Abstract
A novel and quantitative high-throughput screening approach was explored as a tool for the identification of novel compounds that inhibit chlamydial growth in mammalian cells. The assay is based on accumulation of a fluorescent marker by intracellular chlamydiae. Its utility was demonstrated by screening 42,000 chemically defined compounds against Chlamydia caviae GPIC. This analysis led to the identification of 40 primary-hit compounds. Five of these compounds were nontoxic to host cells and had similar activities against both C. caviae GPIC and Chlamydia trachomatis. The inhibitory activity of one of the compounds, (3-methoxyphenyl)-(4,4,7-trimethyl-4,5-dihydro-1H-[1,2]dithiolo[3,4-C]quinolin-1-ylidene)amine (MDQA), was chlamydia specific and was selected for further study. Selection for resistance to MDQA led to the generation of three independent resistant clones of C. trachomatis. Amino acid changes in SecY, a protein involved in Sec-dependent secretion in Gram-negative bacteria, were associated with the resistance phenotype. The amino acids changed in each of the resistant mutants are located in the predicted central channel of a SecY crystal structure, based on the known structure of Thermus thermophilus SecY. These experiments model a process that can be used for the discovery of antichlamydial, anti-intracellular, or antibacterial compounds and has led to the identification of compounds that may have utility in both antibiotic discovery and furthering our understanding of chlamydial biology.
INTRODUCTION
Chlamydiae are a successful group of obligate intracellular bacteria that cause serious diseases in a wide range of hosts and have a developmental cycle that is unique among prokaryotes. Infection is initiated by a metabolically inactive infectious elementary body (EB) that is taken up into the host cell, where it differentiates into a metabolically active noninfectious reticulate body (RB). Chlamydial RBs replicate in a membrane-bound vacuole (the inclusion) until about 18 h postinfection, when asynchronous dedifferentiation to infectious EBs can first be observed (1). Golgi-derived vesicles are trafficked to the inclusion where chlamydiae direct the modification of the inclusion membrane through secretion of proteins that facilitate vacuolar modification and interaction with host proteins (18, 21, 22, 27, 30).
Chlamydiae are known to sequester a variety of host-generated lipids from their intracellular environments (8, 32). One of these is sphingomyelin, a phospholipid involved in lipid trafficking from the Golgi apparatus to the plasma membrane. In cells infected with chlamydiae, a biologically active fluorescent lipid, 6{N-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl)-amino]caproylsphingosine} (C6-NBD-ceramide), is converted to sphingomyelin by the host and then recruited to the chlamydial inclusion (14, 15, 23, 29, 37), a process that can be exploited to fluorescently label chlamydiae in live infected cells. In this study, we demonstrate that C6-NBD-ceramide can be used in a robust, reproducible, and simple high-throughput assay for identifying inhibitors of chlamydial growth, using Chlamydia caviae as a model organism.
Although stable clinical antibiotic resistance has not been documented within human-pathogenic chlamydiae, resistance can evolve in vitro in response to antibiotic stressors through the accumulation of point mutations or via horizontal gene transfer and homologous recombination (11, 12, 28, 35). In vitro spontaneous resistance to a broad range of antibiotic classes has been demonstrated in chlamydiae by culturing the bacteria in the presence of subinhibitory concentrations of antibiotics. This process selects for mutations in the genes targeted by the antibiotic (e.g., rifampin and ofloxacin resistance) or in genes associated with resistance to the antibiotic (2–6, 13, 19, 25, 26, 33, 38). Here, we have shown that resistant mutants can be selected for with novel uncharacterized inhibitors whose mechanisms of action are unknown. Mutations correlating with the resistance phenotype identified by genome sequencing indicate genetic resistance determinants or possible pathways that are perturbed by treatment with the inhibitors. Spontaneous resistant mutants were selected for, cloned, and sequenced to a hit identified in the high-throughput screen. Three independently generated resistant mutants had mutations in secY, which encodes SecY, a structural protein required for the secretion of sec-dependent protein substrates across the bacterial inner membrane (20). MIC values varied depending on the position of the nucleotide change in each clone. These data suggest that SecY is a resistance determinant in Chlamydia trachomatis L2-434 and that specific mutations in this gene can confer resistance to a novel antichlamydial compound identified in a high-throughput screen.
