Abstract
HCV NS3/4a protease inhibitors are proven therapeutic agents against chronic hepatitis C virus infection, with boceprevir and telaprevir having recently received regulatory approval as add-on therapy to pegylated interferon/ribavirin for patients harboring genotype 1 infections. Overcoming antiviral resistance, broad genotype coverage, and a convenient dosing regimen are important attributes for future agents to be used in combinations without interferon. In this communication, we report the preclinical profile of MK-5172, a novel P2-P4 quinoxaline macrocyclic NS3/4a protease inhibitor currently in clinical development. The compound demonstrates subnanomolar activity against a broad enzyme panel encompassing major hepatitis C virus (HCV) genotypes as well as variants resistant to earlier protease inhibitors. In replicon selections, MK-5172 exerted high selective pressure, which yielded few resistant colonies. In both rat and dog, MK-5172 demonstrates good plasma and liver exposures, with 24-h liver levels suggestive of once-daily dosing. When administered to HCV-infected chimpanzees harboring chronic gt1a or gt1b infections, MK-5172 suppressed viral load between 4 to 5 logs at a dose of 1 mg/kg of body weight twice daily (b.i.d.) for 7 days. Based on its preclinical profile, MK-5172 is anticipated to be broadly active against multiple HCV genotypes and clinically important resistance variants and highly suited for incorporation into newer all-oral regimens.
INTRODUCTION
Chronic hepatitis C virus (HCV) infection afflicts more than 170 million people worldwide and is the major etiological cause of fibrosis, liver cirrhosis, and hepatocellular carcinoma (20, 53). Current treatment relies on a backbone of interferon and ribavirin, a regimen with poor tolerability and toxicity (31, 34). Efforts to develop novel therapies to improve treatment have focused largely on direct acting antiviral agents (DAAs) (19), which therapeutically intervene with virally encoded components essential for HCV replication.
Hepatitis C virus, a member of the Flaviviridae family of viruses in the Hepacivirus genus, is encoded by a 9.6-kb positive-strand RNA genome (8). It is initially translated into a single polypeptide that is subsequently cleaved into individual protein components by a combination of both host- and virally encoded proteases (2, 38). HCV protease inhibitors currently in clinical development span a variety of structural classes. The most advanced of these are keto amide compounds, which covalently bind to the active-site serine of the protease in a slowly reversible manner. Boceprevir (29) and telaprevir (37), both from this class, recently received regulatory approval as add-on therapy to pegylated interferon/ribavirin in the treatment of genotype 1-infected patients. A number of rapidly reversible NS3/4a protease inhibitors, including the P1-P3 constrained macrocyclic inhibitors TMC 435 (23) and danoprevir (45), the P2-P4 constrained macrocyclic inhibitor vaniprevir (33), the linear inhibitors BI 201335 (52), BMS650032 (47), and ABT-450 (51), and others of undisclosed structure, including GS 9451 and ACH-1625, are currently in mid- to late-stage development.
Previously low-nanomolar protease inhibitors utilizing a P2-P4 macrocyclic constraint were described (25). The most advanced compound from this series, vaniprevir (24, 33), is currently being evaluated in clinical trials in combination with pegylated interferon/ribavirin. Unlike the keto amide inhibitors, macrocycles do not derive potency from covalent linkage. While potent, the structural constraints limit their ability to be broadly active and effective outside genotype 1 (24). However, through a concerted structure-based design effort, we have generated compounds in this series which demonstrate increased potency against a broader range of HCV genotypes as well as resistant variants identified in ongoing clinical studies with earlier protease inhibitors (13, 14). This communication focuses on the preclinical profile of the most advanced compound of this new series, MK-5172, which demonstrates potent activity in vitro across genotypes and common resistant variants (1, 9, 18, 42, 44), improved pharmacokinetics in preclinical animal species, and efficacy in the chimpanzee model of HCV infection.
MATERIALS AND METHODS
Compound.
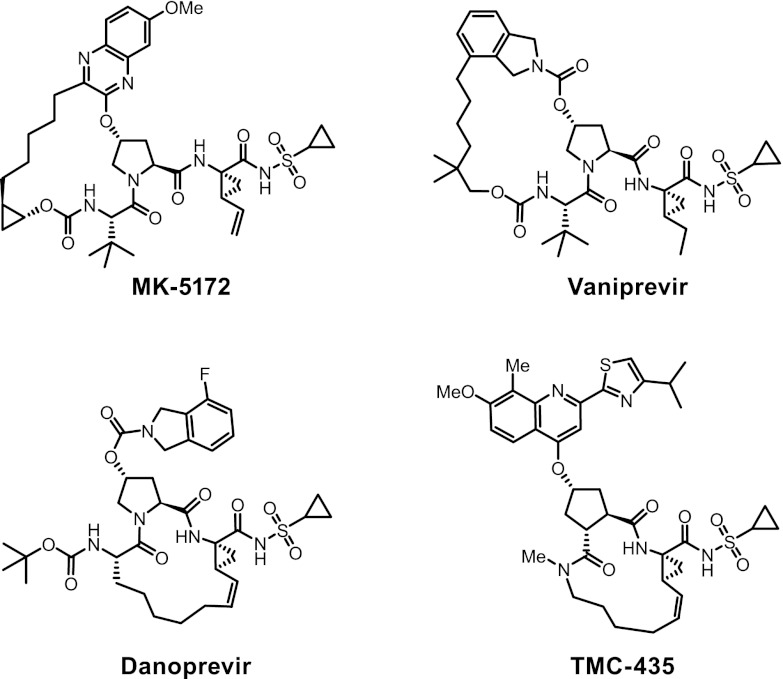
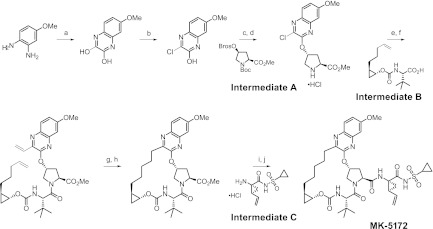
MK-5172, (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide (Fig. 1), was prepared using the synthetic scheme shown in Fig. 2. A more detailed description of the synthetic procedures and structure activity data leading to MK-5172 are published (13). Vaniprevir (33), danoprevir (46), and TMC435 (41) were synthesized internally.
