Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) isolates that are susceptible to vancomycin but are tolerant to its killing effect may present a potential challenge for effective treatment. This study compared the microbiologic characteristics of clinical vancomycin-tolerant (VT-MRSA) and vancomycin-susceptible (VS-MRSA) strains using phenotypic and gene regulation studies. MRSA isolates collected from vancomycin-treated patients with bacteremia over a 5-year period were analyzed for vancomycin, daptomycin, and telavancin susceptibility, as well as accessory gene regulator (agr) group and function. Vancomycin tolerance was defined by a minimum bactericidal concentration (MBC)/minimum inhibitor concentration (MIC) ratio of ≥32 mg/liter. VT-MRSA isolates were compared to VS-MRSA isolates for differences in antimicrobial susceptibility, time-kill activity, and gene expression of key cell envelope response genes vraSR, dltA, and mprF. All 115 isolates evaluated were susceptible to vancomycin, daptomycin, and telavancin. Seven isolates (6%) were VT-MRSA. agr group II was more prevalent in isolates with vancomycin MBC/MIC ratios of ≥8. In time-kill analyses, VT-MRSA had reduced vancomycin killing, but daptomycin and telavancin activities were maintained. Significantly greater gene expression was observed in VT-MRSA after 72 h of subinhibitory antibiotic exposures. Vancomycin most notably increased vraSR expression (P = 0.002 versus VS-MRSA strains). Daptomycin and telavancin increased expression of all genes studied, most significantly mprF expression (P < 0.001). Longer durations of antibiotic exposure (72 h versus 24 h) resulted in substantial increases in gene expression in VT-MRSA. Although the clinical impact of VT-MRSA is not fully recognized, these data suggest that VT-MRSA strains, while still susceptible, have altered gene regulation to adapt to the antimicrobial effects of glyco- and lipopeptides that may emerge during prolonged durations of exposure.
INTRODUCTION
The treatment of serious methicillin-resistant Staphylococcus aureus (MRSA) infections is often limited to antibiotics within the glycopeptide or lipopeptide class. Vancomycin has been the mainstay of this treatment for several decades, but clinical MRSA strains have developed a number of reduced-susceptibility phenotypes that continue to challenge clinical treatment. This phenomenon of glycopeptide “MIC creep” may vary considerably based on methods used to analyze organism susceptibility (24). However, a number of studies have associated an increased risk of vancomycin treatment failure with vancomycin Etest MICs of 1.5 to 2 mg/liter (15, 17). Antistaphylococcal β-lactam treatment failure in patients with methicillin-susceptible S. aureus (MSSA) bacteremia was also more likely in strains with vancomycin MICs of >1.5 mg/liter, suggesting that additional organism or host factors, not solely vancomycin treatment, may drive this response (8).
Vancomycin tolerance, commonly defined as having a minimum bactericidal concentration (MBC)/MIC ratio of ≥32, is known to occur in staphylococci. However, the clinical and microbiological features of these strains remain relatively uncharacterized. The prevalence of vancomycin-tolerant isolates in wild-type MRSA strains can vary widely, with most studies reporting tolerance in fewer than 20% of strains (24). Notably, vancomycin tolerance is more often present in strains with reduced vancomycin susceptibility, such as vancomycin-intermediate S. aureus (VISA) and heterogeneous VISA (hVISA) (12). hVISA and VISA have been associated with persistent bloodstream infections, but have not necessarily been implicated in increased mortality (10). This effect in tolerant strains may be due to a combination of reduced antibiotic killing and immune evasion characteristics of S. aureus resulting in altered host recognition and pathogen clearance (11).