MATERIALS AND METHODS
Reagents and antibodies.
C6-NBD-ceramide was purchased from Life Technologies (Grand Island, NY; catalog number N-1154). The original high-throughput screen was conducted at SIGA Technologies (Corvallis, OR), who supplied micromolar amounts of the original compounds. Milligram amounts of (3-methoxyphenyl)-(4,4,7-trimethyl-4,5-dihydro-1H-[1,2]dithiolo[3,4-C]quinolin-1-ylidene)amine (MDQA) were purchased from a commercial source (Chembridge, San Diego, CA; catalog number 5678481). Cells infected with Chlamydia spp. or Coxiella burnetii were labeled with monoclonal or polyclonal primary antibodies specific to chlamydial lipopolysaccharide (LPS) (34) or whole C. burnetii (16). Species- and isotype-specific secondary antibodies were purchased from Life Technologies (Alexa Fluor 488 [green] and 594 [red]).
Host cells, chlamydial strains, and cultures.
McCoy cells were propagated in minimum essential medium supplemented with 10% fetal bovine serum (MEM-10) and 10 μg/ml gentamicin. The following chlamydia strains were used in this study; C. caviae GPIC, C. trachomatis L2-434, C. trachomatis J6276, Chlamydia psittaci CP3, and Chlamydia muridarum MOPN. Other bacteria used to evaluate broad-spectrum activity of compounds were Coxiella burnetii Nine Mile phase II (obtained from Robert Heinzen, Rocky Mountain Laboratories), Staphylococcus aureus (ATCC number 25923), methicillin-resistant S. aureus (MRSA; ATCC 43300), and Escherichia coli (ATCC 25922).
Primary screen.
Over 40,000 compounds were assayed from the SIGA chemical compound library for antichlamydial activity. The chemical library owned by SIGA Technologies consists of approximately 250,000 compounds, representing a broad chemical structural and property diversity space. Screening of this library has identified unique and potent drug-like compounds representing diverse and novel chemical structural classes that are selectively active against phylogenetically unrelated viral families.
We tested 4,500 compounds each week using 48 96-well clear-bottom, black-walled plates. A total of 5 × 104 McCoy cells were plated in 100 μl of MEM into wells and incubated overnight to a confluence of 100%. A Perkin-Elmer MultiPROBE II HT Plus robotic system was used to deliver individual structurally defined candidate inhibitory compounds from the SIGA library to 80 wells of each plate at a concentration of 5 μM. Wells in the first column of each plate were mock-infected controls, and the last column of each plate contained infected wells not treated with any compound. Controls in each assay also included wells incubated with chlamydiae plus tetracycline at 10 μg/ml. Gradient-purified (7) C. caviae GPIC cells were inoculated onto wells of the plates at a multiplicity of infection of between 1 and 2 inclusion-forming units per cell. Chlamydiae were centrifuged onto cells at 1,200 × g for 1 h at 37°C and incubated under standard culture conditions for 24 h.
At 24 h postinfection, medium was aspirated from cells and replaced with 50 μl of a 10 μM C6-NBD-ceramide in phosphate-buffered saline (PBS). Plates were incubated for 1 h at 37°C, and then the fluorescent compound was aspirated from each well. Cells were then overlaid with MEM-10 and incubated at 37°C for an additional 3 h. At 28 h postinfection, the medium was replaced with PBS and fluorescence intensity was quantified on a Wallac Envision plate reader using an excitation wavelength of 485 nm and emission wavelength of 535 nm, with the instrument programmed to take readings from the bottom of the well. Positive inhibitor compounds were identified by a reduction in fluorescence intensity of 50% or greater relative to that of untreated but infected control wells. All wells identified as positive by the plate reader were evaluated visually on the fluorescence microscope in order to eliminate wells with culture artifacts and to identify compounds that were acutely toxic to the cells.
Immunofluorescence.
A total of 1 × 105 cells were plated onto 12-mm coverslips in 24-well trays, infected as described above, fixed with 100% methanol for 10 min, and washed twice with PBS. Fixed cells were labeled with monoclonal mouse anti-MOMP or anti-LPS primary antibodies at a 1:1,000 dilution followed by appropriate secondary antibodies at a 1:1,000 dilution.
