Abstract
AbaR resistance islands in Acinetobacter baumannii isolates from South Korea were investigated. AbaR4-type resistance islands, including blaOXA-23-containing Tn2006, interrupted the comM gene in A. baumannii ST75 isolates. However, Tn2006 was not identified within AbaR resistance islands of ST138 isolates, although the blaOXA-23 gene was detected in them. The similar structures of resistance islands suggest that most carbapenem-resistant A. baumannii isolates in South Korea have originated from the same ancestor with a globally disseminated clone, GC II.
TEXT
Acinetobacter baumannii is an opportunistic pathogen that primarily causes nosocomial infections (5). Such infections are often associated with resistance to many antimicrobial agents, including carbapenems (12). In addition to carbapenem resistance, a resistance island, termed AbaR1, was identified by comparison of whole-genome sequences (6). AbaR1 is integrated into the ATPase gene (now called comM) and contains a large cluster of antimicrobial and heavy-metal resistance genes. Further studies have revealed that there is significant diversity among the AbaR-type resistance islands in the same region of comM in A. baumannii isolates (1, 8–10, 13, 14), and AbaR transposons have been found in A. baumannii since the 1970s (10). Although AbaR-type resistance islands have been reported in A. baumannii isolates from many regions, little is known regarding the composition of different AbaRs in isolates from East Asia. In this study, we investigated the structural variations of AbaR-type resistance islands in multidrug-resistant (MDR) A. baumannii isolates from South Korea and their evolution in related clones during a short period.
Forty-four isolates of A. baumannii were analyzed (Table 1). All of them were isolated from blood obtained from patients in South Korean hospitals from 2003 to 2010. Antimicrobial susceptibility testing was performed by the broth microdilution method according to the CLSI guidelines (4). Multilocus sequence typing (MLST) according to the Oxford scheme was performed as previously described (2). Of the clones, 19 ST92, seven ST75, five ST138, nine ST69, two ST71, one ST70, and one ST220 clone were included (Table 1). The ST69, ST71, and ST70 clones included only carbapenem-nonsusceptible A. baumannii isolates, but ST220 included all carbapenem-susceptible isolates. In the ST92, ST75, and ST138 clones, both carbapenem-nonsusceptible and -susceptible isolates were included (Table 1).
Table 1.
A. baumannii isolates analyzed in this study
| Sequence type (allelic profilea) | Isolate | Isolation yr | Resistance (MIC, mg/liter)b |
|||||
|---|---|---|---|---|---|---|---|---|
| Imipenem | Meropenem | Tetracycline | Amikacin | Cefepime | Ampicillin-sulbactam | |||
| ST92 (1-3-3-2-2-7-3) | C003 | 2003 | S (2) | S (2) | R (>64) | R (>128) | R (32) | R (64/32) |
| C011 | 2003 | I (8) | R (16) | R (>64) | R (>128) | R (64) | R (64/32) | |
| C022 | 2004 | S (1) | S (2) | R (>64) | R (>128) | R (64) | R (64/32) | |
| C032 | 2004 | S (1) | S (2) | R (>64) | R (>128) | R (32) | R (>64/32) | |
| C043 | 2005 | S (1) | S (2) | R (>64) | R (>128) | R (64) | R (>64/32) | |
| C052 | 2005 | S (2) | S (4) | R (>64) | R (>128) | R (32) | R (64/32) | |