S. aureus possesses a number of genetic regulators that are differentially expressed in strains with reduced susceptibility to glyco- and lipopeptides. Isolates that contain a dysfunctional accessory gene regulator (agr) group II element, which controls δ-hemolysin, have reduced in vitro vancomycin and teicoplanin killing and correspond to treatment failure in patients with severe MRSA infections (21, 22, 27). Alterations in S. aureus membrane surface charge, phosphatidylglycerol turnover, and cell wall architecture are known to affect the activity of daptomycin (13). Increased expression of genes that control these functions, most notably mprF and/or dltABCD, has been identified in most S. aureus strains with daptomycin nonsusceptibility (6, 31, 32). Additionally, mutations in the two-component regulatory system VraSR, which controls cell wall peptidoglycan production, contribute to reduced vancomycin and daptomycin activity. None of these gene changes is essential for in vivo reduced susceptibility to occur (6, 11, 31), which highlights the complexities of S. aureus regulation during exposure to glyco- and lipopeptides.
The phenotypic and genotypic differences between susceptible (yet vancomycin-tolerant) MRSA compared to nontolerant MRSA strains have not been evaluated. This study compared the microbiologic characteristics between these two strain types and demonstrates novel differences in antimicrobial activity and gene regulation upon exposure to vancomycin, daptomycin, and telavancin. For purposes of this study, we term nontolerant strains as “VS-MRSA” (vancomycin-susceptible MRSA) and tolerant strains as “VT-MRSA” (vancomycin-tolerant MRSA).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Isolates collected from patients with MRSA bacteremia from 2005 to 2010 at the University of Wisconsin Hospital and Clinics were analyzed. Vancomycin tolerance was defined as an MBC/MIC ratio of ≥32. The initial positive blood culture was collected for analysis of antibiotic susceptibility and agr group and function. From this cohort, five VT-MRSA strains (MBC, 32 mg/liter; MIC, 1 mg/liter) were compared to five VS-MRSA strains (MBC, 1 mg/liter; MIC, 1 mg/liter) for differences in time-kill activity and gene regulation as described below.
Antibiotics and susceptibility testing.
Vancomycin was purchased from Sigma (Sigma-Aldrich, St. Louis, MO). Daptomycin (Cubist Pharmaceuticals, Lexington, MA) and telavancin (Astellas Pharma US, Inc., Deerfield, IL) powders were provided by each manufacturer. MIC testing was performed by broth microdilution in Mueller-Hinton II broth (BD) supplemented with 12.5 mg/liter magnesium and 25 mg/liter calcium (for vancomycin and telavancin) or 50 mg/liter calcium (for daptomycin) according to the methods of the Clinical and Laboratory Standards Institute (2).
agr group and function.
The S. aureus agr group was determined for all isolates using a previously described multiplex PCR method with control isolates for agr groups I, II, III, and IV (16). The function of the agr locus was determined with a semiquantitative δ-hemolysin assay and categorized as functional (present) or dysfunctional (absent) (27). The presence of δ-hemolysin was determined on a tryptic soy agar plate with 5% sheep blood by streaking the S. aureus test isolate adjacent to a β-hemolysin disk (Remel, Lenexa, KS). Following overnight incubation at 37°C, synergistic hemolysis within the β-hemolysin zone indicated the presence of δ-hemolysin and a functional agr locus, while lack of hemolysis indicated a dysfunctional strain. S. aureus isolates RN6607 and RN9120 were used as positive and negative controls, respectively, for each assay (27).
In vitro time-kill curve.
Time-kill analysis was performed as described previously using an initial inoculum of 106 CFU/ml in the presence of vancomycin, daptomycin, and telavancin at 4-fold the organism MIC. Maximum bacterial kill and percentage of bactericidal activity (99.9% kill) were compared between the five strains described for each group above. Two independent experiments were performed for each antibiotic exposure.
Analysis of vraSR, dltA, and mprF gene expression by real-time RT-PCR.
The same cohort of vancomycin-tolerant and vancomycin-nontolerant strains used in the time-kill analysis was used in the gene expression studies. Isolates were exposed to the respective antibiotics at one-half the MIC for vancomycin, daptomycin, or telavancin in duplicate experiments. This antibiotic concentration eliminated the confounding effect of bacterial kill on gene expression. Analysis of gene expression was conducted after two separate exposure duration experiments of 24 h and 72 h. At the end of each antibiotic exposure, a 1:1,000 dilution of the sample was inoculated in fresh medium with the same antibiotic concentration. The new cultures were monitored for turbidity using a DEN-1 densitometer and were harvested for RNA isolation once they reached a McFarland turbidity standard of 0.25 in order to obtain cultures during the exponential phase of growth.