Infections.
A total of 4.5 × 105 cells were plated in 24-well trays and incubated overnight to a confluence of 100%. Bacterial strains were suspended in MEM-10 and infected onto cells at a multiplicity of infection of 0.5 to 1 bacteria per cell. Plates were centrifuged at 1,200 × g at 37°C for 1 h. Media were aspirated, and then compounds were suspended in fresh MEM-10 and added to plates in 0.5-ml aliquots and transferred to a 37°C incubator.
Quantification of bacteria from infected cell culture using TaqMan quantitative real-time PCR.
At the appropriate time points, infected cells were briefly sonicated to lyse and release bacteria. The lysates were collected in microcentrifuge tubes and stored at −20°C prior to analysis. DNA was extracted using the Qiagen DNeasy blood and tissue kit protocol with the addition of 5 mM dithiothreitol (DTT) to the initial lysis buffer. Quantitative PCR (qPCR) was run on all samples in triplicate using the TaqMan universal PCR master mix (Applied Biosystems), with an input of 5 μl of template DNA from each treatment for C. caviae GPIC and C. trachomatis L2-434-infected cells. Primers and probes were specific to C. caviae ompA (F-CCCTGCGCGGATGCT, R-CAGGCGATCCTTGTGATCCT, probe 6-carboxyfluorescein [FAM]-CATCACACCAAGTAGAGC-MGBNFQ) and C. trachomatis ompA (F-CATGGTATCTCCGAGCTGACC, R-ACTGTCTTTGATGTTACCACTCTGAAC, probe 6FAM-CTAGCTTTCACATCGCC-MGBNFQ). Tenfold serial dilutions of quantitated plasmid standards containing each targeted gene were included at copies ranging from 1 × 109 to 1 × 103. Genome copy number and standard error were calculated using an ABI StepOne real-time PCR machine with standard curve settings.
Acid sensitivity and cell cytotoxicity.
The acid lability of MDQA was assessed by suspending the compound at 10 mM in a citric acid buffer (10 mM citric acid, 10 mM KCl, 135 mM NaCl, pH 3) for 10 min prior to dilution to 10 μM and the addition to C. trachomatis L2-434-infected cells. Chlamydial growth was assayed 40 h postinoculation using qPCR. The MTT reagent was used as a measure of cytotoxicity for compounds, which were tested at concentrations up to 50 μM, using a standard assay system (Life Technologies). Rabbit kidney, murine McCoy, human HeLa, and bovine embryonic kidney immortalized cell lines were tested in these assays.
Secondary confirmation and MIC analysis.
MICs of each compound were determined using the Hewlett Packard D300 digital dispenser (Tecan Systems, San Jose, CA) that delivered compound in half-log dilutions to 96-well plates in concentrations that ranged from 20 μM to 0.0063 μM. Chlamydiae were then infected onto cells as described above under “Primary screen” and incubated until they were fixed for fluorescent antibody analysis (C. trachomatis) or labeled with C6-NBD-ceramide (C. caviae GPIC) to determine the MIC. When immunofluorescence was used to measure inhibition, the MIC was determined microscopically and scored as the inhibitory concentration that reduced observable inclusion formation by at least 90%. In assays where the C6-NBD-ceramide assay was used to examine inhibition, the MIC was determined by a quantitative measure of reduction of fluorescence to levels at or below the background. This generally translated to a reduction of fluorescence to below 12,500 on an Envision multiplate fluorescence spectrometer.
Selection for resistance.
Strains of C. trachomatis L2-434 were inoculated onto McCoy cells in 24-well trays using a multiplicity of infection between 1 and 10, depending on the strain. Infections were centrifuged at 1,200 × g for 1 h at 37°C, overlaid with subinhibitory concentrations (1/2 of MIC) of the appropriate drug, and incubated at 37°C for 24 to 48 h depending on the species inoculated. The cell monolayers were then disrupted by −80°C/37°C freeze-thaw or sonication, and then the lysate was centrifuged at 1,500 rpm for 10 min to pellet cell debris. Supernatants from disrupted cell monolayers were used to infect fresh McCoy cell monolayers in 24-well plates, and the concentration of compound was increased incrementally in sequential cultures until the MIC exceeded that of the original wild-type parental strains by 2-fold or greater. Emerging resistance was monitored using immunofluorescence. As resistant chlamydiae emerged in the treated population, they were cloned by limiting dilution.