| C100 | 2006 | S (1) | S (2) | R (>64) | R (>128) | R (32) | R (>64/32) | |
| H06-454 | 2006 | S (2) | R (16) | R (>64) | R (>128) | R (32) | R (64/32) | |
| H07-129 | 2007 | S (2) | R (16) | I (8) | R (>128) | R (>64) | R (>64/32) | |
| H07-332 | 2007 | R (32) | R (16) | R (>64) | R (>128) | R (32) | R (64/32) | |
| H07-395 | 2007 | R (32) | I (8) | R (>64) | R (>128) | R (32) | R (>64/32) | |
| E07-111 | 2007 | S (2) | S (4) | R (>64) | R (>128) | R (64) | R (64/32) | |
| H08-796 | 2008 | R (16) | R (>64) | R (>64) | R (>128) | R (>64) | R (>64/32) | |
| E08-46 | 2008 | R (16) | R (>64) | R (>64) | R (>128) | R (>64) | R (>64/32) | |
| H09-169 | 2009 | R (16) | R (>64) | R (>64) | R (>128) | R (>64) | R (>64/32) | |
| H09-256 | 2009 | S (4) | I (8) | I (8) | R (>128) | R (>64) | R (64/32) | |
| H09-421 | 2009 | R (32) | R (64) | R (>64) | R (>128) | R (>64) | R (>64/32) | |
| H09-422 | 2009 | R (16) | R (64) | R (>64) | R (>128) | R (>64) | R (>64/32) | |
| E09-228 | 2009 | S (2) | S (4) | R (>64) | R (>128) | R (>64) | I (16/8) | |
| ST69 (1−46-3-2-2-58-3) | C018 | 2004 | R (16) | R (>64) | R (16) | R (64) | R (>64) | R (64/32) |
| C026 | 2004 | R (32) | R (64) | R (16) | R (64) | R (>64) | R (64/32) | |
| C046 | 2005 | I (8) | R (16) | R (16) | R (64) | R (>64) | R (32/16) | |
| C067 | 2005 | R (32) | R (32) | I (8) | I (32) | R (>64) | R (>64/32) | |
| C093 | 2006 | I (8) | R (16) | R (16) | S (16) | R (>64) | R (>64/32) | |
| H06-855 | 2006 | R (16) | R (16) | R (16) | R (>128) | R (>64) | R (>64/32) | |
| H07-130 | 2007 | R (32) | R (>64) | R (16) | R (>128) | R (>64) | R (64/32) | |
| E07-334 | 2007 | R (16) | R (>64) | R (16) | R (128) | R (64) | I (16/8) | |
| H09-347 | 2009 | R (32) | R (64) | R (16) | R (>128) | R (>64) | R (64/32) | |
| ST75 (1-3-3-2-2-11-3) | E08-600 | 2008 | R (64) | R (64) | R (>64) | I (32) | R (>64) | R (>64/32) |
| E08-650 | 2008 | R (>64) | R (>64) | R (>64) | I (32) | R (>64) | R (>64/32) | |
| H09-94 | 2009 | R (>64) | R (>64) | R (>64) | I (32) | R (>64) | R (>64/32) | |
| H09-146 | 2009 | R (>64) | R (>64) | R (>64) | I (32) | R (>64) | R (>64/32) | |
| H09-504 | 2009 | R (>64) | R (>64) | R (>64) | R (128) | R (>64) | R (>64/32) | |
| H09-526 | 2009 | R (>64) | R (>64) | R (>64) | I (32) | R (>64) | R (>64/32) | |
| E10-93 | 2010 | S (0.25) | S (≤0.0625) | R (64) | S (8) | S (0.5) | R (>64/32) | |
| ST138 (1-3-3-2-2-50-3) | H09-400 | 2009 | R (>64) | R (>64) | S (4) | S (8) | R (>64) | R (>64/32) |
| H09-401 | 2009 | R (>64) | R (>64) | S (4) | S (8) | R (>64) | R (>64/32) | |
| H09-454 | 2009 | R (>64) | R (>64) | S (4) | R (>128) | R (>64) | R (>64/32) | |
| H09-465 | 2009 | R (>64) | R (>64) | S (4) | R (>128) | R (>64) | R (>64/32) | |
| H10-103 | 2009 | S (0.25) | S (≤0.0625) | R (>64) | R (>128) | R (>64) | R (>64/32) | |
| ST71 (1-46-3-2-2-58-4) | C077 | 2005 | I (8) | R (16) | R (16) | S (16) | R (>64) | R (>64/32) |
| C099 | 2006 | R (16) | R (64) | I (8) | I (32) | R (>64) | R (>64/32) | |
| ST70 (1-46-3-2-1-58-3) | C083 | 2005 | R (16) | R (32) | I (8) | R (64) | R (>64) | R (>64/32) |
| ST220 (1-3-3-2-1-7-3) | H07-988 | 2007 | S (2) | S (4) | I (8) | I (8) | R (>64) | R (32/16) |
gltA-gyrB-gdhB-recA-cpn60-gpi-rpoD.
R, resistant; I, intermediate; S, susceptible.
