Abstract
We identified a clinical isolate of Candida glabrata (CG156) exhibiting flocculent growth and cross-resistance to fluconazole (FLC), voriconazole (VRC), and amphotericin B (AMB), with MICs of >256, >256, and 32 μg ml−1, respectively. Sterol analysis using gas chromatography-mass spectrometry (GC-MS) revealed that CG156 was a sterol 14α-demethylase (Erg11p) mutant, wherein 14α-methylated intermediates (lanosterol was >80% of the total) were the only detectable sterols. ERG11 sequencing indicated that CG156 harbored a single-amino-acid substitution (G315D) which nullified the function of native Erg11p. In heterologous expression studies using a doxycycline-regulatable Saccharomyces cerevisiae erg11 strain, wild-type C. glabrata Erg11p fully complemented the function of S. cerevisiae sterol 14α-demethylase, restoring growth and ergosterol synthesis in recombinant yeast; mutated CG156 Erg11p did not. CG156 was culturable using sterol-free, glucose-containing yeast minimal medium (glcYM). However, when grown on sterol-supplemented glcYM (with ergosta 7,22-dienol, ergosterol, cholestanol, cholesterol, Δ7-cholestenol, or desmosterol), CG156 cultures exhibited shorter lag phases, reached higher cell densities, and showed alterations in cellular sterol composition. Unlike comparator isolates (harboring wild-type ERG11) that became less sensitive to FLC and VRC when cultured on sterol-supplemented glcYM, facultative sterol uptake by CG156 did not affect its azole-resistant phenotype. Conversely, CG156 grown using glcYM with ergosterol (or with ergosta 7,22-dienol) showed increased sensitivity to AMB; CG156 grown using glcYM with cholesterol (or with cholestanol) became more resistant (MICs of 2 and >64 μg AMB ml−1, respectively). Our results provide insights into the consequences of sterol uptake and metabolism on growth and antifungal resistance in C. glabrata.
INTRODUCTION
The prevalence of infections caused by Candia glabrata among elderly and immunocompromised individuals is increasingly well documented (8, 27, 28, 29). C. glabrata is intrinsically less susceptible to triazole antifungals than other species of Candida (30), and polyene agents (e.g., amphotericin B) constitute one treatment alternative. In contrast to azoles, which disrupt fungal ergosterol biosynthesis through inhibition of sterol 14α-demethylase activity (Fig. 1), polyenes intercalate directly with membrane ergosterol, forming channels that result in leakage of monovalent ions and cellular lysis (6). Today, evidence for the emergence of multidrug, including polyene, resistance in C. glabrata (18) has highlighted the need for a better understanding of the mechanisms that mediate resistance in this species so that more effective antifungals and treatment regimens can be developed.
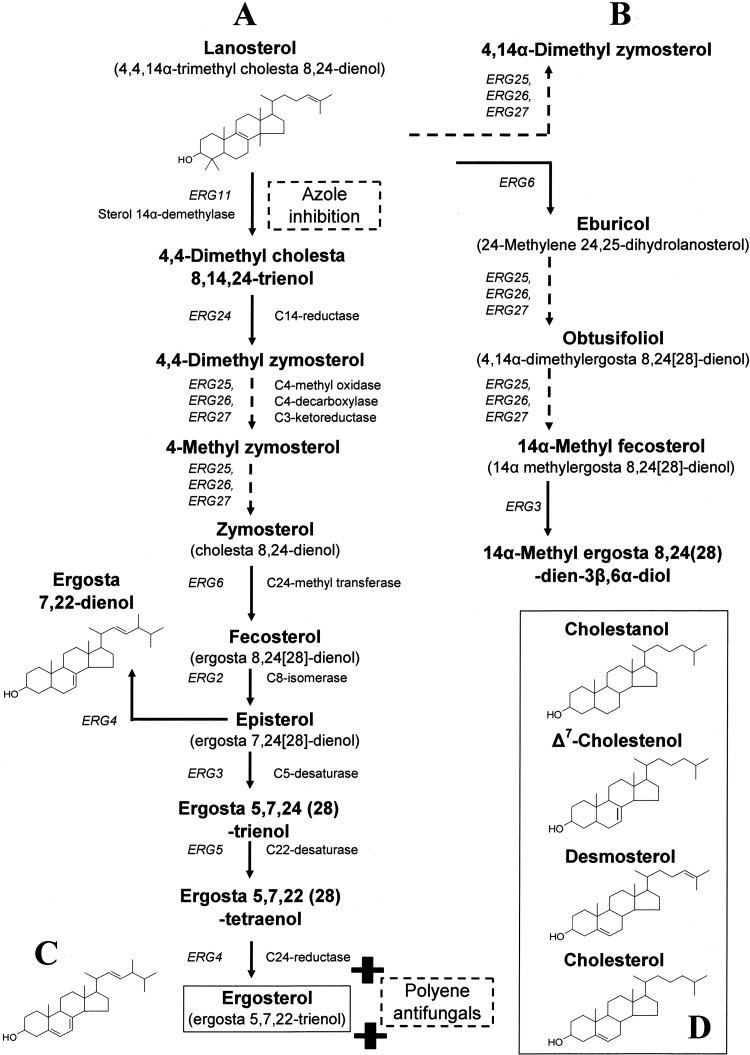
Fig 1.
Schematic representation of the ergosterol biosynthetic pathway in C. glabrata showing the structure of sterols used in the present study. (A) Classical pathway; (B) sterol intermediates that accumulate following azole inhibition of sterol 14α-demethylase (ERG11 protein); (C) end product ergosterol, the binding target of polyene antifungals; (D) cholesterol and related sterol intermediates.
Studies of azole resistance in clinical isolates of C. glabrata have demonstrated the importance of drug efflux mechanisms (4, 7, 34). Elevated in vitro resistance to azoles, concomitant with increased susceptibility to polyenes, has also been associated with mitochondrial DNA deficiency (petite mutation) in this species (3, 33). However, the universality of this mechanism is unclear given that petite mutants of C. glabrata exhibiting hypersusceptibility to azoles and polyenes have also been identified (39). It has been proposed that aerobic uptake of exogenous sterol (e.g., host cholesterol) contributes to azole (1, 26) and polyene (12) resistance in vivo, and a putative sterol transporter gene for C. glabrata (AUS1) has been identified (26). The genetic controls and biological consequences of obligate and facultative sterol uptake in C. glabrata require further investigation, not least because the potential exists for similarities with other opportunistic pathogens, such as Aspergillus fumigatus (41) and Pneumocystis jirovecii (14).
The present study focuses on a clinical isolate of C. glabrata (CG156) that persisted under treatment with high doses of fluconazole, voriconazole, and amphotericin B (MIC values on yeast-extract-peptone-dextrose [YEPD] of >256, >256, and 32 μg ml−1, respectively). CG156 was found to be an ergosterol-deficient erg11 (sterol 14α-demethylase) mutant, exhibiting flocculent growth, low efflux, and facultative sterol uptake that supported, but was not essential for, growth. Four additional clinical isolates (CG26, CG44, CG388, and CG1012) were selected for comparative studies on the basis of their total cellular sterol composition (>75% ergosterol); this composition indicates sterol 14α-demethylase functionality and is typical of wild-type C. glabrata reference strains (e.g., ATCC 2001) (25).
Ergosterol-deficient, polyene-resistant strains of C. glabrata possessing mutations in ERG1 (squalene epoxidase) (37) and ERG6 (C24-methyl transferase) (38) have been reported; however, unlike CG156, both isolates exhibited increased sensitivity to azoles. Results from this study highlight the potential consequences of sterol uptake on both azole and polyene resistance in C. glabrata and reveal how this opportunistic species might exploit a wide spectrum of exogenous sterols, including several previously uninvestigated cholesterol precursors (Fig. 1D), for survival.
MATERIALS AND METHODS
Media.