Paired-end Illumina genome sequencing.
Purified elementary bodies were prepared for sequencing as previously described (35). Draft genomes were assembled using the reference-guided assembly software Maq (http://maq.sourceforge.net/) and aligned to the published C. trachomatis L2-434 (GenBank accession no. AM884177) or C. muridarum (GenBank accession no. AE002160) genome sequences. Single nucleotide polymorphisms (SNPs) identified by the genome alignment were confirmed by standard PCR sequencing. Any necessary manual sequence analysis was performed using MacVector sequence analysis software (MacVector, Cary, NC).
SecY structure prediction and modeling.
The amino acid sequence of C. trachomatis SecY was submitted to the Phyre prediction server on the Web to predict the three-dimensional (3D) structure (17). The mutated residues in the resistant SecY genes were mapped onto the predicted SecY three-dimensional structure using the UCSF Chimera modeling program (24).
RESULTS
Identification and characterization of antichlamydial compounds.
The NBD-cer assay allowed screening of 42,000 compounds at a rate of approximately 5,000 compounds per week. The images in Fig. 1 show qualitatively the difference between complete inhibition, no inhibition, and partial inhibition in the assay. A quantitative analysis of these differences demonstrated that wells treated with noninhibitory compounds had an average fluorescence reading that was 95% of the fluorescence of untreated, infected wells, while wells carrying infected cells treated with tetracycline as a positive control for inhibition had average fluorescence readings that were 22% of infected, untreated cells. This was virtually identical to the readings for uninfected cells treated with the fluorescent compound. The Z-factor for this assay was determined to be 0.74, with a signal to background ratio of 4.7.
Fig 1.
Labeling of infected host cells with C6-NBD-ceramide can be used to discriminate between productively infected, uninfected, or nonproductively infected cells. McCoy cells infected with C. caviae GPIC labeled with C6-NBD-ceramide at 28 h postinfection. Panel A shows cells treated with 10 μg/ml tetracycline. Panel B shows infected cells not treated with inhibitor. Panel C shows cells treated with one compound in the SIGA library that partially blocks the development of C. caviae GPIC within cells at 10 μM. Scale bar = 10 μm.
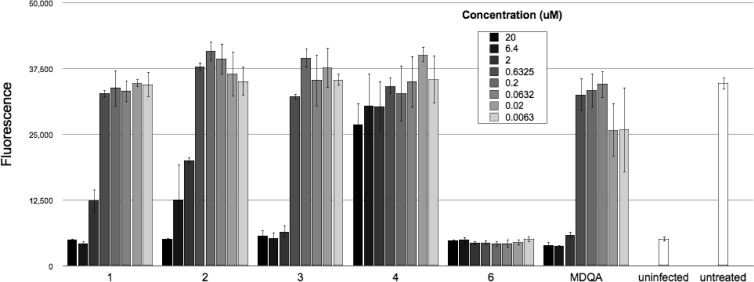
The assay allowed us to identify 40 compounds out of 42,000 tested compounds (hit rate of <0.1%) that inhibited C. caviae GPIC growth and were not inherently toxic to host cells. A secondary screen was done to evaluate the inhibitory properties of these 40 compounds against both C. caviae GPIC and the human pathogen, C. trachomatis. Five compounds that inhibited both chlamydial strains and were nontoxic to host cells were further evaluated (Fig. 2). MICs were established for each of these compounds against C. trachomatis L2-434, C. trachomatis J6276, C. muridarum, and C. caviae GPIC to show their cross-chlamydia antimicrobial activity (Table 1). All 5 compounds were also effective at a concentration of 10 μM against the category B select agent, C. psittaci (Fig. 3), although MIC values were not determined for this organism. Substructure searches of the compound library identified several structurally related analogs for each compound class, but no clear structure activity relationships could be identified. The antichlamydial compound MDQA (molecular weight, 369) (Fig. 4) was selected for further study.
Fig 2.