Fig 1.
Structures of HCV NS3/4A protease inhibitors.
Fig 2.
Synthetic route and chemical structure of MK-5172. The reagents used are as follows: (a) diethyloxalate, triethylamine, 150°C; (b) thionyl chloride, dimethylformamide, 110°C; (c) cesium carbonate, 1-methyl-2-pyrrolidinone, intermediate A (Boc, t-butoxycarbonyl; Bros, 4-bromophenyl sulfonyl); (d) HCl, dioxane; (e) O-(7-azabenzotriazol-7-yl)-N,N,N′,N'-tetramethyluronium hexafluorophosphate, diisopropylethylamine, dimethylformamide, intermediate B; (f) potassium vinyltrifluoroborate, triethylamine, dichloro[1,1-bis(diphenylphosphino)ferrocene]palladium (II) chloride, ethanol; (g) Zhan 1B catalyst, 1,3-bis(2,4,6-trimethylphenyl)-4,5-dihydroimidazol-2-ylidene[2-(i-propoxy)-5-(N,N-dimethylaminosulfonyl)phenyl] methyleneruthenium (II) dichloride (CAS 918870-76-5), 1,2-dichloroethane; (h) hydrogen, 10% Pd/C, methanol, dioxane; (i) lithium hydroxide, tetrahydrofuran, tetrahydrofuran, water; (j) O-(benzotriazol-1-yl)-N,N,N′,N'-tetramethyluronium tetrafluoroborate, diisopropylethylamine, 4-dimethylamino pyridine, dichloromethane, intermediate C.
In vitro assays.
Recombinant HCV NS3/4A enzymes were expressed and purified from Escherichia coli as previously described (24). Enzyme sequences were derived from genotype 1a (gt1a) H77 (GenBank accession no. AF09606), gt1b con1 (GenBank accession no. AJ238799), gt2a JFH1 (GenBank accession no. AB047639), gt2b HCJ8 (GenBank accession no. D10988), and gt3a NZL1 (GenBank accession no. D17763). Inhibition of HCV NS3/4A protease activity in reaction mixtures containing MK-5172, vaniprevir, or the reference compounds danoprevir and TMC435 (Fig. 1) was determined in a time-resolved fluorescence assay (32). Cell-based HCV replicon assays were conducted in genotype 1b (con1) stable cell line HB1 (26) or a gt2a cell line (JFH) (17) in the presence of either 10% fetal bovine serum (FBS) or 40% normal human serum (NHS) (7). Determinations of 50% effective concentrations (EC50s) against the panel of genotype or mutant replicon cell lines were conducted using a TaqMan-based assay (24). The 50% cytotoxic concentration (CC50) was determined in the HCV replicon cell line with the use of an MTS assay according to the manufacturer's protocol (Cell Titer Aqueous One; Promega, Madison, WI). Potency determinations against clinical genotype 1 NS3/4A sequences were made using a transient cell-based phenotype assay (28). The NS3/4A patient isolates were cloned from human plasma infected with HCV (28). Broad counterscreening, in which MK-5172 was evaluated for its inhibitory potency at a concentration of 10 μM, was conducted at MDS Pharma Services (Taipei, Taiwan).
For in vitro resistance selections, 100,000 HB1 cells were seeded into a T162 Z-top flask and cultured in the presence of 0.5 mg/ml G418 and the desired concentration of MK-5172. Cells were cultured for approximately 3 weeks with regular exchanges of medium until sufficient cell death had occurred to enable distinct colonies to form. After expansion, total RNA was isolated, used as a template to generate NS3/4a cDNA, and sequenced using conventional molecular biology techniques. Mutations were identified through comparison with the sequence generated from untreated cells.
For the 2-week potency evaluations, 30,000 HB1 cells were seeded per well of a 6-well tissue culture plate per drug concentration. The next day (day 0), the medium was replenished with fresh medium and MK-5172 at the appropriate drug concentration. Cells from a single well per drug concentration were harvested on days 0, 1, and 2, washed, and stored frozen until evaluation. The fourth well was similarly harvested on day 3.5 except that 30,000 cells were reseeded with fresh medium and MK-5172 at the appropriate drug concentration. For additional time points, cells were passaged and harvested every one-half week for 2 weeks. For the third week, cells were similarly treated except that cells received replenishing medium which contained 0.5 mg/ml G418 without protease inhibitor.
Pharmacokinetic studies.
Studies were performed in both rats and dogs. The study protocols were reviewed and approved by the Institutional Animal Care and Use Committees at Merck Research Laboratories. For studies in which MK-5172 was dosed intravenously to rats or dogs, the compound was formulated in polyethylene glycol 200 (PEG200) and administered as a bolus at either 2 mg/kg of body weight (rat) or 0.5 mg/kg (dog). For oral studies, the crystalline potassium salt of the compound was dosed as a solution in PEG400 at 5 mg/kg (rat) or 1 mg/kg (dog). For all studies, blood samples were collected in EDTA-containing tubes at appropriate times and plasma was separated by centrifugation and stored at −70°C until analysis. Quantitation of MK-5172 levels was conducted by high-performance liquid chromatography/mass spectroscopy (LC/MS/MS) following protein precipitation. Liver samples were obtained from rat studies at the termination of the experiment. For dog, liver biopsy samples (∼20 μl) were collected following sedation. Tissue samples were homogenized in four volumes of deionized water, and drug concentrations were determined by LC/MS/MS after protein precipitation.