RNA extraction for reverse transcription-PCR (RT-PCR) began with pelleting samples by centrifugation (10,000 × g for 5 min) followed by resuspension in 1 ml TRIzol (Invitrogen, Carlsbad, CA). Cell walls were mechanically disrupted via bead beating in order to access total RNA. Sample vials were then allowed to warm to room temperature for 15 min and were centrifuged at 10,000 × g for 2 min. The supernatants were collected, and total RNA was isolated according to the manufacturer's instructions. RNA was resuspended in diethyl pyrocarbonate (DEPC)-water and stored at −80°C. The concentration and purity for each sample were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). cDNA was synthesized using the Omniscript RT kit (Qiagen, Valencia, CA).
The quantitative PCR (qPCR) primer sequences, specific to the vraSR, dltA, and mprF genes, were constructed using PrimerQuest software and obtained from Integrated DNA Technologies (Coralville, IA) as follows (5′→3′): vraSR, (forward, TGCTTACAGAACGAGAAATGGAAA, and reverse, CGTTTTAATAGTAATATGCGATGCA; dltA, forward, CCGTTTCCCAAGTGCGACGATTTA, and reverse, ATCTTGCGCCTAGTCTTTAAACGC; and mprF, forward, GTGCCGGTGTAAGAGCAATGGTTT, and reverse, AGCATAAAGTACCCATCTCACCCACG. 16S primers were used for the endogenous control (forward, GGCAACCGTTATCCGGAATT; reverse, GTTTCCAATGACCCTCCACG). Reaction mixtures comprised 12.5 μl Go Taq qPCR Master mix (Promega, Madison, WI), 1 μl each of forward and reverse primers (10 μM), 2 μl cDNA sample, and 8.5 μl sterile double-distilled water (ddH2O) for a total of 25 μl. qPCRs were carried out on an ABI 7500 series thermocycler using standard ABI cycling conditions with a melt curve added. The reporter dye filter was set to SYBR, and the reference dye filter was set to ROX. Relative gene expression to the control (growth without antibiotic exposure for each isolate) was determined using the threshold cycle (ΔΔCT) method.
Statistics.
All analyses compared characteristics of interest in VT-MRSA versus VS-MRSA strains. Comparisons of agr group and function and antibiotic bactericidal activity were conducted by chi-square analysis or Fisher's exact test where appropriate. Comparisons of antibiotic kill and fold change in gene regulation for each treatment were conducted by Student's t test.
RESULTS
One hundred fifteen isolates from individual patients with confirmed MRSA bacteremia were collected from 2005 to 2010 and analyzed in this study. Bacteremia sources in these patients were diverse and included skin or wound (27%), intravenous catheter (26%), endovascular or endocarditis (20%), bone or joint (11%), or pneumonia (8%).
Antibiotic susceptibility.
All isolates were susceptible to vancomycin, daptomycin, and telavancin. Seven isolates (6%) were vancomycin tolerant that were obtained from skin/wound (n = 5) or pneumonia (n = 2) infection sources. An additional 21 isolates had an MBC/MIC ratio of ≥16 when the MBC was in the resistant range (≥16 mg/liter). The antibiotic MIC50 and range for these isolates classified by vancomycin MBC/MIC ratio are shown in Table 1. No differences were detected in vancomycin, daptomycin, or telavancin MIC50 values in VT-MRSA compared to VS-MRSA strains.
Table 1.