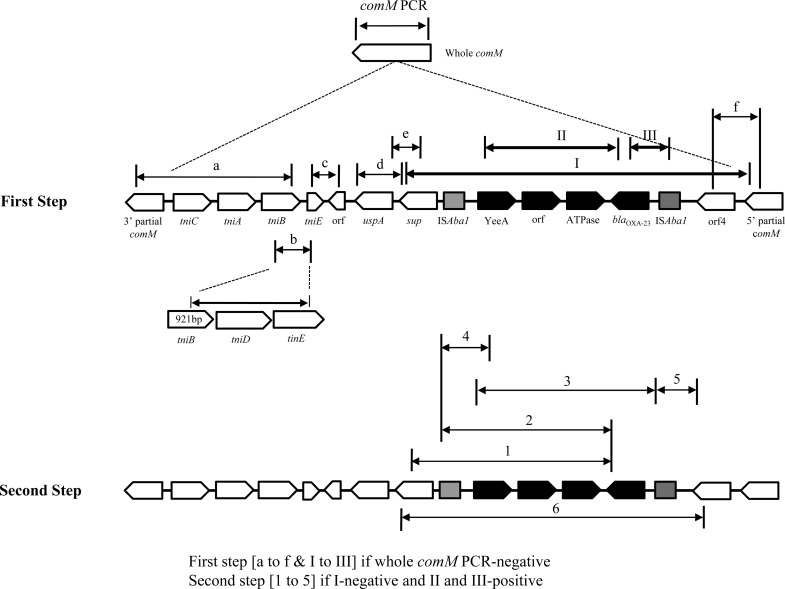
The insertions of a transposon into the comM gene were investigated using previously published primers for all A. baumannii isolates, including both carbapenem-nonsusceptible and -susceptible isolates (14, 15). AbaR mapping was performed using two steps (Fig. 1). The first step included nine primer sets, including those for detection of the insertion of AbaR in the comM gene. When a yeeA-blaOXA-23 or blaOXA-23-ISAba1 fragment was amplified in the first step (primer sets II and III), the second step of PCR, including five primer sets (1 to 5), was performed (Fig. 1). If the resistance island was I positive and II negative in the first step, primer set 6 in the second step was used as a confirmation.
Fig 1.
Scheme of PCR mapping used to detect the AbaR-type resistance island and its structure. The first step was applied for all A. baumannii isolates. In the second step, primer sets 1 to 5 were used if the first step was I negative and II positive or III positive, and primer set 6 was used if the first step was I positive and II negative.
All A. baumannii isolates, including carbapenem-susceptible isolates of ST92, ST75, ST138, and ST220, failed to produce a comM amplicon, indicating the interruption of the comM gene. In addition, all isolates with an interrupted comM gene produced an amplicon for the uspA gene, indicating that the gene has not been interrupted by an ISPpu12 composite transposon as in other AbaR types (14).
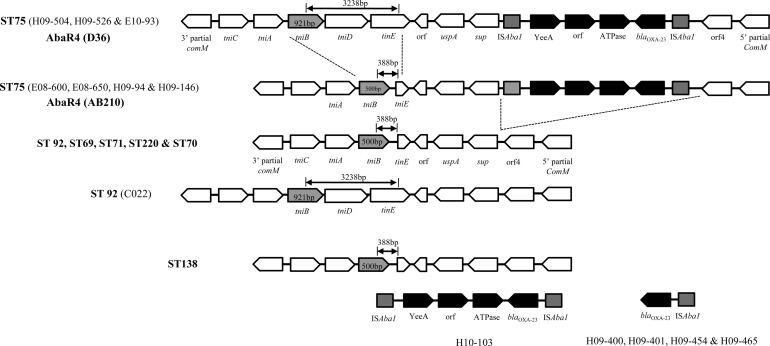
The structure of the AbaR-type resistance islands indicated that all comM inserts of carbapenem-resistant isolates corresponded largely to two types (Fig. 2). While isolates of the ST75 clone showed a typical AbaR4 island (7), isolates of the other clones, such as ST92, ST69, ST71, ST220, ST70, and ST138, possessed resistance islands similar to AbaR4, which lacks Tn2006. AbaR4 was initially found in a GC1 isolate, but it was not inserted into the comM gene but rather found in a different location (1). Interestingly, Tn2006 including blaOXA-23 was not present in the resistance island but Tn2006 or the blaOXA-23 gene was present in other regions of ST138 isolates (Fig. 2). Isolate H10-103 was positive for primer set I in the first step and negative for primer sets 1 and 5 in the second step but gave amplicons for primer sets 2 to 4 in the second step. In addition, four isolates of ST138 were positive for primer set III in the first step but were negative with primer sets 1 to 5 in the second step.