Concentration-dependent inhibition of chlamydial infections by different compounds in the SIGA library. C. caviae GPIC-infected cells were treated with half-log dilutions of each of 5 compounds identified as chlamydial inhibitors in the high-throughput screen and quantitated at 24 h postinfection (hpi) using C6-NBD-ceramide. Compound 4 is included as an example of a compound with no effect in the assay. Fluorescence readings (y axis) are plotted against each compound at different concentrations (x axis).
Table 1.
MIC of each compound identified in the high-throughput screen against four different chlamydial strains and speciesa
| Strain | MIC |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 6 | MDQA | |
| C. trachomatis L2-434 | 5 | 10 | 0.3125 | <0.156 | 5 |
| C. trachomatis J6276 | 5 | 10 | 0.3125 | <0.156 | 10 |
| C. muridarum Mopn | 2.5 | 10 | 2.5 | <0.156 | 10 |
| C. caviae GPIC | 6.4 | 20 | 2 | <0.0063 | 2 |
MICs for C. trachomatis and C. muridarum were tested in 2-fold dilutions starting at 10 μM and assayed by the fluorescent antibody test. Infections were fixed at 40 hpi (C. trachomatis) or 20 hpi (C. muridarum), and MICs were determined by the concentration that resulted in >90% inhibition. C. caviae GPIC MICs were determined using the robotic delivery system and the fluorescent C6-NBD-ceramide assay in half-log dilutions starting at 20 μM.
Fig 3.
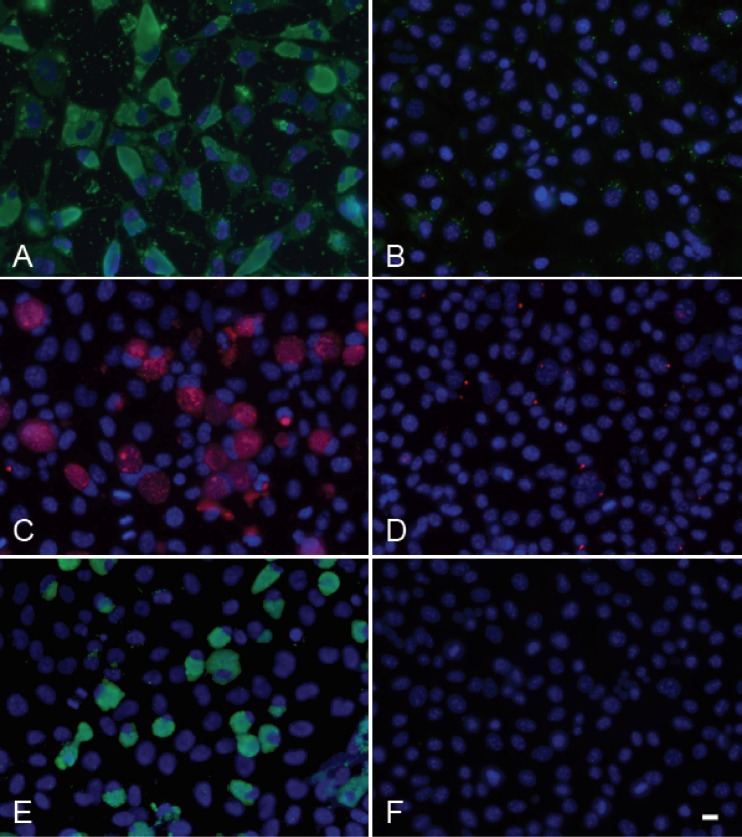
Immunofluorescence images of MDQA-treated C. caviae GPIC-, C. psittaci CP3-, and C. trachomatis L2-434-infected cells. McCoy cells were infected with each of the following strains and grown in the presence of either 1% DMSO as a vehicle control (A, C, E) or with MDQA at 10 μM (B, D, F) before fixation at each indicated time point. Panels A and B are C. psittaci CP3-infected cells fixed at 72 h. Panels C and D are C. trachomatis L2-434-infected cells fixed at 42 h. Panels E and F are C. caviae-infected cells fixed at 42 h. Scale bar = 10 μm.
Fig 4.

Molecular structure of MDQA.