Antibiotic susceptibility results of isolates from patients with MRSA bacteremia
| Antibiotic | MIC50 (mg/liter) for vancomycin MBC/MIC ratio ofa: |
|||||
|---|---|---|---|---|---|---|
| 1 (n = 16) | 2 (n = 31) | 4 (n = 25) | 8 (n = 15) | 16 (n = 21) | 32 (n = 7) | |
| Vancomycin | 1 (0.5–2) | 1 (0.5–2) | 1 (0.5–2) | 1 (0.5–2) | 1 (1–2) | 1 (1–2) |
| Daptomycin | 0.125 (0.06–0.19) | 0.125 (0.06–0.19) | 0.09 (0.03–0.25) | 0.125 (0.06–0.38) | 0.125 (0.06–0.19) | 0.125 (0.06–0.38) |
| Telavancin | 0.25 (0.13–0.38) | 0.25 (0.25–0.38) | 0.25 (0.13–0.38) | 0.38 (0.13–0.38) | 0.38 (0.25–0.38) | 0.38 (0.25–0.38) |
Susceptibilities to vancomycin, daptomycin, and telavancin, shown as MIC50s with ranges in parentheses, are presented for each vancomycin MBC/MIC ratio to compare susceptibility with increasing vancomycin tolerance.
Analysis of agr group and function.
Most MRSA isolates in this study (74%) contained the agr II genotype, most commonly from an endovascular or catheter source. The remaining isolates were classified into agr I (25%) and agr III (1%) groups. Thirty-three percent of all isolates possessed a dysfunctional agr locus identified by lack of δ-hemolysin production on sheep blood agar. Isolates with high vancomycin MBC/MIC ratios were more likely to possess the agr group II genotype. As displayed in Fig. 1, 86% of isolates with vancomycin MBC/MIC ratios of ≥8 contained agr II type compared to 67% of isolates with a vancomycin MBC/MIC ratio of <8 (P = 0.028). The percentage of isolates with a dysfunctional agr locus was slightly lower in strains with ratios of ≥8, but this was not a statistically significant finding.
Fig 1.
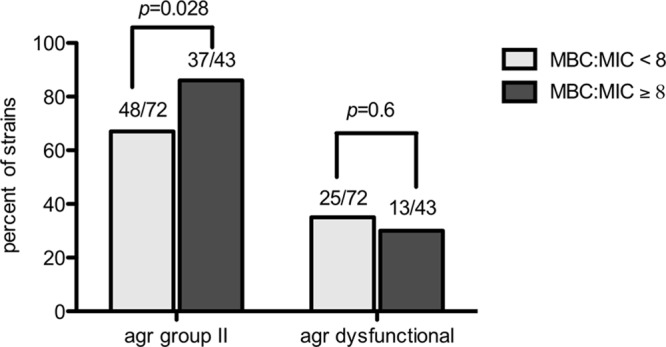
Comparison of accessory gene regulator (agr) group II and agr dysfunction in strains with high versus low vancomycin MBC/MIC ratios from patients with MRSA bacteremia.
In vitro time-kill analysis.
Vancomycin, daptomycin, and telavancin activities at concentrations of 4-fold the MIC in VT-MRSA and VS-MRSA strains are shown in Table 2. Overall, there was a trend for reduced vancomycin activity in VT-MRSA compared to susceptible strains. A lower maximum bacterial kill (P = 0.090) and lower percentage of bactericidal activity (P = 0.520) occurred in strains classified as VT-MRSA. Daptomycin achieved the greatest level of bacterial kill and was bactericidal in all strains. The presence of vancomycin tolerance in the strains tested did not impact daptomycin or telavancin activity.
Table 2.
Vancomycin, daptomycin, and telavancin activities in a 24-h time-kill curve at 4-fold the MIC for each antibiotic in vancomycin-susceptible (n = 5) and -tolerant (n = 5) strains from patients with MRSA bacteremia
| Antibiotic treatment | Classification (vancomycin MBC/MIC ratio) | Mean ± SD log-CFU/ml kill (median) | Bactericidal activity (% of strains) |
|---|---|---|---|
| Vancomycin | Susceptible (1) | 3.1 ± 0.2 (3.1) | 80 |
| Tolerant (32) | 2.7 ± 0.4 (2.4) | 40 | |
| Daptomycin | Susceptible (1) | 3.9 ± 0.3 (4.0) | 100 |
| Tolerant (32) | 3.8 ± 0.2 (4.0) | 100 | |
| Telavancin | Susceptible (1) | 3.5 ± 0.7 (4.0) | 80 |
| Tolerant (32) | 3.4 ± 0.8 (3.5) | 80 |
Differential gene expression in VT-MRSA with glyco- and lipopeptide exposure.