All study isolates (Table 1) were routinely maintained at 30°C on YEPD medium containing 2% (wt/vol) glucose, 2% (wt/vol) Bacto peptone, and 1% (wt/vol) yeast extract (with or without 2% agar). Glucose-based yeast minimal medium (glcYM) containing 1.34% yeast nitrogen base without amino acids (Difco Laboratories) and 2% glucose was prepared as a sterol-free (by gas chromatography-mass spectrometry [GC-MS]) basal medium for use in antifungal susceptibility assays and sterol supplementation experiments. Abbreviations for the sterol supplements used in this study are ergosta 7,22-dienol (7,22), ergosterol (ERG), lanosterol (LAN), cholestanol (CAN), cholesterol (CER), Δ7-cholestenol (CEN), and desmosterol (DES) (medium containing one of these supplements is indicated as, e.g., glcYM + 7,22, glcYM + ERG, etc.). All supplements except cholestenol (Steraloids Inc.) were purchased from Sigma-Aldrich (Poole, United Kingdom). The supplements were dissolved in (1:1) Tween-80-ethanol and added to glcYM to achieve a final concentration of 10 μg sterol ml−1. The morphology of all study isolates was examined using light microscopy following growth on YEPD and glcYM.
Table 1.
Typical growth parameters, antifungal MIC, rhodamine 6G efflux, and sterol data for C. glabrata study isolates cultured using YEPDa
| Isolate | Growth parameter |
MIC (μg ml−1) |
R6G assay efflux ratio | Sterol distribution (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔOD600 | Lag phase (h) | T half max (h) | Max DT (h) | FLC | VRC | AMB | Lanosterol | 14α-Methylatedb | Ergosterol | 14α-Demethylatedc | ||
| CG26 | 1.30 | 6.3 | 9.0 | 1.0 | 256 | 16 | 2 | 12.66 | 4.0 | — | 80.5 | 15.5 |
| CG44 | 1.32 | 6.5 | 9.5 | 1.0 | 128 | 2 | 2 | 1.2 | 3.6 | — | 75.5 | 20.9 |
| CG388 | 1.49 | 6.0 | 8.0 | 1.0 | 128 | 2 | 2 | 2.7 | 2.5 | — | 82.7 | 14.8 |
| CG1012 | 1.55 | 6.0 | 8.5 | 1.0 | 128 | 2 | 2 | 2.5 | 3.3 | — | 77.6 | 19.1 |
| CG156 | 1.34 | 14.5 | 18.5 | 2.0 | >256 | >256 | 32 | 1.5 | 81.5 | 18.5 | — | — |
ΔOD600, maximum minus minimum optical density reading (at 600 nm); lag phase, length of time culture remains at <10% of maximum OD; T half max, time taken to achieve half-maximal culture growth [(maximum OD minus minimum OD) times 0.5]; max DT, maximum observed doubling time; FLC, fluconazole; VRC, voriconazole; AMB, amphotericin B. R6G efflux values indicate ratios of fluorescence to forward scatter (FSC/FL-2) for 1.2 × 104 cells. The highest values are in boldface; —, not detected.
Other (not lanosterol) 14α-methylated sterols.
Other (not ergosterol) 14α-demethylated sterols.
The S. cerevisiae (YUG37:erg11) yeast transformants constructed for heterologous expression studies were selected and maintained on glcYM+Trp agar (2% agar) supplemented with 100 μg ml−1 tryptophan. For complementation experiments (Table 2), medium to induce plasmid expression (gal/rafYM+dox) was prepared as previously described (24) using 2% galactose and 1% raffinose instead of glucose and with the addition of 5 μg ml−1 (final concentration) doxycycline.
Table 2.
Heterologous expression of C. glabrata wild-type (pCGWT) and mutated (pCG156) ERG11 (encoding sterol 14α-demethylase) proteins in an S. cerevisiae erg11 mutant
| Construct | Medium | Sterol distributiona (%) |
|||
|---|---|---|---|---|---|
| Lanosterol | 14α-Methylatedb | Ergosterol | 14α-Demethylatedc | ||
| YUG37-pCGWT | gal/rafYM+dox | 5.5 (2.4) | 9.9 (1.1) | 54.4 (3.1) | 30.2 (1.4) |
| YUG37-pCG156 | gal/rafYM+dox | 43.0 (3.4) | 56.5 (2.7) | 0.5 (0.7) | — |
| YUG37-CTRL | gal/rafYM+dox | 43.8 (3.9) | 55.3 (3.3) | 0.9 (0.8) | — |
| YUG37-CTRL | gal/rafYM−dox | 5.0 (1.3) | — | 80.0 (3.3) | 15.5 (1.1) |
Mean (n = 3) values (±SD) are provided; successful complementation is indicated in boldface; —, not detected.
Other (not lanosterol) 14α-methylated sterols.
Other (not ergosterol) 14α-demethylated sterols.
Culture growth.
Growth measurements were made using a Bioscreen C (Oy Growth Curves Ab Ltd., Finland) to record changes in optical density at 600 nm (OD600) over time. Uniform initial culture densities (5 × 105 cells ml−1) were achieved by diluting each sample to 1 × 107 cells ml−1 with filter-sterilized water and adding this cell dilution to the growth medium of interest (50 μl cells plus 950 μl medium). The resulting 1-ml volumes were vortexed, aliquoted into microplate wells (four 200-μl replicates per treatment), and incubated at 30°C in the Bioscreen. Optical density readings (at 600 nm) were taken once every 30 min following the automated (10-s) agitation of the microplate, exported from the Bioscreen in ASCII format, and analyzed using Excel (Microsoft Office 2003). Growth parameters were derived according to the methodology of Weiss et al. (40). Briefly, ΔOD values describe the maximum OD minus the minimum OD; the lag phase is defined as the length of time a culture spends at <10% of the maximum OD; T half max values are equivalent to the time taken to reach half the maximum increase in growth of a culture (ΔOD × 0.5). Maximum doubling times were estimated by dividing the natural logarithm of 2 by the fastest culture growth rates (μ) that were derived from linear trend lines (y = m · x + c, where m
 μ) fitted to log-transformed OD data.
μ) fitted to log-transformed OD data.
Antifungal susceptibility assays.
The susceptibilities of CG26, CG44, CG156, CG388, and CG1012 to fluconazole (FLC), voriconazole (VRC), and amphotericin B (AMB) were determined using standard broth dilution methodology (5). Interpretive breakpoints for susceptibility assays are the following: (i) for FLC, ≤32 μg ml−1 indicates dose-dependent sensitivity and ≥64 μg ml−1 indicates resistant; (ii) for VRC, ≤1 μg ml−1 indicates sensitive and ≥4 μg ml−1 indicates resistant; (iii) for AMB, ≤1 μg ml−1 indicates sensitive. Visual readings for all isolates (except CG156) were made following 48 h of incubation at 30°C; readings for CG156 were made after 72 h. Assays were initially performed using YEPD as the basal growth medium. To investigate the potential contribution of sterol uptake to the resistance phenotypes observed, susceptibility assays for CG156 and CG388 were also conducted using sterol-free and sterol-supplemented glcYM.
Rhodamine 6G efflux assay.
The intracellular accumulation of rhodamine 6G (Sigma-Aldrich) in growing isolates was measured according to standard methodology (20). Briefly, cells from an overnight culture (YEPD) were inoculated into fresh medium to an OD600 of ∼0.1 and grown under shaking at 30°C for 4 h. For staining, 10 μM rhodamine 6G (R6G) was added to the culture, and cells were grown for another 2 h, after which they were diluted (1:100) using NaCl. The fluorescence (FL-2) of 1.2 × 104 cells was quantified using a FACScalibur flow cytometer (BD, Heidelberg, Germany). To exclude any bias toward larger cells or agglomerates, efflux values were calculated as the ratio of forward scatter to total fluorescence (FSC/FL-2).
Sterol analysis.
