Abstract
We report the use of a known pyridochromanone inhibitor with antibacterial activity to assess the validity of NAD+-dependent DNA ligase (LigA) as an antibacterial target in Staphylococcus aureus. Potent inhibition of purified LigA was demonstrated in a DNA ligation assay (inhibition constant [Ki] = 4.0 nM) and in a DNA-independent enzyme adenylation assay using full-length LigA (50% inhibitory concentration [IC50] = 28 nM) or its isolated adenylation domain (IC50 = 36 nM). Antistaphylococcal activity was confirmed against methicillin-susceptible and -resistant S. aureus (MSSA and MRSA) strains (MIC = 1.0 μg/ml). Analysis of spontaneous resistance potential revealed a high frequency of emergence (4 × 10−7) of high-level resistant mutants (MIC > 64) with associated ligA lesions. There were no observable effects on growth rate in these mutants. Of 22 sequenced clones, 3 encoded point substitutions within the catalytic adenylation domain and 19 in the downstream oligonucleotide-binding (OB) fold and helix-hairpin-helix (HhH) domains. In vitro characterization of the enzymatic properties of four selected mutants revealed distinct signatures underlying their resistance to inhibition. The infrequent adenylation domain mutations altered the kinetics of adenylation and probably elicited resistance directly. In contrast, the highly represented OB fold domain mutations demonstrated a generalized resistance mechanism in which covalent LigA activation proceeds normally and yet the parameters of downstream ligation steps are altered. A resulting decrease in substrate Km and a consequent increase in substrate occupancy render LigA resistant to competitive inhibition. We conclude that the observed tolerance of staphylococcal cells to such hypomorphic mutations probably invalidates LigA as a viable target for antistaphylococcal chemotherapy.
INTRODUCTION
NAD+-dependent DNA ligase (LigA) has been identified by numerous authors as an attractive potential target for broad-spectrum antibacterial chemotherapy (7, 23). LigA is well conserved among eubacterial species, is architecturally and biochemically distinct from the ATP-dependent DNA ligases of eukaryotic cells, and has been found to be essential for bacterial viability wherever examined (13, 14, 15, 17, 31). Moreover, the DNA ligation reaction has been dissected mechanistically, mutationally, and structurally (8, 20, 25, 26, 33, 34, 35), and screening assays have been reported for the complete reaction cycle and for individual component steps (2, 11, 18).
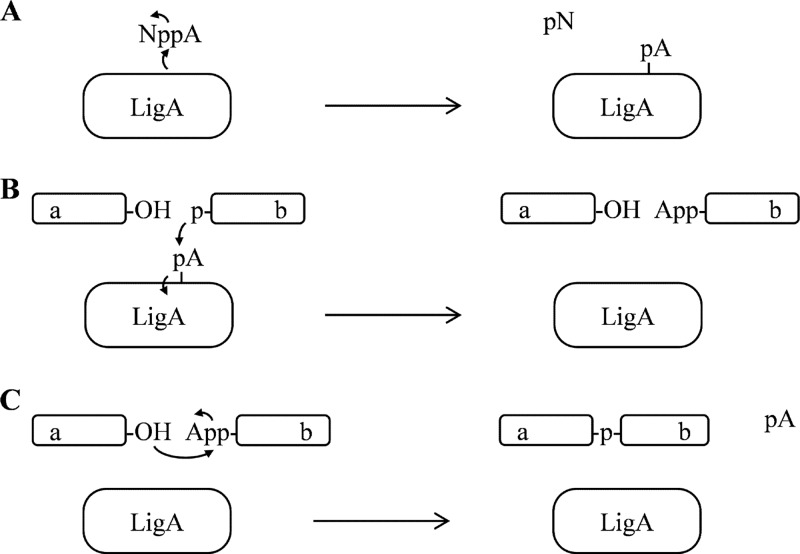
DNA ligation activities are essential for multiple DNA processes in replication and repair, including the joining of Okazaki fragments into a continuous strand during chromosomal DNA replication. Enzymatically, DNA ligation proceeds via three successive adenylyl transfer steps (Fig. 1) (32): first, DNA-independent covalent adenylation of the catalytic lysine by the NAD+ substrate; second, adenylyl transfer to the free 5′ phosphate at the nicked DNA ligation site; and third, the covalent sealing of the DNA nick with concomitant AMP release. Biochemical functions of distinct domains in the modular enzyme structure have been assigned