S. aureus genes vraSR, dltA, and mprF, which are known to affect glyco- and lipopeptide susceptibility, were evaluated pre- and post-in vitro antibiotic treatment. Five VT-MRSA strains and five VS-MRSA strains from our study cohort were exposed to vancomycin, daptomycin, and telavancin, each individually at one-half the MIC. Gene expression changes were first compared after 24 h, and no significant differences between VS-MRSA and VT-MRSA strains were observed (data not shown).
In contrast, at 72 h of antibiotic exposure, gene expression was significantly greater in VT-MRSA compared to VS-MRSA strains. We identified distinct changes in gene expression that correlate to specific glyco- or lipopeptide treatment, as shown in Fig. 2A to C. In VT-MRSA exposed to vancomycin, the highest increase in gene expression occurred in vraSR (P = 0.002 versus VS-MRSA strains), while smaller but still significant increases were observed for mprF. Of note, dltA gene expression after vancomycin exposure was lower in VT-MRSA (P = 0.023). Following daptomycin exposure, VT-MRSA had the greatest increase in mprF expression (P < 0.001 versus VS-MRSA strains), followed by lower but still highly significant increases in expression of vraSR and dltA (P < 0.001). With exposure to telavancin, VT-MRSA had an upregulated gene expression profile parallel to daptomycin; however, the overall fold changes in vraSR and mprF gene expression were not as elevated.
Fig 2.
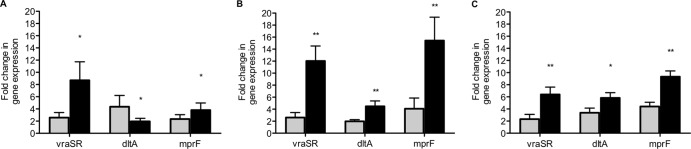
Fold change in gene expression (mean ± standard deviation) of five vancomycin-tolerant MRSA strains (black bars) compared to five vancomycin-nontolerant MRSA strains (gray bars) following 72-h exposures to vancomycin (A), daptomycin (B), and telavancin (C).*, P < 0.05 between tolerant and susceptible strains; **, P < 0.001.
We also compared the effects of the duration of antibiotic exposure in VT-MRSA at 24 h versus 72 h of in vitro treatment. Longer durations of exposure to vancomycin, daptomycin, and telavancin resulted in substantial increases in gene expression in VT-MRSA isolates that were not observed among the VS-MRSA isolates. As displayed in Fig. 3A to C, less than a 4-fold mean change in vraSR, dltA, or mprF expression occurred after 24 h in VT-MRSA. Continued exposures of vancomycin, daptomycin, and telavancin up to 72 h, however, resulted in significantly greater increases in gene expression in most genes. Overall, vraSR and mprF displayed the greatest upregulation in expression from 24 to 72 h with all three antibiotic exposures.
Fig 3.

Fold change in gene expression in five vancomycin-tolerant strains after 24 h and 72 h of exposure to vancomycin (A), daptomycin (B), and telavancin (C). Upregulation of vraSR, dltA, and mprF correlated with the duration of antimicrobial exposure. *, P < 0.05 between gene expression at 72 h versus 24 h; **, P < 0.001.
DISCUSSION
While a number of studies have associated the vancomycin MIC with clinical outcome, the clinical and microbiologic characteristics of reduced vancomycin minimum bactericidal concentration in MRSA strains remain relatively undefined. Clinical studies have reported conflicting results in associating poor vancomycin treatment response with VT-MRSA (9, 29, 33), but the overall low percentage of tolerant isolates in these studies may affect these discrepancies. In our study of MRSA bloodstream isolates collected over a 5-year period, only 6% of strains were VT-MRSA. This low prevalence prevented an assessment of VT-MRSA as independent risk factors in vancomycin treatment outcomes. Further evaluation of the microbiological and genetic characteristics of VT-MRSA will help to more clearly associate their clinical impact.