Single colonies were used to inoculate 15-ml volumes of media to obtain cultures for sterol analysis. Cells were harvested by centrifugation and washed three times with sterile water prior to sterol extraction and derivatization using a previously optimized methodology (22). Trimethylsilane-derivatized sterols were analyzed using GC-MS and identified with reference to retention times and fragmentation spectra for known standards (23). GC-MS data were analyzed using Agilent software (MSD Enhanced ChemStation; Agilent Technologies Inc.) to determine the sterol composition of all isolates and for derivation of integrated peak areas.
ERG11 gene sequencing.
Full-length ERG11 genes for all study isolates were amplified from genomic DNA (single-colony extraction: 0.2% SDS 90°C, 10 min) using forward (F) and reverse (R) primers optimized for use in subsequent heterologous protein expression studies: ERG11F, 5′-ATGTCCACTGAAAACACT-3′; ERG11R, 5′-CTAGTACTTTTGTTCTGG-3′. DNA polymerase with proofreading capacity (High Fidelity Expand; Roche) was used for all PCRs, and sequencing results (Eurofins MWG Operon, United Kingdom) were screened for ambiguous signals prior to translation and alignment against a C. glabrata sterol 14α-demethylase (Erg11p) reference protein (Uniprot accession number P50859).
Heterologous expression of sterol 14α-demethylase (Erg11p) proteins.
ERG11 genes for CG26 and CG156 were ligated directly into the yeast expression vector pYES2.1 TOPO (Invitrogen), yielding wild-type and mutated ERG11 plasmid DNA (pCGWT and pCG156, respectively). An S. cerevisiae host (YUG37:erg11), wherein native ERG11 expression is controlled using a doxycycline-regulatable promoter (11, 24), was transformed with pCGWT, pCG156, or empty vector pYES2.1 to create experimental and control strains (YUG37-pCGWT, YUG37-pCG156, and YUG37-pCTRL, respectively), which were selected and maintained on glcYM+Trp agar.
For complementation studies, single colonies from YUG37-pCGWT, YUG37-pCG156, and YUG37-pCTRL transformation plates (all constructs in triplicate) were used to inoculate 15-ml volumes of gal/rafYM+dox; these were incubated for 72 h (30°C, 180 rpm) prior to checks for successful Erg11p complementation, which is evidenced by ergosterol synthesis (GC-MS analysis) concomitant with cell growth.
Sterol uptake experiments.
Clinical isolates CG156 and CG388 were cultured using sterol-free and sterol-supplemented glcYM. Optical density data from studies performed with the Bioscreen were used to derive comparative growth parameters for both strains. In addition to antifungal susceptibility assays, GC-MS was used to determine the sterol composition of CG156 and CG388 following growth of 15-ml cultures on supplemented and unsupplemented glcYM (with or without FLC at concentrations equivalent to ×0.5 MIC).
RESULTS
Study isolates.
The isolates of C. glabrata included in the present study were retrieved from a collection of pathogenic clinical specimens established as part of an EU FP6 project, the European Resistance Fungal Network (EURESFUN). Specific specimen information is limited; however, it is known that all were isolated from different patients: CG26 and CG388 from blood cultures, CG44 from peritoneal cavity fluid, CG1012 from a central venous catheter, and CG156 from a urine sample.
Growth characteristics and cell morphology.
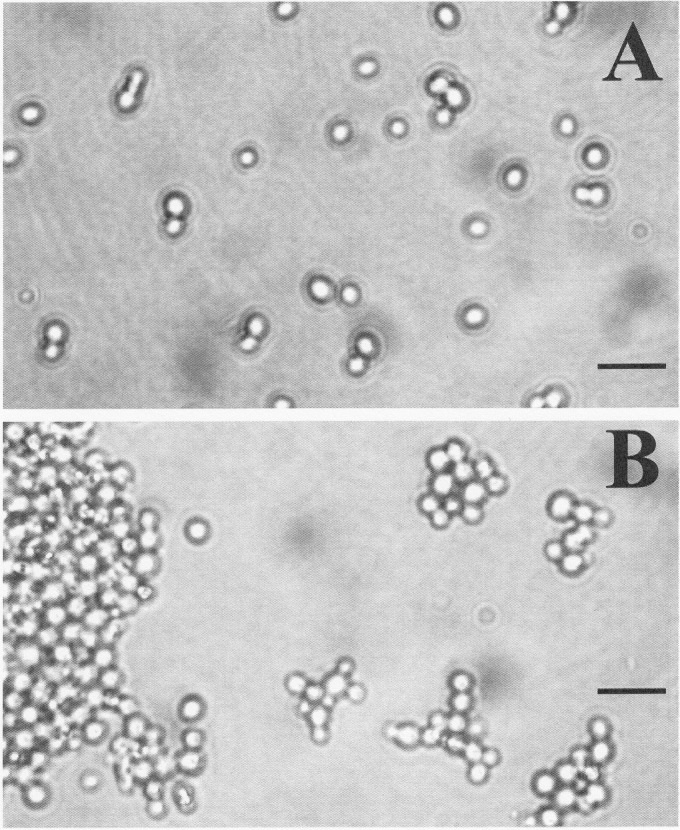
During routine culture maintenance using YEPD, CG156 exhibited slow growth, forming small (<1-mm diameter), flat colonies that were only visible 36 to 48 h after each subculture. All other study isolates were readily culturable, forming larger (typically >2-mm diameter) colonies in <18 h. Bioscreen growth experiments indicate that the lag phases and maximum doubling times for CG156 cultured on YEPD broth were at least double the length of those recorded for CG26, CG44, CG388, and CG1012 (Table 1). Microscope examination of cells from these experiments revealed that CG26, CG44, CG388, and CG1012 all displayed yeast-like growth characterized by solitary blastoconidia and normal cell budding (Fig. 2A). CG156 cultures were highly flocculent and exhibited abnormal cellular budding (Fig. 2B).
Fig 2.
Cell morphology of C. glabrata study isolates cultured using YEPD medium and glcYM (including sterol-supplemented media). (A) Yeast-like growth of CG388 showing solitary blastoconidia and normal budding; (B) flocculent growth of CG156. Scale bars represent 10 μm.
The typical growth rates and culture densities supported by glcYM were lower for CG156 than those recorded using complex YEPD (Table 1 and 3). It was noted that CG388 and CG156 were both culturable using sterol-free glcYM (Table 3; also see Fig. 4). CG26, CG44, CG388, and CG1012 exhibited normal and CG156 abnormal cellular budding regardless of the growth medium used (i.e., including when cultured on sterol-free and sterol-supplemented glcYM).
Table 3.
Mean (n = 3) growth parameters and MICs for CG156 and CG388 cultured using unsupplemented and supplemented glcYMa
| Growth parameter | Result according to medium supplement and strain |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No supplement (glcYM only) |
7,22 |
ERG |
LAN |
CAN |
CER |
CEN |
DES |
|||||||||
| CG156 | CG388 | CG156 | CG388 | CG156 | CG388 | CG156 | CG388 | CG156 | CG388 | CG156 | CG388 | CG156 | CG388 | CG156 | CG388 | |
| ΔOD600 | 0.83 | 1.28 | 1.19 | 1.28 | 1.18 | 1.28 | 0.85 | 1.27 | 1.06 | 1.24 | 1.19 | 1.26 | 1.02 | 1.24 | 1.12 | 1.24 |
| Lag phase (h) | 62.7 | 17.0 | 30.0 | 23.3 | 34.0 | 31.0 | 61.0 | 28.3 | 44.3 | 29.0 | 34.0 | 23.3 | 43.0 | 28.0 | 38.7 | 27.0 |
| T half max (h) | 68.7 | 22.0 | 33.0 | 30.0 | 42.3 | 36.7 | 68.3 | 35.3 | 52.3 | 36.3 | 40.3 | 27.3 | 52.0 | 37.7 | 48.3 | 34.3 |
| Max DT (h) | 2.83 | 2.01 | 2.34 | 2.02 | 3.39 | 2.80 | 2.86 | 2.85 | 3.84 | 3.38 | 2.44 | 2.24 | 3.62 | 3.76 | 3.91 | 3.51 |
| MIC (μg ml−1) | ||||||||||||||||
| Fluconazole | >256 | 128 | >256 | >256 | >256 | >256 | >256 | 128 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Voriconazole | >256 | 2.0 | >256 | >256 | >256 | >256 | >256 | 2.0 | >256 | 64 | >256 | >256 | >256 | >256 | >256 | >256 |
| Amphotericin B | 16 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 16 | 2.0 | 64 | 2.0 | 64 | 2.0 | 4.0 | 2.0 | 4.0 | 2.0 |
All sterols were added to a final concentration of 10 μg ml−1 and are the following: 7,22, ergosta 7,22-dienol; ERG, ergosterol; LAN, lanosterol; CAN, cholestanol; CER, cholesterol; CEN, Δ7-cholestenol; and DES, desmosterol. ΔOD600, maximum minus minimum optical density reading (at 600 nm); lag phase, length of time culture remains at <10% of maximum OD; T half max, time taken to achieve half-maximal culture growth [(maximum OD minus minimum OD) times 0.5]; max DT, maximum observed doubling time.