Studies in HCV-infected chimpanzees.
The housing, maintenance, and care of the chimpanzees (Pan troglodytes) used in the study were in compliance with requirements at both Merck Research Laboratories and New Iberia Research Center (University of Louisiana at Lafayette) (6). The study protocol was reviewed and approved by the Institutional Animal Care and Use Committee at both Merck Research Laboratories and New Iberia Research Center (University of Louisiana at Lafayette), where the experiments were conducted, to ensure compliance with all federal regulations. The HCV genotype was determined by a line probe assay (Versant HCV genotype assay [LiPa]; Bayer Diagnostics/Innogenetics) and confirmed by reverse transcription-PCR (RT-PCR) rescue and sequencing of HCV genetic material (6). HCV-infected chimpanzees were dosed orally at 1 mg per kg twice daily (b.i.d.) for 7 days by the voluntary ingestion of MK-5172 (in a Tang vehicle) or vaniprevir (in a milk vehicle). Viral load determinations were performed on plasma samples using the HCV TaqMan assay (Cenetron Diagnostics, Austin, TX). MK-5172 drug concentrations in plasma or liver biopsy specimens were conducted as described above (under “Pharmacokinetic studies”). Viral resistance analysis was conducted according to a previously published protocol (6).
RESULTS
In vitro activity and selectivity.
In biochemical assays, MK-5172 was effective against a panel of major genotypes and variants engineered with common resistant mutations observed in clinical studies with other NS3/4a protease inhibitors (Table 1). Data for MK-5172 are shown in comparison to data for vaniprevir and two other macrocyclic protease inhibitors currently in clinical development, danoprevir and TMC435. MK-5172 demonstrated subnanomolar potency against all genotypes tested, including proteases of genotypes 2 and 3. It also demonstrated subnanomolar potency against a panel of protease resistance mutants frequently observed in clinical studies of other protease inhibitors (9, 10, 18, 21, 35, 42, 44, 46). Mutations at residue 168 are commonly elicited by macrocyclic inhibitors, such as danoprevir and vaniprevir, while mutations at residues 155 and 156 have been observed in studies with all developmental protease inhibitors evaluated to date, with R155K especially prominent in some viral breakthrough populations due to its high fitness (44). MK-5172 demonstrated an improved mutant potency profile across all variants compared to vaniprevir and especially pronounced inhibition against mutations at R155 and D168.
Table 1.
In vitro potency of MK-5172 and other macrocyclic protease inhibitors against a genotype/mutant panel in an HCV NS3/4A protease enzymatic assay
| Genotype/mutant |
Ki (nM)a |
|||
|---|---|---|---|---|
| MK-5172 | Vaniprevir | Danoprevir | TMC435 | |
| gt1b | 0.01 ± <0.01 | 0.05 ± 0.01 | 0.03 ± <0.01 | 1.49 ± 0.73 |
| gt1a | 0.01 ± 0.01 | 0.07 ± 0.03 | 0.06 ± 0.02 | 1.98 ± 0.23 |
| gt2a | 0.08 ± 0.02 | 0.85 ± 0.31 | 0.17 ± 0.07 | 31.7 ± 5.0 |
| gt2b | 0.15 ± 0.06 | 1.44 ± 0.52 | 0.79 ± 0.07 | 232 + 62 |
| gt3a | 0.90 ± 0.2 | 54 ± 4 | 11.1 ± 2.2 | 850 ± 58 |
| gt1b R155K | 0.07 ± 0.01 | 19 ± 2 | 4.3 ± 0.4 | 136 ± 15 |
| gt1b A156T | 5.3 ± 0.9 | 22 ± 2 | 0.36 ± 0.06 | 35.6 ± 6.2 |
| gt1b A156V | 12 ± 2 | 88 ± 15 | 1.52 ± 0.33 | 390 ± 6 |
| gt1b D168V | 0.14 ± 0.03 | 77 ± 27 | 0.67 ± 0.14 | 598 ± 71 |
| gt1b D168Y | 0.30 ± 0.04 | 120 ± 60 | 0.74 ± 0.20 | 169 ± 53 |
Values are means and standard deviations.
In the replicon assay, MK-5172 demonstrated subnanomolar to low-nanomolar EC50s against genotypes 1a, 1b, and 2a (Table 2), a modest potency shift in high serum concentration, and no evidence for cellular cytotoxicity. When assessed against a replicon panel of protease mutants (Table 2), subnanomolar to low-nanomolar potencies are observed against the R155K and D168Y mutations. The potency against the A156T mutation was less robust, as also observed in the enzyme assays described above. Both in vitro and clinical studies suggest that A156T mutants are debilitated for replication (15, 44).
Table 2.
MK-5172 potency against a genotype/mutant panel of stable replicon cell lines
| Genotype/mutant | MK-5172 EC50 (nM)a |
|---|---|
| gt1b con1 | 0.5 ± 0.1 |
| gt1b con1 40% NHS | 7.0 ± 3 |
| gt1b Q41R | 1.8 ± 1.0 |
| gt1b F43S | 1.3 ± 0.1 |
| gt1b R155K | 0.3 ± 0.1 |
| gt1bA156T | 131 ± 35 |
| gt1b D168Y | 4.0 ± 0.8 |
| gt1a | 2.0 ± 1 |
| gt2a | 8.0 ± 4 |
| CC50 | >50,000 |
Values are means and standard deviations except for CC50, for which the value is only the mean.