S. aureus contain a number of quorum-sensing regulators that contribute to its pathogenicity and antibiotic resistance. Among these, agr has been identified as a significant factor in the development of reduced vancomycin susceptibility and associated treatment failure (21, 26, 27). Schweizer et al. reported the first independent association between agr dysfunction and mortality among severely ill patients with S. aureus bacteremia (28). Pharmacodynamic models have shown reduced glycopeptide activity in agr dysfunctional isolates, particularly those within the agr group II genotype (30). Isolates with agr dysfunction also have a higher proclivity for resistance to innate host defense cationic peptides, such as thrombin-induced platelet microbicidal proteins (tPMPs) (25). In our study, isolates that were more tolerant to the effects of vancomycin (MBC/MIC ratio, ≥8) were not associated with agr dysfunction. Interestingly, these isolates were more likely to possess the agr group II genotype than strains with low vancomycin bactericidal concentrations. This finding is supported by a recent clinical analysis correlating persistent infections to isolates with agr group II genotype and resistance to tPMPs (20). In conjunction with prior studies, our present results support the link between reduced vancomycin killing and agr group II S. aureus, which may contribute to reduced vancomycin activity in vivo and associated persistent infections.
Increased expression and mutations of select regulatory genes involved in cell wall synthesis and turnover have been previously found in strains with reduced susceptibility to vancomycin and daptomycin (4). In VISA strains, these genetic changes are linked to increased peptidoglycan and cell wall teichoic acid synthesis and thickening of the cell wall (3). Using a pharmacodynamic model, we have previously shown that vancomycin doses of at least 1,500 mg every 12 h are needed in order to prevent significant cell wall thickening in hVISA (23). Recently, Cafiso et al. reported that vancomycin exposure in hVISA reduces autolysis, downregulates the autolysin genes atl and lytM, and upregulates dltA. These changes were distinctive for hVISA and did not occur in the comparator vancomycin-susceptible S. aureus strain (1), but the relatively short duration of exposure may have been unable to select for these changes.
We hypothesized that differences in gene regulation may emerge between two types of vancomycin-susceptible MRSA, VT versus nontolerant strains, with prolonged durations of subinhibitory vancomycin exposures. Indeed, we found significantly greater upregulation in vraSR and mprF genes in VT-MRSA that were differentially expressed only with a longer duration of vancomycin exposure. Upregulations of these genes in VISA strains have been linked to increased expression of dltA (1); however, this did not occur in our study with VT-MRSA strains that were susceptible to vancomycin. We did not detect the emergence of the VISA phenotype with these genetic changes, but this finding suggests that VT-MRSA, while still susceptible, alters cell envelope gene regulation to adapt to the antimicrobial effects of vancomycin.
Daptomycin nonsusceptibility is thought to emerge via the accumulation of multiple genetic mutations, but upregulation of or a gain-of-function mprF mutation is a common finding in many in vitro and clinically derived daptomycin-nonsusceptible strains. Recently, Mehtra et al. reported that point mutations in mprF contribute to reduced daptomycin susceptibility but do not correlate with changes in expression levels of mprF (19). Meanwhile, upregulation of the vraSR two-component regulator may contribute to a second resistance mechanism involved in alterations of cell wall synthesis (19). All strains in our study were daptomycin susceptible, and therefore we did not test for mutations in mprF. Daptomycin was the most bactericidal agent evaluated, and the killing effects were similar between VT-MRSA and nontolerant strains. However, all three genes of interest were comparatively upregulated in VT-MRSA after daptomycin exposure. It should be noted that antibiotic concentrations were different in the kill-curve (4-fold MIC) versus gene expression studies (0.5-fold MIC), which limits comparison between these results. We would anticipate future studies that directly compare these relationships.