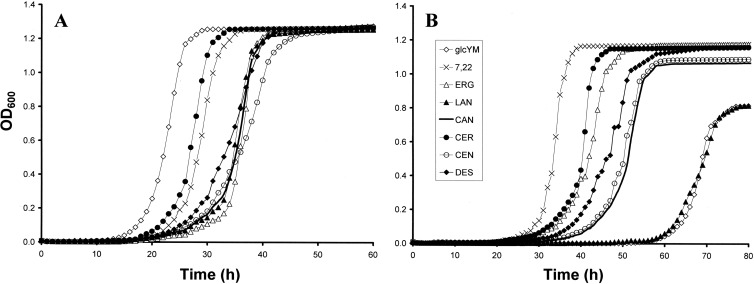
Fig 4.
Growth curves for CG388 (A) and CG156 (B) cultured using sterol-free and sterol-supplemented glcYM. 7,22, ergosta 7,22-dienol; ERG, ergosterol; LAN, lanosterol; CAN, cholestanol; CER, cholesterol; CEN, Δ7-cholestenol; DES, desmosterol.
Antifungal resistance phenotypes: YEPD media.
When YEPD was used as the basal growth medium (Table 1), CG44, CG388, and CG1012 displayed identical sensitivity to FLC, VRC, and AMB (MICs of 128, 2, and 2 μg ml−1, respectively). CG26 was relatively more resistant to FLC and VRC but not to AMB (MICs of 256, 16, and 2 μg ml−1, respectively). These results are consistent with R6G efflux values for these isolates (Table 1), which indicate that CG26 is a high-efflux strain. Despite being the most azole-resistant isolate (MICs of >256, >256, and 32 μg ml−1 for FLC, VRC, and AMB, respectively), CG156 exhibited lower R6G efflux than all other study isolates (except CG44). This suggested that additional or alternative (nonefflux) mechanisms contribute to its antifungal resistance phenotype.
Sterol composition: YEPD media.
Initial GC-MS analysis of YEPD-grown cultures indicated that CG26, CG44, CG388, and CG1012 all had a sterol composition similar to that of wild-type strains (possessing functional sterol 14α-demethylase [Erg11p]), wherein 14α-demethylated sterols accounted for >95% of the total cellular sterol fraction and ergosterol was always the most abundant sterol (Table 1, boldface values). Conversely, CG156 was found to be an erg11 mutant (lacking functional Erg11p), wherein 14α-methylated sterol intermediates (lanosterol was >80% of the total) were the only cellular sterols detectable.
ERG11 sequence and complementation studies.
A single missense mutation (G944A) which resulted in the replacement of glycine 315 with aspartate was detected in the Erg11p sequence for CG156 (Fig. 3A). No amino acid substitutions were identified in the translated Erg11p sequences for CG26, CG44, CG388, or CG1012.
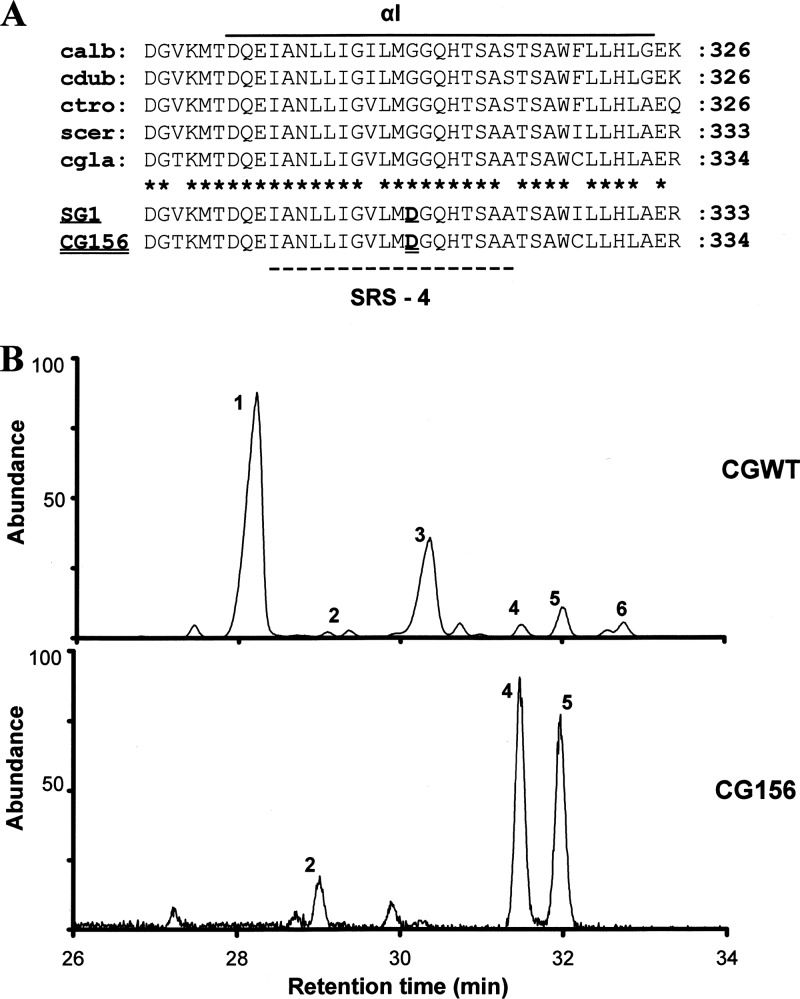
Fig 3.
ERG11 protein sequence analysis and complementation studies. (A) Alignment of sterol 14α-demethylase sequences (αI helix only) for Candida albicans (calb; accession number P10613), C. dubliniensis (cdub; AAK57519), C. tropicalis (ctro; P14263), Saccharomyces cerevisiae (scer; AAA34379), and C. glabrata (cgla; P50859). SRS-4, substrate recognition site 4. Asterisks indicate conserved residues. Note that a glycine-to-aspartate alteration previously reported to nullify the function of sterol 14α-demethylase in SG1, an S. cerevisiae erg11 mutant (13), was the sole amino acid substitution detected in CG156 (this study). (B) Sterol chromatograms for S. cerevisiae constructs heterologously expressing C. glabrata wild-type (CGWT) and mutated (CG156) sterol 14α-demethylase. Peak 1, ergosterol; 2, 4,14α-dimethyl zymosterol; 3, 4-methyl zymosterol; 4,14α-methyl ergosta-8,24(28)-dien-3β,6α-diol; 5, lanosterol; and 6, 4,4-dimethyl zymosterol.