MK-5172 potency was retained against common mutations elicited by inhibitors against NS5A (Y93H) (12), NS5B nucleosides (S282T) (36), and NS5B nonnucleoside inhibitors (C316Y) (16) (Table 3). Combination testing (40) with either interferon or ribavirin demonstrates additive to synergistic effects with interferon and little to no effect with ribavirin (data not shown). Potency was also assessed against a panel of NS3/4A sequences from plasma from untreated HCV-infected patients (Table 4) (28). EC50s ranged narrowly between 0.3 and 5.9 nM, suggesting that MK-5172 will be efficacious across the genetically diverse range of genotype 1 infections encountered in clinical settings (9, 18, 39, 42).
Table 3.
MK-5172 potency against a panel of HCV replication mutants
| Target | Genotype or mutation | EC50 (nM)a |
|||
|---|---|---|---|---|---|
| MK-5172 PI | HCV-796 NS5B NNI | MK-0608 Nucleoside | BMS790052 NS5a | ||
| None | gt1b con1 WT | 0.4 ± 0.1 | 4.7 ± 1.6 | 507 ± 106 | 0.002 ± 0.001 |
| NS5A | Y93H | 0.7 ± 0.3 | 9.6 ± 6.9 | 490 ± 360 | >3.5 |
| NS5B | C316Y | 0.4 ± 0.2 | 1,020 ± 240 | 220 ± 74 | 0.005 ± 0.004 |
| NS5B | S282T | 0.3 ± 0.1 | 9.4 ± 6.7 | >40,000 | 0.002 ± 0.001 |
Values are means and standard deviations. PI, protease inhibitor; NNI, nonnucleoside inhibitor. Bolded values indicate a potency shift.
Table 4.
MK-5172 potencies against GT1 NS3/4a patient isolates in an NS3/4a phenotype assay
| NS3/4a sequence | Genotype | EC50 (nM)a |
|---|---|---|
| Con1 | 1b | 0.9 ± 0.7 |
| H77 | 1a | 1.1 ± 0.6 |
| ps20 | 1b | 1.8 ± 0.2 |
| ps29 | 1a | 0.4 ± 0.3 |
| ps30 | 1b | 5.9 ± 1.4 |
| ps31 | 1b | 0.3 ± 0.1 |
| ps32 | 1b | 0.6 ± 0.3 |
| ps33 | 1b | 4.9 ± 0.4 |
| ps36 | 1a | 1.1 ± 0.4 |
| ps38 | 1a | 2.8 ± 0.7 |
| ps40 | 1a | 0.8 ± 0.1 |
| ps41 | 1a | 1.4 ± 0.2 |
Values are means and standard deviations.
To address selectivity, the inhibitory potency of MK-5172 was assessed against a panel of human serine proteases. MK-5172 exhibited modest potency against chymotrypsin (50% inhibitory concentration [IC50] = 1.5 μM), yielding a selectivity index of >150,000, while demonstrating no inhibitory activity against either elastase or trypsin (IC50 > 100 μM).
Pharmacokinetics.
Optimization of compound potency occurred in conjunction with improvements in the pharmacokinetics to provide an overall favorable pharmaceutical profile. The preclinical pharmacokinetic profile was characterized in rat and dog. MK-5172 demonstrated low to moderate clearance and a modest half-life in both rat and dog (Table 5). Upon oral administration, MK-5172 demonstrated modest bioavailability of 12 to 13%, with moderate plasma exposure in both species (Table 5). Significant liver concentrations were achieved in both rat and dog. The 24-h trough liver concentrations were 0.2 μM in rat and 1.4 μM in dog (1 mg per kg), yielding exposure multiples of 27- to 200-fold over the serum-adjusted replicon EC50.
Table 5.
Pharmacokinetic profile of MK-5172 in rat and doga
| Dosing method and species | Dose (mg/kg) | CLp (ml/min/kg) | V (liters/kg) | t1/2 (h) | AUC0-∞ (μM · h) | Cmax (μM) | C24h (μM) | Tmax (h) | %F | 24-h liver concn (μM) |
|---|---|---|---|---|---|---|---|---|---|---|
| i.v. | ||||||||||
| Rat | 2 | 28 ± 22 | 3.1 ± 2.6 | 1.4 ± 0.6 | 2.3 ± 1.6 | |||||
| Dog | 0.5 | 5 ± 2 | 0.7 ± 0.1 | 3.0 ± 0.8 | 2.0 ± 0.7 | |||||
| p.o. | ||||||||||
| Rat | 5 | 0.7 ± 0.3 | 0.3 ± 0.1 | < 0.005 | 4.0 ± 0.0 | 13 | 0.2 | |||
| Dog | 1 | 0.4 ± 0.3 | 0.1 ± 0.1 | 0.04 | 4.7 ± 1.2 | 12 ± 9 | 1.4 |
Values are means and standard deviations. i.v., intravenous; p.o., oral; CLp, plasma clearance; V, volume of distribution; t1/2, half-life; AUC0-∞, area under the concentration-time curve from 0 h to infinity; Cmax, maximum concentration of the drug; C24h, concentration of the drug at 24 h; Tmax, time to maximum concentration of the drug; %F, oral bioavailability.
In vitro resistance and long-term potency assessment.
Genotype 1b replicon cells were cultured for 3 weeks in the presence of G418 and MK-5172 at 6 nM or 30 nM. MK-5172 elicited 26 or 6 drug-resistant colonies, respectively.
Sequence data were generated for 16 colonies (Table 6). The most-common mutations affected amino acids 156 and 168, as observed for other macrocyclic NS3/4a inhibitors (22, 46, 50). Mutations at residues 41 or 43, both of which reside close to the P1 region of the binding pocket, were also noted (21, 25). Subsequent phenotypic analysis demonstrated that these mutations confer little loss in potency (Table 2; additional data not shown).
Table 6.