Previous S. aureus transcriptional profiling reported that multiple cell wall stress stimulon member genes are expressed upon exposure to daptomycin, correlating with daptomycin's antimicrobial actions on peptidoglycan biosynthesis as well as membrane depolarization (5). Notably, in our study, mprF, and to a lesser extent vraSR, displayed increases in gene expression with daptomycin exposure in VT-MRSA compared to VS-MRSA strains. It will be important to determine if upregulation of these genes in VT-MRSA results in reduced daptomycin susceptibility with exposures exceeding the duration used in our study.
Telavancin is structurally derived from vancomycin but exhibits a dual mechanism of action by inhibiting phosphatidylglycerol synthesis and disrupting cell membrane function (7). This lipoglycopeptide molecule has broad Gram-positive activity, including against isolates with reduced vancomycin and daptomycin susceptibility (14, 18). Televancin resistance in S. aureus has not been described in the clinical setting, but in vitro pressure with subinhibitory telavancin concentrations has resulted in resistant mutants after prolonged exposure (14). No mechanism for telavancin resistance has been investigated, but we surmise that mutations or upregulation in cell wall stress stimulon member genes would contribute to this development. Upregulation in mprF, vraSR, and to a lesser extent, dltA occurred in VT-MRSA after 3-day exposures to subinhibitory concentrations of telavancin, although telavancin susceptibility was unchanged. This gene regulation profile more similarly paralleled that of VT-MRSA after exposure to daptomycin as opposed to vancomycin.
In summary, the prevalence of VT-MRSA was low in our study of MRSA bloodstream isolates, and therefore any inferences from the study results require additional analyses of this trait in MRSA. Nonetheless, some important characteristics of strains with reduced vancomycin bactericidal concentrations were identified. Isolates with high MBC/MIC ratio values (≥8 mg/liter) were associated with the agr group II genotype and reduced vancomycin bactericidal activity in a time-kill curve. A particularly novel finding of this work is that VT-MRSA strains were associated with increased expression of key cell envelope response genes after exposure to cell wall- and/or membrane-active anti-MRSA agents. Additionally, this finding was only significant with longer exposures of these antimicrobials. It will be important to determine if these differences begin to emerge between the endpoints not tested in this study (within 24 to 72 h). We recognize that other genetic response pathways exist that control the adaptation of S. aureus to these antimicrobials, but the effects observed in our study highlight a distinctive response in VT-MRSA that may have important therapeutic implications as reduced vancomycin susceptibilities continue to increase.
ACKNOWLEDGMENT
This study was supported by a grant from the Society of Infectious Diseases Pharmacists' Focus on Gram-Positive Resistance Award Program made possible by Astellas.
Footnotes
Published ahead of print 11 June 2012
REFERENCES
- 1. Cafiso V, et al. 2012. Modulating activity of vancomycin and daptomycin on the expression of autolysis cell-wall turnover and membrane charge genes in hVISA and VISA strains. PLoS One 7:e29573 doi:10.1371/journal.pone.0029573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clinical and Laboratory Standards Institute 2010. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard, 10th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 3. Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui L, Neoh HM, Shoji M, Hiramatsu K. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1231–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer A, et al. 2011. Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J. Antimicrob. Chemother. 66:1696–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higgins DL, et al. 2005. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49:1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holmes NE, et al. 2011. Antibiotic choice may not explain poorer outcomes in patients with Staphylococcus aureus bacteremia and high vancomycin minimum inhibitory concentrations. J. Infect. Dis. 204:340–347 [DOI] [PubMed] [Google Scholar]