Cell cytotoxicity analysis demonstrated that MDQA had a cytotoxic concentration (CC50) above 50 μM for Vero and McCoy cells. MDQA was freely soluble in 100% dimethyl sulfoxide (DMSO) and soluble to greater than 10 mM in 10% DMSO. Incubation of MDQA in a low pH buffer led to a significant increase in the MIC of the compound, suggesting that it is acid labile. MDQA was not inhibitory to E. coli or S. aureus grown in broth culture up to 100 μM or C. burnetii grown in Vero cells up to 10 μM.
Selection for chlamydial resistance to MDQA.
Chlamydial strains were serially passaged in increasing concentrations of compound until the MIC exceeded that of the original wild-type parental strains. The number of passages to generate a fully resistant mutant varied with each attempt and each strain. A concentration curve of C. trachomatis L2-434 wild-type and resistant strains shows that the mutant strain grows equally well in the presence and absence of MDQA and similarly to the wild-type strain (Fig. 5). The MDQA-resistant C. trachomatis strains did not exhibit cross-resistance to chloramphenicol, tetracycline, or rifampin, (not shown). The complete genome sequence of this resistant strain was assembled, and a single nucleotide change was identified that resulted in an amino acid substitution secY (C. trachomatis CT 510). To ensure that the mutation in secY was not a random occurrence, two additional C. trachomatis L2-434 mutants were generated and a PCR-amplified fragment containing secY was sequenced. In each case, mutations in secY were identified that resulted in amino acid substitutions (Fig. 6). The three-dimensional structure of the C. trachomatis L2-434 SecY protein was predicted using I-TASSER threading software and shown to be similar to the crystallized SecY protein from Thermus thermophilus (Fig. 7). The mutated amino acid residues from C. trachomatis were mapped onto the 3D structure of T. thermophilus SecY, revealing localization to the inner channel of the translocon.
Fig 5.
qPCR of sensitive and resistant C. trachomatis L2-434 strains grown in the presence of MDQA. C. trachomatis L2-434-sensitive and C. trachomatis L2-434-resistant (clone 1) strains were cultured in McCoy cell monolayers in the presence or absence of MDQA at various concentrations (x axis). Infections were lysed at 40 hpi, and genome copies/ml were determined by qPCR (y axis).
Fig 6.
Amino acid changes in SecY that are associated with MDQA resistance in C. trachomatis L2-434. Resistant strains were maintained in culture until the concentration exceeded the sensitive strain MIC by at least 2-fold before whole-genome sequencing or gene-specific (secY) sequencing. Three different mutations leading to amino acid substitutions in SecY were identified in 3 independently generated resistant mutants.
Fig 7.
Predicted 3D structure of the chlamydial SecY translocon. The amino acid sequence of C. trachomatis L2-434 SecY was submitted to I-TASSER threading software to obtain a predicted 3D image. A top and side view of the predicted C. trachomatis SecY structure (A) is shown next to the crystallized SecY protein from T. thermophilus (B). The mutated amino acids of the MDQA-resistant C. trachomatis strains (plus each flanking residue) are highlighted on each structure, as well as those residues homologous to the C. trachomatis mutations on the crystallized T. thermophilus protein. Amino acid substitutions are highlighted as follows: alanine-to-serine substitution at amino acid 45 (red), alanine-to-proline substitution at amino acid 246 (blue), and alanine-to-proline substitution at amino acid 420 (orange).
Nucleotide sequence analysis found different structurally significant amino acid changes in SecY in each of the three characterized resistant strains. This includes one strain in which the entire genome was sequenced, and the mutation in secY was the only identified mutation. While this implicates SecY as having a role in the resistance phenotype, it is not clear whether SecY is the target of MDQA or whether changes in the protein function as suppressors of the sensitive, wild-type phenotype. The resulting amino acid changes (Ala → Pro, Ala → Pro, Ala → Ser) (Fig. 6) map to the central channel of the SecY translocon, perhaps altering the structure of this channel. There is precedent for changes in the SecY inner channel conferring a suppressor phenotype toward lethal mutations in other systems. A set of E. coli secY mutant strains, termed “prlA suppressors,” carry single-amino-acid substitutions that are similarly localized to the SecY translocon inner channel. The prlA mutants retain Sec activity and allow translocation of proteins with inactive or nonexistent signal sequences (31, 36). We are currently working toward the definition of the activity of MDQA and the nature of the resistance phenotype in the secY mutants.