Mutations elicited by MK-5172
| Drug concn | Mutation(s) |
|||||
|---|---|---|---|---|---|---|
| F43 | A156 | D168 | Q41:A156 | A156:D168 | None | |
| 6 nM | 1 S | 1 T, 2 S | 1A, 1V, 1G | 1 156V:168V, 1 156S:168A | ||
| 30 nM | 1 S, 2 F/S | 1 T, 1 T/S/A | 1 Q41H:A156V | 1 | ||
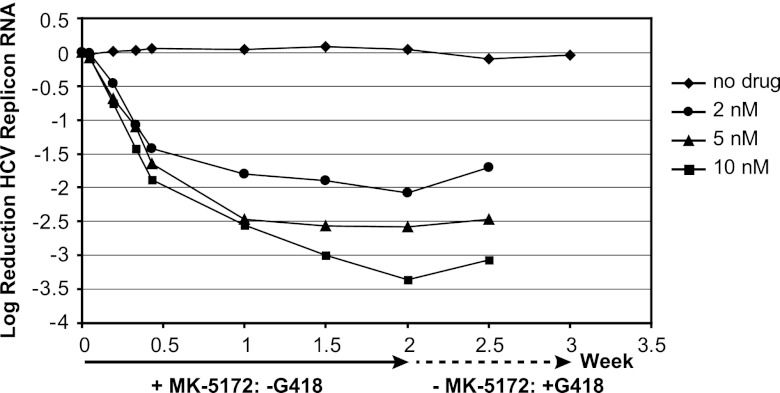
Two-week potency assessments were conducted in the presence of 2, 4, or 10 nM MK-5172 or a dimethyl sulfoxide (DMSO) control without G418 (Fig. 3). Replicon cells were subsequently cultured for an additional week without MK-5172 but with G418 to assess replicon RNA recovery. The initial rates of replicon RNA decline were similar among all MK-5172 concentrations. After approximately one-half week, there was a second slower decline and, for the two lower MK-5172 concentrations, an apparent plateau in replicon RNA reduction. The magnitude of this second decline was drug concentration dependent. After 2 weeks, cells were cultured with medium that contained G418 but not MK-5172. For cells previously treated with MK-5172, there was little apparent cell growth and limited recovery of replicon RNA levels. Cells from the parallel DMSO control arm exhibited no growth inhibition following readdition of G418, and replicon RNA levels were constant throughout the experiment. Resistance was not detected among any of the samples by population-based sequencing of NS3/4a generated from replicon RNA isolated at either the 0- or 2-week time point. Collectively, the data suggest that MK-5172 will maintain antiviral pressure with continued dosing.
Fig 3.
Two-week in vitro potency of MK-5172 against gt1b replicon cells. Stable genotype 1b replicon cells were incubated in the presence of MK-5172 at 2, 5, or 10 nM or with a no-drug DMSO control for 2 weeks and in the presence of G418 (0.5 mg/ml) without MK-5172 for a third week. At the indicated time points, cells were harvested and replicon RNA was quantified by quantitative RT-PCR (qRT-PCR).
In vivo efficacy.
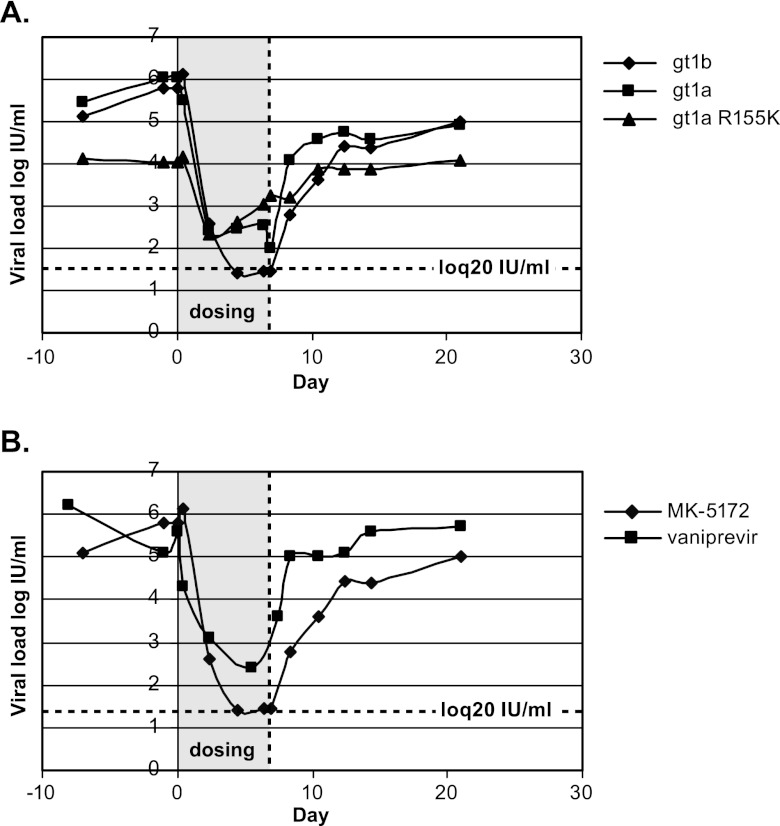
To demonstrate in vivo efficacy, MK-5172 was administered orally to three chronically HCV-infected chimpanzees at a dose of 1 mg per kg twice daily for 7 days. Two of the chimpanzees had wild-type (WT) gt1a or gt1b infections with high viral titers (∼106 IU/ml). A third chimpanzee had a modest viral titer (∼104 IU/ml) that was gt1a NS3 R155K virus. This chimpanzee maintained a chronic R155K viral infection in the absence of prior experimental treatment with an HCV small molecule inhibitor (J. Fontenot, personal communication). Pharmacodynamic responses to MK-5172 are shown in Fig. 4A.