The S. cerevisiae YUG37-pCGWT yeast transformant (expressing the ERG11 gene [encoding wild-type Erg11p] from CG26) was readily culturable using gal/rafYM+dox; mutant YUG37-pCG156 and negative control YUG37-pCTRL cultures were not. Real-time PCR (data not shown) verified that both wild-type and mutant C. glabrata ERG11 genes were expressed at comparable levels in YUG37-pCGWT and YUG37-pCG156 cultured using gal/rafYM+dox. GC-MS sterol chromatograms revealed that ergosterol comprised >50% of the total sterol fraction in gal/rafYM+dox-cultured YUG37-pCGWT. In contrast, neither ergosterol nor any other 14α-demethylated sterols was detectable in YUG37-pCG156 and YUG37-pCTRL transformants (Fig. 3B). That the fungistatic sterol 14α-methyl ergosta 8,24(28)-dien-3β-6α-diol comprised >50% of the sterol fraction in the YUG37-pCG156 construct is consistent with the failure of mutated (G315D) CG156 Erg11p to complement the function of S. cerevisiae sterol 14α-demethylase.
Influence of sterol supplementation on growth kinetics.
Little variation was seen between the final culture densities reached by CG388 grown using sterol-free and sterol-supplemented glcYM (typical ΔOD values of 1.28). However, some differences between the maximum doubling times and shortest lag phases for CG388 were observed (Table 3 and Fig. 4A). It is possible that the presence of extracellular sterols does affect the regulation of growth and endogenous ergosterol production; however, any specific or mechanistic inferences regarding growth rate variability in CG388 are beyond the scope of the current study.
CG156 grew relatively slowly when cultured using sterol-free glcYM and glcYM + LAN (Fig. 4B), with lag phases (typically ≥60 h) and doubling times (≥2.8 h) being similar on both media. Optimal growth parameters (i.e., shortest lag phases, fastest doubling times, and highest cell densities) were achieved for CG156 cultured using glcYM + CER or glcYM + 7,22 (Table 3). Despite reaching optimal final densities (ΔOD of ∼ 1.20), CG156 samples cultured using glcYM supplemented with ERG, CAN, CEN, or DES had slower maximum doubling times (typically >3.5 h) than those grown using glcYM + CER or glcYM + 7,22 (both ≤2.5 h).
Influence of sterol supplementation on antifungal resistance phenotypes.
When sterol-free glcYM or sterol-supplemented glcYM + LAN was used as the basal growth medium, MICs (for FLC and VRC) identical to those recorded using YEPD were observed for both CG156 and CG388. CG388 was equally sensitive to AMB when grown using YEPD, glcYM, and glcYM + LAN. Conversely, YEPD-cultured CG156 was found to be less sensitive to AMB-cultured than glcYM- and glcYM + LAN-cultured cells (MICs of 32 and 16 μg ml−1, respectively).
The azole MICs recorded for CG388 cultured using all sterol-supplemented media (except glcYM + LAN and glcYM + CAN) increased from 128 and 2 μg ml−1 to >256 and >256 μg ml−1 for FLC and VRC, respectively (Table 3). In contrast, there were no differences between the AMB MICs (2 μg ml−1) recorded for CG388 on any media. CG156 retained strong cross-resistance to FLC and VRC (MICs of >256 μg ml−1 for both azoles) regardless of the growth medium used; however, variation was observed in its sensitivity to AMB. Compared to those cultured using YEPD (AMB MIC of 32 μg ml−1), glcYM, or glcYM + LAN (AMB MICs of 16 μg ml−1), CG156 grown on glcYM + CAN and glcYM + CER exhibited elevated resistance to AMB (MIC, 64 μg ml−1). Conversely, CG156 samples grown on glcYM + DES and glcYM + CEN were more sensitive (MICs of 4 μg AMB ml−1); CG156 samples grown using glcYM + ERG and glcYM + 7,22 were again more sensitive to AMB, exhibiting MICs (2 μg ml−1) identical to those of CG388.
Validation of sterol uptake by GC-MS.
The sterol composition of CG156 and CG388 cultured using sterol-free glcYM (and glcYM + LAN) is consistent with that of YEPD-grown cells, except for a lower ergosterol content (60 to 65% of the total sterol fraction) and higher proportions of 14α-demethylated sterol intermediates in CG388 (Tables 1 and 4). This result is indicative of the effects of nutrient limitation on cell growth.
Table 4.
Sterol composition of CG156 and CG388 after 48 h culture in glcYMa
| Strain and parameter | Sterol composition (%) in glcYM with: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No supplement |
7,22 |
ERG |
LAN |
CAN |
CER |
CEN |
DES |
|||||||||
| FLC− | FLC+ | FLC− | FLC+ | FLC− | FLC+ | FLC− | FLC+ | FLC− | FLC+ | FLC− | FLC+ | FLC− | FLC+ | FLC− | FLC+ | |
| CG156 | ||||||||||||||||
| Cholesterol | [2.8] | [2.0] | 30.7 | 27.8 | ||||||||||||
| Δ7-Cholestenol (lathosterol) | 1.6 | 9.5 | ||||||||||||||
| Cholestanol | 19.6 | 22.7 | ||||||||||||||
| Desmosterol | 2.8 | 1.9 | ||||||||||||||
| Ergosta 5,22-dienol | [16.3] | [15.4] | ||||||||||||||
| Zymosterol | ||||||||||||||||
| Ergosterol | [14.3] | [12.8] | 25.2 | 29.2 | ||||||||||||
| Ergosta 7,22-dienol | 0.8 | 0.3 | ||||||||||||||
| Ergosta 5-enol | [10.7] | [5.7] | ||||||||||||||
| 14α-Methyl fecosterol | 3.4 | 3.6 | 1.7 | 3.9 | 1.2 | 3.6 | 1.4 | 4.4 | 1.3 | 1.5 | 2.3 | 3.8 | 1.2 | 2.2 | 1.5 | 3.6 |
| Fecosterol | ||||||||||||||||
| Ergosta 8-enol | ||||||||||||||||
| 4,14α-Dimethylzymosterol | 11.5 | 12.1 | 4.4 | 6.1 | 2.3 | 2.9 | 7.0 | 10.3 | 2.7 | 4.9 | 1.4 | 3.5 | 6.4 | 8.7 | 7.4 | 4.9 |
| Ergosta 5,7-dienol | ||||||||||||||||
| Episterol | ||||||||||||||||
| Ergosta 7-enol | ||||||||||||||||
| 14α-Methyl 3β,6α-diolb | 7.5 | 8.2 | 1.6 | 5.2 | 0.7 | 1.5 | 4.8 | 7.6 | 0.8 | 1.5 | 1.0 | 2.8 | 2.6 | 1.7 | 1.9 | 1.5 |
| Lanosterol | 74.0 | 71.1 | 75.8 | 69.5 | 70.0 | 60.9 | 85.3 | 77.6 | 71.1 | 66.7 | 62.7 | 59.8 | 86.3 | 76.6 | 59.4 | 66.3 |
| 4,4-Dimethylzymosterol | ||||||||||||||||
| Eburicol | 0.7 | 1.2 | 0.2 | 1.1 | 0.8 | 0.4 | 1.0 | 1.2 | 0.6 | 0.6 | ||||||
| Unknown | 3.5 | 5.0 | 0.7 | 0.9 | 0.4 | 0.8 | 1.5 | 0.8 | 0.4 | 1.0 | 1.0 | 1.3 | 1.4 | 0.2 | ||
| CG388 | ||||||||||||||||
| Cholesterol | [3.2] | 35.3 | ||||||||||||||
| Δ7-Cholestenol (lathosterol) | 8.5 | |||||||||||||||
| Cholestanol | 22.7 | |||||||||||||||
| Desmosterol | 2.2 | |||||||||||||||
| Ergosta 5,22-dienol | [16.7] | |||||||||||||||
| Zymosterol | 3.7 | 0.7 | 3.6 | 0.8 | 4.0 | 3.2 | 0.6 | 4.2 | 4.0 | 3.3 | 1.7 | 4.5 | ||||
| Ergosterol | 59.7 | 37.0 | 60.4 | 45.5 | 63.1 | 43.3 | 65.4 | 1.7 | 61.7 | 0.9 | 60.7 | 3.3 | 63.4 | 1.3 | 62.0 | 7.9 |
| Ergosta 7,22-dienol | 7.6 | 7.0 | 0.7 | 6.7 | 0.8 | 6.3 | 6.4 | 7.2 | 7.7 | 7.5 | ||||||
| Ergosta 5-enol | [8.7] | |||||||||||||||
| 14α-Methyl fecosterol | 1.0 | 0.3 | 1.5 | 0.5 | 0.5 | 0.9 | 2.3 | 0.4 | ||||||||
| Fecosterol | 3.2 | 3.5 | 2.4 | 2.0 | 2.8 | 3.1 | 2.1 | 2.2 | ||||||||
| Ergosta 8-enol | 1.7 | 2.0 | 1.5 | 1.6 | 2.0 | 2.0 | 1.9 | 2.2 | ||||||||
| 4,14α-Dimethylzymosterol | 2.0 | 1.1 | 1.5 | 3.7 | 6.4 | 4.4 | 9.1 | 4.3 | ||||||||
| Ergosta 5,7-dienol | 5.2 | 5.5 | 5.8 | 4.7 | 5.0 | 5.1 | 5.4 | 5.7 | ||||||||
| Episterol | 6.1 | 5.8 | 6.4 | 6.1 | 6.1 | 6.0 | 5.8 | 6.0 | ||||||||
| Ergosta 7-enol | 1.2 | 1.6 | 1.4 | 1.6 | 1.3 | 1.2 | 1.1 | 1.5 | ||||||||
| 14α-Methyl 3β,6α-diol | 1.5 | 1.0 | 1.1 | 3.8 | 2.5 | 3.2 | 4.1 | 1.6 | ||||||||
| Lanosterol | 2.3 | 55.1 | 2.6 | 47.8 | 2.2 | 50.1 | 2.5 | 89.7 | 3.0 | 63.9 | 3.3 | 52.5 | 3.0 | 73.0 | 2.5 | 57.3 |
| 4,4-Dimethylzymosterol | 9.4 | 1.9 | 8.0 | 6.5 | 6.6 | 7.5 | 7.4 | 6.3 | 5.9 | |||||||
| Eburicol | 1.9 | 1.3 | 0.2 | 0.4 | ||||||||||||
| Unknown | 0.0 | 0.7 | 0.8 | 0.5 | 0.2 | 0.3 | ||||||||||
The most abundant sterol in each treatment is in boldface; exogenously supplied sterols are underlined; metabolites of exogenously supplied sterols are in brackets. FLC was added at ×0.5 MIC.