DISCUSSION
High-throughput screens are an effective way to identify novel inhibitors against a wide range of organisms and can be a very powerful approach for new antibiotic discovery. With the use of sophisticated robotics to streamline workflow, we completed a high-throughput screen in which we identified and characterized 5 potent antichlamydial compounds that display a broad spectrum of activity against chlamydia species. We also show that the C6-NBD-ceramide assay with C. caviae GPIC is a useful model system that can be used to screen and identify narrow and broad-spectrum antibiotics that target intracellular and extracellular organisms.
We used chemical genetics and next-generation sequencing technology to identify potential drug targets or resistance determinants that were associated with resistance to MDQA. The MDQA-resistant C. trachomatis L2-434 strains harbor amino acid substitutions in secY, which encode the membrane translocon of the Sec-dependent secretion system. Sec-dependent secretion is a critical pathway utilized by Gram-negative bacteria to secrete proteins across the inner membrane, and knockouts of secY in other bacterial systems have proven to be lethal (10). Homologs to components of the Sec-dependent secretion system are encoded by each chlamydial genome. The ability of MDQA to inhibit chlamydial growth, and subsequent identification of mutations in secY that confer resistance, supports a logical hypothesis that an active Sec pathway is essential for chlamydial growth and life cycle progression (9). Moreover, MDQA-resistant C. trachomatis was still sensitive to chloramphenicol, tetracycline, and rifampin, suggesting that resistance was specific for MDQA and not the result of common mutations that lead to multidrug resistance. It is still unknown if MDQA directly targets the chlamydial sec-dependent secretion pathway or if chlamydiae mutate to regulate interactions of substrates with their secretion systems that allow them to circumvent growth restrictions imposed by treatment with MDQA.
Although chlamydiae remain sensitive to several classes of antibiotics and have not yet shown evidence of antibiotic resistance in clinical settings, many other pathogens that coinfect the genital and respiratory tracts are associated with emerging antibiotic resistance. The identification of chlamydia-specific inhibitory compounds may be clinically significant, as the use of such compounds might reduce the overall application of broad-spectrum antibiotics in clinics and reduce the emergence of antibiotic resistance to broad-spectrum antibiotics in other organisms. We continue to explore the utility of this compound as a potential drug candidate in preclinical experiments and to evaluate the antimicrobial properties of other compounds identified in our screen.
ACKNOWLEDGMENTS
We acknowledge Jeff Burnett and Kelsey Quinn for their help during the high-throughput screen, Kayla Kickner, Erin Brown, and the many individuals employed at SIGA technologies who provided ongoing technical support throughout the study. We also thank Katja Zellmer for assistance with real-time PCR. We thank R. Heinzen of the Rocky Mountain Laboratories, NIH, for antibodies to C. burnetii.
This research is supported by grants AI088540-02 and AI086469-01 from the National Institutes of Health.
Dennis E. Hruby and Robert Jordan declare financial interests as shareholders with SIGA Technologies; however, MDQA has been released to the public domain and is not a product to be controlled by SIGA.