Fig 2.
The structures of resistance islands identified in this study. AbaR4 islands of ST75 are subdivided into two subtypes, the D36 type and the AB210 type, according to the presence or absence of the tniD gene, resulting in different fragment sizes of primer set b in the first step of PCR mapping. Although AbaR4-like islands of ST138 isolates lacked the blaOXA-23-containing Tn2006, they were positive for the blaOXA-23 gene and one isolate (H10-103) possessed the complete structure of Tn2006.
So far, two subtypes of AbaR4 have been reported. One, produced by primer set b in the first step, is a 3,238-bp amplicon that spans tniB to tniE (referred to as the D36 type) (15). The other, produced by the same primer set, is a 388-bp amplicon referred to as the AB210 type (Fig. 1 and 2) (7). In this study, three isolates of clone ST75 and one isolate of ST92 showed the D36-type AbaR4, and the AbaR4 of the remaining clones was the AB210 type (Fig. 2). In addition to the findings that blaOXA-23 is found in carbapenem-resistant GC1 isolates (11), the diverse pattern of blaOXA-23 in this study may support a previous suggestion that Tn2006-mediated acquisition of blaOXA-23 may occur as independent events or that Tn2006 is a mobile structure in a given genome (11). However, it is now unclear whether clones with a blaOXA-23-positive AbaR4 resistance island have been introduced recently in South Korea or whether the transposon or a portion of it has inserted into the genomes of ST75 and ST138.
One of the interesting results in this study is the similarity of AbaR4-type resistance islands between carbapenem-nonsusceptible and -susceptible A. baumannii isolates belonging to the same clone. An isolate of an antimicrobial-susceptible clone, ST220, was found to have the same resistance island structure as that of isolates of several resistant clones, including ST92, ST69, ST71, and ST70. In addition, both carbapenem-nonsusceptible and -susceptible isolates were included in ST92, ST75, and ST138, but the structure of a resistance island was not correlated with the carbapenem susceptibility. Tn6021 forming an AbaR backbone has been identified in a susceptible A. baumannii reference strain, ATCC 17978 (6). Our result also indicates that the identification of an AbaR4-like element is not always correlated with carbapenem resistance or MDR, in confirmation of a previous report (3).
In summary, an AbaR4-type resistance island or resistance islands similar to it were identified in A. baumannii isolates from South Korea. ST75 isolates possessed AbaR4 islands containing a Tn2006 insert, but isolates of other clones lacked Tn2006 in their resistance islands. ST138 isolates may have the blaOXA-23 gene in loci other than the resistance island. The AbaR4-like island or a portion of it was found even in susceptible clones with an intact comM gene.
ACKNOWLEDGMENTS
Some Acinetobacter isolates used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID, Seoul, South Korea), and the others were provided by Sook-In Jung (Chonnam National University Medical School, South Korea).
This study was supported by a Samsung Biomedical Research Institute grant (no. BA-90012).
Footnotes
Published ahead of print 5 June 2012
REFERENCES
- 1. Adams MD, et al. 2008. Comparative genomics sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartual SG, et al. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonnin RA, Poirel L, Nordmann P. 2012. AbaR-type transposon structures in Acinetobacter baumannii. J. Antimicrob. Chemother. 67:234–236 [DOI] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing: 21st informational supplement. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in the hospital: multidrug resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 6. Fournier PE, et al. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7 doi:10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J. Antimicrob. Chemother. 66:2484–2491 [DOI] [PubMed] [Google Scholar]
- 8. Iacono M, et al. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krizova L, Dijkshoorn L, Nemec A. 2011. Diversity and evolution of AbaR genomic resistance island in Acinetobacter baumannii strains of European clone I. Antimicrob. Agents Chemother. 55:3201–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krizova L, Nemec A. 2010. A 63 kb genomic resistance island found in a multidrug-resistant Acinetobacter baumannii isolate of European clone I from 1977. J. Antimicrob. Chemother. 65:1915–1918 [DOI] [PubMed] [Google Scholar]
- 11. Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perez F, et al. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Post V, Hall RM. 2009. AbaR, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:2667–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:1162–1170 [DOI] [PubMed] [Google Scholar]
- 15. Turton JF, Baddal B, Perry C. 2011. Use of accessory genome for characterization and typing of Acinetobacter baumannii. J. Clin. Microbiol. 49:1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]