14α-Methyl ergosta 8,24(28)-dien-3β-6α-diol is a fungistatic sterol (16).
CG156.
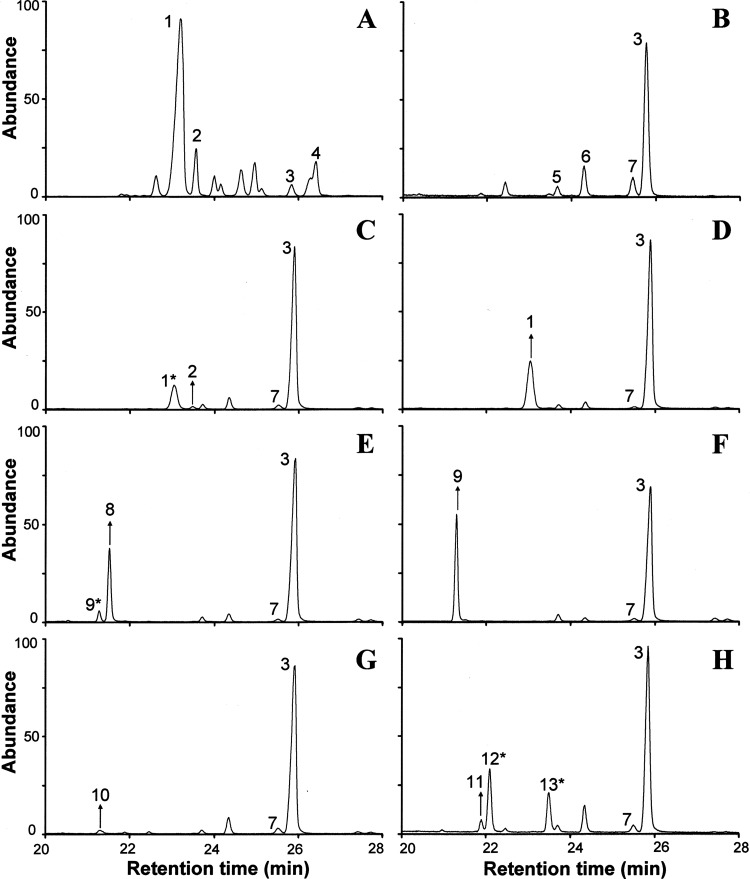
GC-MS chromatograms for CG156 cultured in the absence of FLC (Fig. 5B to H) provide evidence for facultative sterol uptake and the metabolism of exogenously supplied sterols. Negligible (<1% of the total sterol fraction) quantities of ergosta 7,22-dienol were detected in untreated CG156 cultured using glcYM + 7,22; the second most abundant sterol (after lanosterol) identified in these cultures was ergosterol (12.8%). Exogenously supplied ergosterol comprised approximately 25% of the sterol fraction of CG156 cultured using glcYM + ERG (Table 4); ergosterol was not detected in CG156 grown on any other sterol-supplemented medium (except glcYM + 7,22). CG156 cultured using glcYM + CER incorporated relatively large quantities of exogenously supplied cholesterol, such that it comprised approximately 31% of the total sterol fraction in untreated cells (Table 4). Cholesterol was also detected as a sterol metabolite (comprising 2.8% of the total) in CG156 grown on glcYM + CAN; in these cultures the original cholestanol substrate constituted approximately 20% of the total sterol fraction. Lathosterol (Δ7-cholestenol) was only detected in trace quantities (<2% of the total sterol fraction) in CG156 cultured using glcYM + CEN; no potential sterol metabolites of CEN (e.g., cholesterol) were detected in these cells. Conversely, while similarly low levels of desmosterol (2.8% of total) were detected in CG156 cultured using glcYM + DES, metabolites of desmosterol ordinarily not synthesized by C. glabrata (ergosta 5,22-dienol and ergosta 5-enol) were detected in relative abundance, comprising 16.3 and 10.7% of the total sterol fraction, respectively.
Fig 5.
Sterol chromatograms for C. glabrata study isolates following growth on sterol-free and sterol-supplemented glcYM (no azole treatment). (A) Typical sterol profile for CG388 (all media); (B) typical sterol profile for CG156 on sterol-free glcYM and glcYM + LAN; (C) CG156 on glcYM + 7,22; (D) CG156 on glcYM + ERG; (E) CG156 on glcYM + CAN; (F) CG156 on glcYM + CER; (G) CG156 on glcYM + CEN; (H) CG156 on glcYM + DES. Arrows indicate the uptake of the original, exogenously supplied sterol. Asterisks highlight metabolites of exogenously supplied sterols. Peak 1, ergosterol; 2, ergosta 7,22-dienol; 3, lanosterol; 4, 4,4 dimethyl zymosterol; 5, 14α-methyl fecosterol; 6, 4,14α dimethyl zymosterol; 7, 14α-methyl ergosta 8,24(28)-dien-3β-6α-diol; 8, cholestanol; 9, cholesterol; 10, Δ7-cholestenol; 11, desmosterol; 12, ergosta 5,22-dienol; and 13, ergosta 5-enol.
In FLC-treated CG156 cultured using sterol-free glcYM and all sterol-supplemented media (except glcYM + CEN), the proportions of exogenously supplied sterols and their respective sterol metabolites (detailed above) are very similar to those recorded in untreated cells (Table 4, bracketed and underlined values). GC-MS data indicate that FLC-treated CG156 incorporated approximately 5-fold more Δ7-cholestenol (9.5% of total sterol fraction) than untreated CG156 grown on glcYM + CEN; however, as seen in untreated CG156 cultures, potential sterol metabolites of CEN were again absent.
CG388.