- 9. Honda H, Doern CD, Michael-Dunne W, Jr, Warren DK. 2011. The impact of vancomycin susceptibility on treatment outcomes among patients with methicillin resistant Staphylococcus aureus bacteremia. BMC Infect. Dis. 11:335 doi:10.1186/1471-2334-11-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Howden BP, Johnson PD, Ward PB, Stinear TP, Davies JK. 2006. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 50:3039–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Howden BP, et al. 2008. Different bacterial gene expression patterns and attenuated host immune responses are associated with the evolution of low-level vancomycin resistance during persistent methicillin-resistant Staphylococcus aureus bacteraemia. BMC Microbiol. 8:39 doi:10.1186/1471-2180-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones RN. 2006. Microbiological features of vancomycin in the 21st century: minimum inhibitory concentration creep, bactericidal/static activity, and applied breakpoints to predict clinical outcomes or detect resistant strains. Clin. Infect. Dis. 42(Suppl 1):S13–S24 [DOI] [PubMed] [Google Scholar]
- 13. Jones T, et al. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kosowska-Shick K, et al. 2009. Activity of telavancin against staphylococci and enterococci determined by MIC and resistance selection studies. Antimicrob. Agents Chemother. 53:4217–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kullar R, Davis SL, Levine DP, Rybak MJ. 2011. Impact of vancomycin exposure on outcomes in patients with methicillin-resistant Staphylococcus aureus bacteremia: support for consensus guidelines suggested targets. Clin. Infect. Dis. 52:975–981 [DOI] [PubMed] [Google Scholar]
- 16. Lina G, et al. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69:18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lodise TP, et al. 2008. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob. Agents Chemother. 52:3315–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacGowan AP, Noel AR, Tomaselli S, Elliott HC, Bowker KE. 2011. Pharmacodynamics of telavancin studied in an in vitro pharmacokinetic model of infection. Antimicrob. Agents Chemother. 55:867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mehta S, et al. 2012. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 56:92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moise PA, et al. 2010. Factors influencing time to vancomycin-induced clearance of nonendocarditis methicillin-resistant Staphylococcus aureus bacteremia: role of platelet microbicidal protein killing and agr genotypes. J. Infect. Dis. 201:233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moise-Broder PA, et al. 2004. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 38:1700–1705 [DOI] [PubMed] [Google Scholar]
- 22. Rose WE, Kaatz GW, Sakoulas G, Rybak MJ. 2008. Teicoplanin pharmacodynamics in reference to the accessory gene regulator (agr) in Staphylococcus aureus using an in vitro pharmacodynamic model. J. Antimicrob. Chemother. 61:1099–1102 [DOI] [PubMed] [Google Scholar]
- 23. Rose WE, Knier RM, Hutson PR. 2010. Pharmacodynamic effect of clinical vancomycin exposures on cell wall thickness in heterogeneous vancomycin-intermediate Staphylococcus aureus. J. Antimicrob. Chemother. 65:2149–2154 [DOI] [PubMed] [Google Scholar]
- 24. Sader HS, Jones RN, Rossi KL, Rybak MJ. 2009. Occurrence of vancomycin-tolerant and heterogeneous vancomycin-intermediate strains (hVISA) among Staphylococcus aureus causing bloodstream infections in nine USA hospitals. J. Antimicrob. Chemother. 64:1024–1028 [DOI] [PubMed] [Google Scholar]
- 25. Sakoulas G, et al. 2005. Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49:2687–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakoulas G, et al. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187:929–938 [DOI] [PubMed] [Google Scholar]
- 27. Sakoulas G, et al. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schweizer ML, et al. 2011. Increased mortality with accessory gene regulator (agr) dysfunction in Staphylococcus aureus among bacteremic patients. Antimicrob. Agents Chemother. 55:1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sorrell TC, Packham DR, Shanker S, Foldes M, Munro R. 1982. Vancomycin therapy for methicillin-resistant Staphylococcus aureus. Ann. Intern. Med. 97:344–350 [DOI] [PubMed] [Google Scholar]
- 30. Tsuji BT, Rybak MJ, Lau KL, Sakoulas G. 2007. Evaluation of accessory gene regulator (agr) group and function in the proclivity towards vancomycin intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:1089–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang SJ, et al. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200:1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang SJ, et al. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 53:2636–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoon YK, Kim JY, Park DW, Sohn JW, Kim MJ. 2010. Predictors of persistent methicillin-resistant Staphylococcus aureus bacteraemia in patients treated with vancomycin. J. Antimicrob. Chemother. 65:1015–1018 [DOI] [PubMed] [Google Scholar]