Fig 4.
MK-5172 demonstrates efficacy in vivo against chronic-HCV-infected chimpanzees. (A) MK-5172 was dosed orally at 1 mg/kg b.i.d. as a suspension in Tang to chronic-HCV-infected chimpanzees harboring gt1a, gt1b, or gt1a NS3 R155K infections for 7 days. Blood samples were collected periodically, processed to plasma, and evaluated for HCV viral load. (B) A chronic-HCV-infected chimpanzee infected with a gt1b virus was dosed orally with vaniprevir or MK-5172 at 1 mg/kg b.i.d. for 7 days. The experiments were performed approximately 1.5 years apart, during which time viral load had fully rebounded.
All animals experienced an immediate, profound reduction in viral titer. The gt1a (WT) infection was suppressed ∼4 logs within 2 days to ∼100 IU/ml, and viral suppression was maintained throughout dosing. The gt1b infection was suppressed more than 5 logs to the level of quantification (20 IU/ml); there was no genetic evidence for the emergence of resistance either during dosing or postdosing.
The gt1a NS3 R155K-infected chimp experienced a rapid ∼2-log reduction in viral titer. Viral load gradually drifted higher during the remainder of the dosing period and returned to baseline levels only following cessation of dosing. The virus was homogenous for the R155K mutation throughout the study. There were no additional mutations elicited by dosing and no genetic evidence to suggest that fluctuations in viral titer either during or postdosing were due to newly emerging resistant variants.
MK-5172 concentrations were determined from matched plasma and liver biopsy samples collected 12 h after administration of the final dose (Table 7). Drug concentrations were significantly higher in the liver, ranging from 0.85 to 1.99 μM, compared to the low-nanomolar concentrations in plasma. This yields liver-to-plasma ratios of 425 to 785. Viral load reductions at this time point were greater than 4 logs for the gt1a and gt1b infections and 0.8 logs for the gt1a NS3 R155K infection. Although a pharmacokinetic-pharmacodynamic relationship cannot be determined from a single drug dose, the viral load reductions are more reflective of drug concentrations in liver.
Table 7.
MK5172 concentrations in matched liver/plasma samples from HCV-infected chimpanzees treated with MK-5172
| HCV genotype | MK5172 concn (μM) ina: |
Liver/plasma ratio | |
|---|---|---|---|
| Plasma | Liver | ||
| gt1a | 0.002 | 0.85 | 425 |
| gt1b | 0.003 | 1.99 | 663 |
| gt1a NS3 R155K | 0.002 | 1.57 | 785 |
MK-5172 drug concentrations were determined from paired plasma and liver biopsy samples collected 12 h after the final dose.
The in vivo efficacy of MK-5172 is illustrated further by comparing responses of the gt1b-infected chimpanzee to either MK-5172 or vaniprevir under identical dosing regimens (Fig. 4B). The viral titer was suppressed an additional log with MK-5172. Liver drug concentrations 12 h after final doses were also ∼4-fold higher with MK-5172, at 1.97 μM compared to 0.54 μM with vaniprevir, indicating better drug exposure at the site of HCV replication.
On the basis of the greater potency across both genotypes and clinically relevant resistant mutants, the improved pharmacokinetics, the excellent 24-h liver concentrations in preclinical species, and the in vivo efficacy in HCV-infected chimpanzees, MK-5172 was selected for clinical development.
DISCUSSION
A concerted medicinal chemistry effort directed at optimizing the P2-P4 macrocyclic protease series to increase potency across both HCV genotypes and clinically important resistance variants led to the discovery of MK-5172, a novel P2-P4 quinoxaline macrocyclic peptide (13, 14). In biochemical assays, MK-5172 demonstrated subnanomolar Kis against enzymes derived from HCV genotypes 1 to 3 and a broad panel of resistant mutants, including those either preexisting or prominent in viral breakthrough populations of patients enrolled in ongoing clinical studies of other protease inhibitors (1, 9, 18, 42, 44). The in vitro enzyme potency translated in cell-based replication assays to low-nanomolar EC50s against both wild-type replicons and mutant replicons harboring key R155K and D168Y mutations (30). Phenotypic assays showed that MK-5172 maintained potency across a genetically diverse panel of genotype 1a and 1b sequences derived from plasma of HCV-infected patients. In preclinical animal species, MK-5172 demonstrated a favorable pharmacokinetic profile with good plasma concentrations while maintaining the high liver concentrations as previously described with vaniprevir. Importantly, moderate oral doses achieved 24-h liver concentrations in preclinical species that were well above the in vitro EC50s. Resistance selections demonstrated that MK-5172 elicited few colonies even at low concentrations of the inhibitor. MK-5172 proved highly efficacious in vivo at moderate doses against chronic-HCV-infected chimpanzees, including greater viral load suppression than vaniprevir when dosed alternatively to the same animal at an otherwise identical dose and frequency. Collectively, these properties identified MK-5172 as an inhibitor of greater potency than current developmental HCV protease inhibitors with the potential to improve HCV treatment options. Indeed, early phase I studies in both healthy volunteers and HCV-infected patients showed that MK-5172 has a favorable preclinical profile that translates into a clinically efficacious drug, is broadly active across multiple HCV genotypes, and possesses favorable pharmacokinetics suggestive of once-daily (QD) dosing (3, 4).