In the absence of FLC, CG388 exhibited wild-type sterol composition (Fig. 5A) on both sterol-free and sterol-supplemented glcYM; this reflects the continuation of endogenous ergosterol biosynthesis. Conversely, sterol profiles for FLC-treated CG388 indicate azole inhibition of Erg11p (evidenced by decreased ergosterol content and the accumulation of 14α-methylated sterol intermediates) in addition to the uptake and metabolism of exogenously supplied sterols. The ergosterol content of FLC-treated CG388 cultured using sterol-free glcYM decreased to 62% of that of untreated cells, comprising 37 and 59.7% of the total cellular sterol fraction, respectively. The levels of ergosterol detected in FLC-treated CG388 cultured using glcYM + 7,22 and glcYM + ERG were relatively similar (45.5 and 43.3%, respectively). The depletion of ergosterol in all other FLC-treated CG388 cultures was more significant; ERG comprised at most 8% (glcYM + DES) and typically <3% of the total cellular sterol fraction (Table 4).
DISCUSSION
Results from this study of five clinical isolates of C. glabrata (CG26, CG44, CG156, CG388, and CG1012) complement findings from work with engineered laboratory strains (10, 25, 26) and provide novel evidence regarding the versatility and consequences of facultative sterol uptake for growth and antifungal resistance in C. glabrata.
CG156, the most azole- and polyene-resistant isolate (Table 1), was found to be an erg11 (sterol 14α-demethylase) mutant completely lacking 14α-demethylated sterols (including ergosterol) and harboring a single-amino-acid substitution (G315D) which nullified the function of Erg11p. This substitution occurs in the highly conserved αI helix of Erg11p and other cytochromes P450 (Fig. 3A), affecting the structure of substrate recognition site IV (36). An equivalent substitution also results in loss of function in S. cerevisiae sterol 14α-demethylase (13). It was initially suspected that CG156 persists by scavenging trace amounts of ergosterol from the YEPD growth medium on which it was maintained. However, using sterol-free (glcYM) minimal medium, we found that CG156 could grow in the absence of exogenously supplied sterol (Table 3) with 14α-methylated sterol intermediates (lanosterol was >80% of the total), again constituting the entire cellular sterol fraction. This finding is consistent with reports of other erg11 strains of C. glabrata (10) and S. cerevisiae (9), which remain aerobically viable and are able to grow on lanosterol-type sterols. It is possible that changes in overall membrane composition (e.g., proportions of fatty acid, phospholipids, and sphingolipids) compensate for perturbations in sterol content in ergosterol-deficient yeast; this is now an avenue for further investigation. Such plasticity in membrane composition might well enhance the capacity of C. glabrata to withstand extended periods of starvation stress (31) and underscores data showing that Erg11p is not an ideal drug target in C. glabrata (25).
Facultative sterol uptake supported higher cell densities in CG156 samples cultured using all (except glcYM + LAN) sterol-supplemented glcYM than in samples grown on sterol-free glcYM (Table 3), indicating that exogenously supplied sterols could serve bulk functions (35) in C. glabrata. The elevated growth rates of CG156 cultured using cholesterol-containing (glcYM + CER) media demonstrate how this species is well adapted for survival in the human host (31). The ability of both CG156 and CG388 to sequester Δ7-cholestenol (lathosterol) and to further metabolize both cholestanol and desmosterol (Table 4) is also significant. All three cholesterol precursors accumulate within the body in certain inborn human malformation syndromes, including lathosterolosis, desmosterolosis, and cerebrotendinous xanthomatosis (17, 32).
In contrast to CG388, wherein the uptake of exogenously supplied sterols (observed only in azole-treated CG388) (Table 4) correlated with an increase in resistance to fluconazole and voriconazole (Table 3), sterol uptake had no effect on azole MICs recorded for CG156. Furthermore, while clear differences were observed between the sterol composition of untreated and FLC-treated CG388 (Table 4), the sterol content of CG156 grown on all sterol-supplemented media was similar in the absence and presence of FLC (Table 4). It has been proposed that facultative sterol uptake contributes to elevated azole resistance in C. glabrata (1, 26); however, it does not appear to be essential for the high-level azole resistance observed in CG156. Interestingly, the proportions of exogenously supplied cholestanol, cholesterol, Δ7-cholestenol, and desmosterol (and their respective sterol metabolites) detected in FLC-treated CG388 and CG156 are highly comparable (Table 4, bracketed and underlined values). It is possible that these sterols serve a compensatory role (e.g., stabilizing membrane integrity) in azole-treated cells but that they are only required in threshold amounts.
Owing to the continuation of endogenous ergosterol biosynthesis, CG388 exhibited uniform sensitivity to amphotericin B (MIC, 2 μg ml−1) on all media; conversely, the sensitivity of CG156 to AMB was affected by the uptake and metabolism of specific exogenously supplied sterols. CG156 cultured using media containing ergosterol (and ergosta 7,22-dienol) were more sensitive to AMB than CG156 grown on cholesterol (or cholestanol) (MICs of 2 and >64 μg ml−1, respectively). The presence of ergosterol (the binding target of polyene antifungals [6]) and cholesterol (weaker binding to polyenes) in CG156 cultured using glcYM + 7,22 and glcYM + CAN likely reflects the activity of endogenous ERG3 protein (C5-desaturase) on ergosta 7,22-dienol and cholestanol precursors (Fig. 1D) and would account for the decreased sensitivity to AMB we observed. This is the first time that sterol uptake has been shown to alter AMB sensitivity in C. glabrata; it highlights the potential to exploit sterol transport proteins for the uptake and intracellular targeting of antifungal compounds.
Aside from demonstrating the ability of C. glabrata to sequester several previously uninvestigated sterols, results from the present study provide novel insights into endogenous sterol metabolism. That significant (>5%) amounts of Δ7-cholestenol were only sequestered by azole-treated CG388 and CG156 suggests that it is not an ideal bulk sterol. That no potential products of Δ7-cholestenol metabolism (e.g., ergosterol) were detected in glcYM + CEN-grown cells indicates that C. glabrata lacks the enzymatic machinery required to further metabolize Δ7-cholestenol (e.g., to ergosterol or cholesterol). Azole-treated CG156 and CG388 cultured using glcYM + DES contained only negligible amounts of desmosterol (≤3% of the total sterol fraction). Nonetheless, significant proportions of ergosta 5-enol (indicative of endogenous ERG6 protein [C24-methyl transferase] activity on desmosterol) and ergosta 5,22-dienol (indicative of endogenous ERG5 protein [C22-desaturase] activity on ergosta 5-enol) were detected in both isolates.
Sterol metabolism data suggest that (i) despite binding with high affinity to heterologously expressed C. glabrata C22-desaturase (Erg5p) (19), FLC is not a potent inhibitor of Erg5p activity in vivo, and (ii) that CG156 also possesses functional Erg6p. That C22-desaturase activity is apparent in both CG388 and CG156 is important, because Erg5p (CYP61) is a heme-thiolate cytochrome P450 enzyme (19). It remains possible that the clinical isolates, including CG156, harbor additional mutations; however, heme biosynthetic mutations detected in other studies (1, 9, 10) can be excluded for CG156. The functionality of Erg6p is relevant because a clinical isolate of C. glabrata with reduced susceptibility to polyenes, harboring a missense mutation in ERG6 and exhibiting pseudohyphal growth, has been reported (38). The polyene resistance and the flocculent growth of CG156 thus appear to have a different molecular basis. An ace2 mutant of C. glabrata exhibiting clumpy growth and hypervirulence in a murine model of invasive candidiasis has been described (15). This isolate was able to cause systemic infections characterized by fungal escape from the vasculature, tissue penetration, and proliferation in vivo. Although poorly documented, the molecular basis and consequences of phenotypic switching in C. glabrata now require fuller investigation, not least because it is understood to be a key virulence factor in Candida albicans (2).
