI] [PubMed] [Google Scholar]
- 6. Crittenden LB, Hayward WS, Hanafusa H, Fadly AM. 1980. Induction of neoplasms by subgroup E recombinants of exogenous and endogenous avian retroviruses (Rous-associated virus type 60). J. Virol. 33:915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davidson I. 2009. The knowledge that human tumor virology can gain from studies on avian tumor viruses. Adv. Tumor Virol. 1:9–19 [Google Scholar]
- 8. Davidson I. 2005. Veiled and unveiled aspects of infection with oncogenic viruses. Keynote lecture article. Proceedings of the 14th World Veterinary Poultry Congress, Istanbul, Turkey [Google Scholar]
- 9. Davidson I. 2001. Differential diagnosis of avian oncogenic viruses. http://www.poultrymed.com Accessed 6 October 2009
- 10. Davidson I, Borenshtain R. 2001. In vivo events of retroviral LTR integration into MDV in commercial poultry: detection of chimeric molecules as a marker. Avian Dis. 45:102–121 [PubMed] [Google Scholar]
- 11. Deng X, et al. 2010. Development of a loop-mediated isothermal amplification method for rapid detection of reticuloendotheliosis virus. J. Virol. Methods 168:82–86 [DOI] [PubMed] [Google Scholar]
- 12. Dougherty RM, Marucci AA, Distefano HS. 1972. Application of immunohistochemistry to study of avian leukosis virus. J. Gen. Virol. 15:149–162 [DOI] [PubMed] [Google Scholar]
- 13. Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. 2000. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 13:559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fadly AM, Venugopal K. 2008. Leukosis/sarcoma group, p 514–568 In Saif YM, et al. (ed), Diseases of poultry, 12th ed Iowa State Press, Ames, IA [Google Scholar]
- 15. Fadly AM, Zavala G, Witter RL. 2008. Reticuloendotheliosis, p 568–588 In Saif YM, et al. (ed), Diseases of poultry, 12th ed Iowa State Press, Ames, IA [Google Scholar]
- 16. Fadly AM. 2000. Isolation and identification of avian leukosis viruses: a review. Avian Pathol. 29:529–535 [DOI] [PubMed] [Google Scholar]
- 17. Fadly AM, Witter RL. 1998. Oncornaviruses: leukosis/sarcoma and reticuloendotheliosis, p 185–196 In Glisson JR, Jackwood DJ, Pearson JE, Reed WM, Swayne DE. (ed), A laboratory manual for the isolation and identification of avian pathogens, 4th ed American Association of Avian Pathologists, Kennett Square, PA [Google Scholar]
- 18. Fadly AM, Witter RL. 1997. Comparative evaluation of in vitro and in vivo assays for the detection of reticuloendotheliosis virus as a contaminant in a live vaccine of poultry. Avian Dis. 41:695–701 [PubMed] [Google Scholar]
- 19. Frumkin D, et al. 2008. Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues. BMC Biotechnol. 8:17 doi:10.1186/1472-6750-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia M, Narang N, Reed WM, Fadly AM. 2003. Molecular characterization of reticuloendotheliosis virus insertions in the genome of field and vaccine strains of fowlpox virus. Avian Dis. 43:343–354 [DOI] [PubMed] [Google Scholar]
- 21. Handberg KJ, Nielson OL, Jorgensen PH. 2001. The use of serotype 1 and serotype 3 specific PCR for the detection of MDV in chickens. Avian Pathol. 30:243–249 [DOI] [PubMed] [Google Scholar]
- 22. Kim TJ, Tripathy DN. 2001. Reticuloendotheliosis virus integration in the Fowl poxvirus genome: not a recent event. Avian Dis. 45:663–669 [PubMed] [Google Scholar]
- 23. Koyama HY, Suzuki T, Ohwada Y, Saito Y. 1976. Reticuloendotheliosis group virus pathogenic to chicken isolated from material infected with turkey herpesvirus (HVT). Avian Dis. 20:429–434 [PubMed] [Google Scholar]
- 24. Lee SI, Takagi M, Ohashi K, Sugimoto C, Onuma M. 2000. Difference in the meq gene between oncogenic and attenuated strains of Marek's disease virus serotype 1. J. Vet. Med. Sci. 62:287–292 [DOI] [PubMed] [Google Scholar]
- 25. Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. 2006. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 4:283–294 [DOI] [PubMed] [Google Scholar]
- 26. Payne LN, Fadly AM. 1997. Leukosis/sarcoma group, vol IA Iowa State University Press, Ames, IA [Google Scholar]
- 27. Santos VLSL, et al. 2009. Detection of reticuloendotheliosis virus by immunohistochemistry and in situ hybridization in experimentally infected chicken embryo fibroblasts. Braz. J. Vet. Pathol. 2:29–34 [Google Scholar]
- 28. Silva RF, Fadly AM, Taylor SP. 2007. Development of a polymerase chain reaction to differentiate avian leukosis virus (ALV) subgroups: detection of an ALV contaminant in commercial Marek's disease vaccines. Avian Dis. 51:663–667 [DOI] [PubMed] [Google Scholar]
- 29. Smith EJ, Fadly AM. 1988. Influence of congenital transmission of endogenous virus-21 on the immune response to avian leukosis virus infection and the incidence of tumors in chickens. Poult. Sci. 67:1674–1679 [DOI] [PubMed] [Google Scholar]
- 30. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 31. Thapa BR, Omar AR, Arshad SS, Hair-Bejo M. 2004. Detection of avian leukosis virus subgroup J in chicken flocks from Malaysia and their molecular characterization. Avian Pathol. 33:359–363 [DOI] [PubMed] [Google Scholar]
- 32. Venugopal K, Howes K, Flannery DMJ, Payne LN. 2000. Isolation of acutely transforming subgroup J avian leukosis viruses that induce erythroblastosis and myelocytomatosis. Avian Pathol. 29:327–332 [DOI] [PubMed] [Google Scholar]
- 33. Venugopal K, Smith LM, Howes K, Payne LN. 1998. Antigenic variants of J subgroup avian leukosis virus: sequence analysis reveals multiple changes in the env gene. J. Gen. Virol. 79:757–766 [DOI] [PubMed] [Google Scholar]
- 34. Witter RL, Fadly AM. 2003. Reticuloendotheliosis, p 517–535 In Saif YM, et al. (ed), Diseases of poultry, 11th ed Iowa State University Press, Ames, IA [Google Scholar]
- 35. Witter RL, Schat KA. 2003. Marek's disease, p 407–465 In Saif YM, et al. (ed), Diseases of poultry, 11th ed Iowa State University Press, Ames, IA [Google Scholar]
- 36. Xu B, et al. 2004. Occurrence of avian leukosis virus subgroup J in commercial layer flocks in China. Avian Pathol. 33:13–17 [DOI] [PubMed] [Google Scholar]
- 37. Zavala G, Cheng S. 2006. Detection and characterization of avian leukosis virus in Marek's disease vaccines. Avian Dis. 50:209–215 [DOI] [PubMed] [Google Scholar]