Boceprevir and telaprevir have recently gained regulatory approval as the first direct-acting antiviral agents to be combined with current pegylated-interferon/ribavirin treatment to increase rates of sustained virological response in genotype 1 patients. Several other protease inhibitors, including vaniprevir (30), danoprevir (43), ABT450 (21), BI 201335 (5, 39), and TMC435 (10), are progressing through later-stage clinical studies in similar combinations with pegylated interferon/ribavirin with the promise of shortening treatment duration while limiting side effects in this same patient population. As an add-on agent, MK-5172 will likely increase response rates while potentially shortening treatment duration from the current 24- to 48-week paradigm. It is anticipated to also be more effective against the wide array of genotypes and resistant variants that are common among the general patient population (1), including those resistant variants harbored by patients who failed with earlier treatment options which included other protease inhibitors (48, 49, 54).
The long-term vision for HCV therapy is the development of combination DAA regimens that eliminate the requirement for pegylated interferon and its accompanying issues of side effects and patient compliance. Early interferon-sparring combination testing that included a protease inhibitor evaluated short-term efficacy in combinations that included NS5A (27), NS5B nucleoside (11), and NS5B nonnucleoside (55) inhibitors. Whereas promising results were reported, resistance was frequently elicited and in some cases confirmed to be against the protease inhibitor (27, 35, 55). The potency profile of MK-5172 against the resistance mutants commonly observed in clinical settings, coupled with its potential for moderate QD dosing, makes it highly suited for future clinical combination testing with other DAAs.
Footnotes
Published ahead of print 21 May 2012
REFERENCES
- 1. Bartels DJ, et al. 2008. Natural prevalence of hepatitis C virus variants with decreased sensitivity to NS3/4A protease inhibitors in treatment-naive subjects. J. Infect. Dis. 198:800–807 [DOI] [PubMed] [Google Scholar]
- 2. Bartenschlager R, Cosset FL, Lohmann V. 2010. Hepatitis C virus replication cycle. J. Hepatol. 53:583–585 [DOI] [PubMed] [Google Scholar]
- 3. Brainard DM, et al. 2010. Safety, tolerability, and pharmacokinetics after single and multiple doses of MK-5172, a novel HCV NS3/4A protease inhibitor with potent activity against known resistance mutants, in healthy subjects. Hepatology 52(Suppl):1216A20803561 [Google Scholar]
- 4. Brainard DM, et al. 2010. Safety and antiviral activity of MK-5172, a novel HCV NS3/4a protease inhibitor with potent activity against known resistance mutants, in genotype 1 and 3 HCV-infected patients. Hepatology 52(Suppl):706A [Google Scholar]
- 5. Bronowicki J-P, et al. 2011. BMS-650032, an NS3 inhibitor, in combination with PetInterferon alpha-2A and ribavirin in treatment-naïve subjects with genotype 1 chronic hepatitis C infection. J. Hepatol. 54:S472 [Google Scholar]
- 6. Carroll SS, et al. 2009. Robust antiviral efficacy upon administration of a nucleoside analog to hepatitis C virus-infected chimpanzees. Antimicrob. Agents Chemother. 53:926–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carroll SS, et al. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979–11984 [DOI] [PubMed] [Google Scholar]
- 8. Choo QL, et al. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362 [DOI] [PubMed] [Google Scholar]
- 9. Forestier N, et al. 2007. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2α in patients with hepatitis C. Hepatology 46:640–648 [DOI] [PubMed] [Google Scholar]
- 10. Fried MW, et al. 2010. Efficacy and safety of TMC435 in combination with PegInterferon α-2A and ribavirin in treatment-naïve genotype-1 HCV patients: 24-week interim results from the PILLAR study. Hepatology 4(Suppl):LB-5 [Google Scholar]
- 11. Gane EJ, et al. 2010. Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1); a randomized, double-blind, placebo-controlled, dose-escalation trial. Lancet 376:1467–1475 [DOI] [PubMed] [Google Scholar]
- 12. Gao M, et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harper S, et al. 2012. Discovery of MK-5172, a macrocyclic hepatitis C virus NS3/4a protease inhibitor. ACS Med. Chem. 3(4):332–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harper S, Summa V, Liverton NJ, McCauley JA. 2010. Macrocyclic quinoxaline compounds as HCV NS3 protease inhibitors, Int PCT, App. WO/2010/011566. [Google Scholar]
- 15. He Y, et al. 2008. Relative replication capacity and selective advantage profiles of protease inhibitor-resistant hepatitis C virus (HCV) NS3 protease mutants in the HCV genotype 1b replicon system. Antimicrob. Agents Chemother. 52:1101–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howe AY, et al. 2008. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob. Agents Chemother. 52:3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kato T, et al. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817 [DOI] [PubMed] [Google Scholar]
- 18. Kieffer TL, et al. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631–639 [DOI] [PubMed] [Google Scholar]
- 19. Lange CM, Sarrazin C, Zeuzem S. 2010. Specifically targeted anti-viral therapy for hepatitis C—a new era in therapy. Aliment. Pharmacol. Ther. 32:14–28 [DOI] [PubMed] [Google Scholar]
- 20. Lavanchy D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl 1):74–81 [DOI] [PubMed] [Google Scholar]
- 21. Lawitz E, et al. 2011. ABT-450/Ritonavir (ABT-450/R) combined with pegylated interferon alpha-2A and ribavirin (SOC) after 3-day monotherapy in genotype 1 HCV-infected treatment-naïve subjects: 12-week interim efficacy and safety results. J. Hepatol. 54:S482 [Google Scholar]