Taken as a whole, results from this study indicate that some isolates (here, CG156) might persist as slow-growing agents of chronic infections, because they (i) possess mutated Erg11p that has low or no affinity for azoles; (ii) lack cellular ergosterol and thus exhibit high-level resistance to polyenes; (iii) can survive without sterol auxotrophy through growth on lanosterol-type sterols; and/or (iv) can opportunistically exploit a wide spectrum of host/environmental sterols for growth. That CG156 was found to be a low-efflux isolate (Table 1) is particularly interesting. It is possible that the altered cellular sterol composition of CG156 affects intracellular signaling and trafficking pathways, including the distribution and function of membrane-anchored proteins. The latter could include efflux machinery (3, 7, 34) and any transport proteins that are proposed to mediate azole import via facilitated diffusion (21). The investigation of these possibilities represents an avenue for further work.
ACKNOWLEDGMENTS
This research was supported by the EU FP6 project EURESFUN (European Resistance Fungal Network).
Strains CG26, CG44, CG388, and CG1012 were obtained through the German National Reference Center for Systemic Mycoses. CG156 was kindly contributed to the EURESFUN strain collection by E. Mellado (ISCII, Madrid, Spain).
Technical support was from N. J. Rolley, and analytical facilities were provided by the EPSRC National Mass Spectrometry Service Centre (Swansea University, United Kingdom).
Footnotes
Published ahead of print 21 May 2012
REFERENCES
- 1. Bard M, et al. 2005. Sterol uptake in Candida glabrata: rescue of sterol auxotrophic strains. Diagn. Microbiol. Infect. Dis. 52:285–293 [DOI] [PubMed] [Google Scholar]
- 2. Brockert PJ, et al. 2003. Phenotypic switching and mating type switching of Candida glabrata at sites of colonization. Infect. Immun. 71:7109–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brun S, et al. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48:1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chapeland-Leclerc F, et al. 2010. Acquisition of flucytosine, azole, and caspofungin resistance in Candida glabrata bloodstream isolates serially obtained from a hematopoietic stem cell transplant recipient. Antimicrob. Agents Chemother. 54:1360–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts (M27–A3), 3rd ed CLSI, Wayne, PA [Google Scholar]
- 6. Ellis D. 2002. Amphotericin B: spectrum and resistance. J. Antimicrob. Chemother. 49:7–10 [DOI] [PubMed] [Google Scholar]
- 7. Ferrari S, et al. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268 doi:10.1371/journal.ppat.1000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fidel PL, Jr, Vasquez JA, Sobel JD. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gachotte D, et al. 1997. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc. Natl. Acad. Sci. U. S. A. 94:11173–11178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geber A, et al. 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39:2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Groeneveld P, Rolley N, Kell DB, Kelly SL, Kelly DE. 2002. Metabolic control analysis and engineering of the yeast sterol biosynthetic pathway. Mol. Biol. Rep. 29:27–29 [DOI] [PubMed] [Google Scholar]
- 12. Hazen KC, et al. 2005. Isolation of cholesterol-dependent Candida glabrata from clinical specimens. Diagn. Microbiol. Infect. Dis. 52:35–37 [DOI] [PubMed] [Google Scholar]
- 13. Ishida N, et al. 1988. A single amino acid substitution converts cytochrome P45014DM to an inactive form, cytochrome P450SG1: complete primary structures deduced from cloned DNAs. Biochem. Biophys. Res. Commun. 155:317–323 [DOI] [PubMed] [Google Scholar]
- 14. Joffrion TM, Cushion MT. 2010. Sterol biosynthesis and sterol uptake in the fungal pathogen Pneumocystis carinii. FEMS Microbiol. Lett. 311:1–9 [DOI] [PubMed] [Google Scholar]
- 15. Kamran M, et al. 2004. Inactivation of transcription factor gene ACE2 in the fungal pathogen Candida glabrata results in hypervirulence. Eukaryot. Cell 3:546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14α-methyl ergosta-8,24(28)-dien-3β,6α-diol. Biochem. Biophys. Res. Commun. 207:910–915 [DOI] [PubMed] [Google Scholar]
- 17. Krakowiak PA, et al. 2003. Lathosterolosis: an inborn error of human and murine cholesterol synthesis due to lathosterol 5-desaturase deficiency. Hum. Mol. Genet. 12:1631–1641 [DOI] [PubMed] [Google Scholar]
- 18. Krogh-Madsen M, Arendrup MC, Heslet L, Knudsen JD. 2006. Amphotericin B and caspofungin resistance in Candida glabrata isolates recovered from a critically ill patient. Clin. Infect. Dis. 42:938–944 [DOI] [PubMed] [Google Scholar]
- 19. Lamb DC, et al. 1999. Purification, reconstitution, and inhibition of cytochrome P-450 sterol delta(22)-desaturase from the pathogenic fungus Candida glabrata. Antimicrob. Agents Chemother. 43:1725–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maesaki S, Marichal P, Vanden Bossche H, Sanglard D, Kohno S. 1999. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole resistant Candida albicans strains. J. Antimicrob. Chemother. 44:27–31 [DOI] [PubMed] [Google Scholar]
- 21. Mansfield BE, et al. 2010. Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog. 6:e1001126 doi:10.1371/journal.ppat.1001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martel CM, et al. 2010. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14α-demethylase) and ERG5 (encoding C22-desaturase) is cross-resistant to azoles and amphotericin B. Antimicrob. Agents Chemother. 54:3578–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martel CM, et al. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54:4527–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martel CM, et al. 2010. Complementation of a Saccharomyces cerevisiae ERG11/CYP51 (sterol 14 alpha-demethylase) doxycycline-regulated mutant and screening of the azole sensitivity of Aspergillus fumigatus isoenzymes CYP51A and CYP51B. Antimicrob. Agents Chemother. 54:4920–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakayama H, Nakayama N, Arisawa M, Aoki Y. 2001. In vitro and in vivo effects of 14 alpha-demethylase (ERG11) depletion in Candida glabrata. Antimicrob. Agents Chemother. 45:3037–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakayama H, et al. 2007. The Candida glabrata putative sterol transporter gene CgAUS1 protects cells against azoles in the presence of serum. J. Antimicrob. Chemother. 60:1264–1272 [DOI] [PubMed] [Google Scholar]
- 27. Pfaller MA, et al. 2000. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997–1998. Antimicrob. Agents Chemother. 44:747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfaller MA, Diekema DJ. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 42:4419–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfaller MA, Messer SA, Hollis RJ, Jones RN, Diekema DJ. 2002. In vitro activities of ravuconazole and voriconazole compared with those of four approved systemic antifungal agents against 6,970 clinical isolates of Candida spp. Antimicrob. Agents Chemother. 46:1723–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roetzer A, Gabaldon T, Schuller C. 2011. From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 314:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Russell DW. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72:137–174 [DOI] [PubMed] [Google Scholar]
- 33. Sanglard D, Ischer F, Bille J. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanguinetti M, et al. 2005. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 49:668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith SJ, Parks LW. 1997. Requirement of heme to replace the sparking sterol function in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1345:71–76 [DOI] [PubMed] [Google Scholar]
- 36. Strushkevich N, Usanov SA, Park HW. 2010. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 397:1067–1078 [DOI] [PubMed] [Google Scholar]
- 37. Tsai HF, et al. 2004. Candida glabrata erg1 mutant with increased sensitivity to azoles and to low oxygen tension. Antimicrob. Agents Chemother. 48:2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vandeputte P, et al. 2007. Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of Candida glabrata with pseudohyphal growth. Antimicrob. Agents Chemother. 51:982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vandeputte P, et al. 2009. Hypersusceptibility to azole antifungals in a clinical isolate of Candida glabrata with reduced aerobic growth. Antimicrob. Agents Chemother. 53:3034–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weiss A, Delproposto J, Giroux CN. 2004. High-throughput phenotypic profiling of gene-environment interactions by quantitative growth curve analysis in Saccharomyces cerevisiae. Anal. Biochem. 327:23–34 [DOI] [PubMed] [Google Scholar]
- 41. Xiong QB, et al. 2005. Cholesterol import by Aspergillus fumigatus and its influence on antifungal potency of sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 49:518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]