Footnotes
Published ahead of print 29 May 2012
REFERENCES
- 1. Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959 [DOI] [PubMed] [Google Scholar]
- 2. Binet R, Bowlin AK, Maurelli AT, Rank RG. 2010. Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs. Antimicrob. Agents Chemother. 54:1094–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Binet R, Maurelli AT. 2009. The chlamydial functional homolog of KsgA confers kasugamycin sensitivity to Chlamydia trachomatis and impacts bacterial fitness. BMC Microbiol. 9:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binet R, Maurelli AT. 2005. Fitness cost due to mutations in the 16S rRNA associated with spectinomycin resistance in Chlamydia psittaci 6BC. Antimicrob. Agents Chemother. 49:4455–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binet R, Maurelli AT. 2007. Frequency of development and associated physiological cost of azithromycin resistance in Chlamydia psittaci 6BC and C. trachomatis L2. Antimicrob. Agents Chemother. 51:4267–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Binet R, Maurelli AT. 2005. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrob. Agents Chemother. 49:2865–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carabeo RA, Mead DJ, Hackstadt T. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. U. S. A. 100:6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen D, et al. 2010. Secretion of the chlamydial virulence factor CPAF requires the Sec-dependent pathway. Microbiology 156:3031–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dalal K, Duong F. 2011. The SecY complex: conducting the orchestra of protein translocation. Trends Cell Biol. 21:506–514 [DOI] [PubMed] [Google Scholar]
- 11. DeMars R, Weinfurter J. 2008. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J. Bacteriol. 190:1605–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Demars R, Weinfurter J, Guex E, Lin J, Potucek Y. 2007. Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J. Bacteriol. 189:991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dessus-Babus S, Bebear CM, Charron A, Bebear C, de Barbeyrac B. 1998. Sequencing of gyrase and topoisomerase IV quinolone-resistance-determining regions of Chlamydia trachomatis and characterization of quinolone-resistant mutants obtained in vitro. Antimicrob. Agents Chemother. 42:2474–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964–977 [PMC free article] [PubMed] [Google Scholar]
- 15. Hackstadt T, Scidmore MA, Rockey DD. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 92:4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howe D, Heinzen RA. 2006. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell Microbiol. 8:496–507 [DOI] [PubMed] [Google Scholar]
- 17. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 18. Kumar Y, Cocchiaro J, Valdivia RH. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646–1651 [DOI] [PubMed] [Google Scholar]
- 19. Kutlin A, Kohlhoff S, Roblin P, Hammerschlag MR, Riska P. 2005. Emergence of resistance to rifampin and rifalazil in Chlamydophila pneumoniae and Chlamydia trachomatis. Antimicrob. Agents Chemother. 49:903–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee VT, Schneewind O. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15:1725–1752 [DOI] [PubMed] [Google Scholar]
- 21. Mital J, Hackstadt T. 2011. Diverse requirements for SRC-family tyrosine kinases distinguish chlamydial species. mBio 2(2):e00031–11 doi:10.1128/mBio.00031-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol. 12:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore ER, Fischer ER, Mead DJ, Hackstadt T. 2008. The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic 9:2130–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 25. Rothstein DM, Suchland RJ, Xia M, Murphy CK, Stamm WE. 2008. Rifalazil retains activity against rifampin-resistant mutants of Chlamydia pneumoniae. J. Antibiot. (Tokyo) 61:489–495 [DOI] [PubMed] [Google Scholar]
- 26. Rupp J, Gebert A, Solbach W, Maass M. 2005. Serine-to-asparagine substitution in the GyrA gene leads to quinolone resistance in moxifloxacin-exposed Chlamydia pneumoniae. Antimicrob. Agents Chemother. 49:406–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rzomp KA, Moorhead AR, Scidmore MA. 2006. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 74:5362–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sandoz KM, Rockey DD. 2010. Antibiotic resistance in Chlamydiae. Future Microbiol. 5:1427–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scidmore MA, Fischer ER, Hackstadt T. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J. Cell Biol. 134:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scidmore MA, Hackstadt T. 2001. Mammalian 14-3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638–1650 [DOI] [PubMed] [Google Scholar]
- 31. Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. 2005. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J. Bacteriol. 187:6454–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su H, et al. 2004. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 279:9409–9416 [DOI] [PubMed] [Google Scholar]
- 33. Suchland RJ, Bourillon A, Denamur E, Stamm WE, Rothstein DM. 2005. Rifampin-resistant RNA polymerase mutants of Chlamydia trachomatis remain susceptible to the ansamycin rifalazil. Antimicrob. Agents Chemother. 49:1120–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suchland RJ, Rockey DD, Bannantine JP, Stamm WE. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 68:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD. 2009. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob. Agents Chemother. 53:4604–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Van den Berg B, et al. 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44 [DOI] [PubMed] [Google Scholar]
- 37. Wolf K, Hackstadt T. 2001. Sphingomyelin trafficking in Chlamydia pneumoniae-infected cells. Cell Microbiol. 3:145–152 [DOI] [PubMed] [Google Scholar]
- 38. Xia M, et al. 2005. Activities of rifamycin derivatives against wild-type and rpoB mutants of Chlamydia trachomatis. Antimicrob. Agents Chemother. 49:3974–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]