- 22. Lenz O, et al. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob. Agents Chemother. 54:1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin T-I, et al. 2009. In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob. Agents Chemother. 53:1377–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liverton NJ, et al. 2010. MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob. Agents Chemother. 54:305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liverton N, et al. 2008. Molecular modeling based approach to potent P2-P4 macrocyclic inhibitors of hepatitis C NS3/4A protease. J. Am. Chem. Soc. 130:4607–4609 [DOI] [PubMed] [Google Scholar]
- 26. Lohmann V, Korner F, Koch Herian J-OU, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatits C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 27. Lok AS, et al. 2010. Combination therapy with BMS-790052 and BMS-650032 alone or with PEGIFN/RBV results in undetectable HCV RNA through 12 weeks of therapy in HCV genotype 1 null responders. Hepatology 52:LB-8 [Google Scholar]
- 28. Ludmerer SW, et al. 2008. A transient cell-based phenotype assay for hepatitis C NS3/4A protease; application to potency determinations of a novel macrocyclic inhibitor against diverse protease sequences isolated from plasma infected with HCV. J. Virol. Methods 151:301–307 [DOI] [PubMed] [Google Scholar]
- 29. Malcolm BA, et al. 2006. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob. Agents Chemother. 50:1013–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manns MP, et al. 2010. Sustained viral response (SVR) rates in genotype 1 treatment-naïve patients with chronic hepatitis C (CHC) infection treated with vaniprevir (MK-7009), a NS3/4A protease inhibitor, in combination with pegylated interferon alfa-2A and ribavirin for 28 days. Hepatology 4(Suppl):360A [Google Scholar]
- 31. Manns MP, et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomized trial. Lancet 358:958–965 [DOI] [PubMed] [Google Scholar]
- 32. Mao S-S, et al. 2008. A time-resolved, internally quenched fluorescence assay to characterize inhibition of hepatitis C virus nonstructural protein 3-4A at low enzyme concentrations. Anal. Biochem. 373:1–8 [DOI] [PubMed] [Google Scholar]
- 33. McCauley JA, et al. 2010. Discovery of vaniprevir (MK-7009), a macrocyclic hepatitis C virus NS3/4a protease inhibitor. J. Med. Chem. 53:2443–24763 [DOI] [PubMed] [Google Scholar]
- 34. McHutchison JG, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485–1492 [DOI] [PubMed] [Google Scholar]
- 35. McPhee F, et al. 2011. Characterization of virologic escape in HCV genotype 1 null responders receiving a combination of the NS3 protease inhibitor BMS-650032 and NS5A inhibitor BMS-790052. J. Hepatol. 54:S28 [Google Scholar]
- 36. Migliaccio G, et al. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164–49170 [DOI] [PubMed] [Google Scholar]
- 37. Perni RB, et al. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poenisch M, Bartenschlager R. 2010. New insights into structure and replication of the hepatitis C virus and clinical implications. Semin. Liver Dis. 30:333–347 [DOI] [PubMed] [Google Scholar]
- 39. Pol S, et al. 2011. SVR and pharmacokinetics of the HCV protease inhibitor BI201335 with PEGIFN/RBV in HCV genotype-1 patients with compensated liver cirrhosis and non-response to previous PETIFN/RBV. J. Hepatol. 54:S363 [Google Scholar]
- 40. Prichard MN, Shipman C., Jr 1990. A three-dimensional model to analyze drug-drug interaction. Antiviral Res. 14:181–206 [DOI] [PubMed] [Google Scholar]
- 41. Raboisson P, et al. 2008. Structure-activity relationship study on a novel series of cyclopentane-containing macrocyclic inhibitors of the hepatitis C virus NS3/4A protease leading to the discovery of TMC435350. Bioorg. Med. Chem. Lett. 18:4853–4858 [DOI] [PubMed] [Google Scholar]
- 42. Reesink HW, et al. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase 1b, placebo-controlled, randomized study. Gastroenterology 131:997–1002 [DOI] [PubMed] [Google Scholar]
- 43. Rouzier R, et al. 2011. Activity of danoprevir plus low-dose ritonavir (DNV/R) in combination with PegInterferon alfa-2a (40KD) plus ribavirin (PEGINα-2a/RBV) in previous null responders. J. Hepatol. 54:S28 [Google Scholar]
- 44. Sarrazin C, et al. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 45. Seiwert SD, et al. 2008. Preclinical characteristics of the HCV NS3/4A protease inhibitor ITMN-191 (R7227). Antimicrob. Agents Chemother. 52:4432–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seiwert SD, et al. 2007. Sequence variation of NS3/4A in HCV replicons exposed to ITMN-191 concentrations encompassing those likely to be achieved following clinical dosing. J. Hepatol. 46:S244–S245 [Google Scholar]
- 47. Springfield SA, et al. 2010. Process for synthesizing substituted isoquinolines. WO/2010/027889. [Google Scholar]
- 48. Sullivan JC, et al. 2011. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. J. Hepatol. 54:S4. [DOI] [PubMed] [Google Scholar]
- 49. Susser S, et al. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718 [DOI] [PubMed] [Google Scholar]
- 50. Trozzi C, et al. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wagaw S, et al. 2009. Process for making macrocyclic oximyl hepatitis C protease inhibitors. WO/2009/073780. [Google Scholar]
- 52. White PW, et al. 2010. Preclinical characterization of BI 201335, a C-terminal carboxylic acid inhibitor of the hepatitis C virus NS3-NS4A protease. Antimicrob. Agents Chemother. 54:4611–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. World Health Organization 1999. Hepatitis C—global prevalence (update). Wkly. Epidemiol. Rec. 74:425–427 [PubMed] [Google Scholar]
- 54. Zeuzem S, et al. 2011. Boceprevir resistance-associated variants (RAVs) are observed more frequently in HCV (GT1)-infected patients with poor response to PegInterferon alfa-2b/Ribavirin. Gastroenterology 140:S943 [Google Scholar]
- 55. Zeuzem S, et al. 2010. Dual, triple, and quadruple combination treatment with a protease inhibitor (GS-9256) and a polymerase inhibitor (GS-9190) alone and in combination with ribavirin (RBV) or PEGIFN/RBV for up to 28 days in treatment naïve, genotype 1 HCV subjects. Hepatology 52:LB1 [Google Scholar]