to particular reaction steps. The DNA-independent adenylyl transfer activity resides within the amino-terminal adenylation domain, which comprises an amino-terminal Ia region that is specific to NAD+-dependent DNA ligases and a nucleotidyl transferase (NTase) region that is universal among DNA and RNA ligases. The subsequent coupling of adenylation to DNA ligation depends upon downstream DNA-binding domains, which include an oligonucleotide-binding fold (OB fold) and a helix-hairpin-helix (HhH) domain. Structural studies of the adenylation domain have revealed conformational transitions that accompany the adenylation cycle (8), and structural study of the full-length enzyme bound to DNA-adenylate has identified specific contacts between the DNA-binding domains and the DNA duplex substrate near the nicked ligation site (20).
Fig 1.
Reaction scheme depicting the three successive adenylyl transfer steps that underlie the DNA ligation reaction catalyzed by eubacterial NAD+-dependent DNA ligase (LigA). (A) Step 1, DNA-independent adenylation of the catalytic lysine of LigA (depicted as transfer of pA), using NAD+ (NppA) as the substrate and releasing NMN (pN) product. (B) Step 2, covalent transfer of AMP (pA) from LigA to the 5′ phosphate of target DNA strand b. (C) Step 3, ligation of DNA strands a and b with release of AMP from DNA strand b. (A through C) For simplicity, the single DNA strands a and b are depicted without complementary DNA; curved arrows indicate electron movements during the successive adenylyl transfers. Letter designations: A, adenosine nucleoside; N, nicotinamide nucleoside; p, monophosphate; pp, diphosphate.
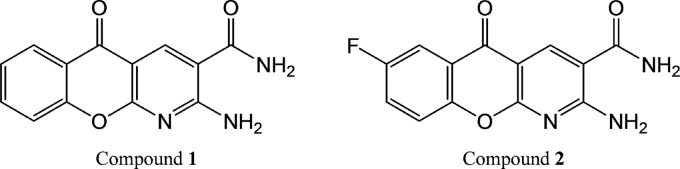
Numerous LigA inhibitors have been reported to date, including arylamino acids, such as chloroquine (4), glycosyl ureides and glycosylamines (27, 28), tetracyclic indoles (29), a pyrimidopyrimidine inhibitor (17), substituted adenosine analogs (19, 30), and the pyridochromanones (1). Pyridochromanones were identified by high-throughput screening as potent competitive inhibitors of DNA ligation by LigA from Staphylococcus aureus (50% inhibitory concentration [IC50] ≤ 0.9 μM) (1). They inhibit LigA from diverse bacteria but are inactive against the ATP-dependent human DNA ligase I (1, 9). Moreover, they show antibacterial activity against S. aureus (MIC ≤ 1 μg/ml) with a bactericidal mode of action; their antibacterial activity in S. aureus has been mapped to a putative resistance lesion in the ligA locus (1).
In this study, we applied the antibacterial activity of a pyridochromanone inhibitor to assess LigA as an antibacterial target in S. aureus. We report the recovery of numerous high-level resistance mutations dispersed across the ligA gene, with an unexpected concentration of mutations in the OB fold domain. We examined the kinetic parameters of several mutant LigA isoforms and report a generalized resistance mechanism in which LigA resistance to competitive inhibitors is achieved via systematic alteration of its kinetic properties. The facility of this mechanism, coupled with the tolerance of the bacteria to broad changes in LigA properties, suggests that LigA makes a poor antibacterial drug target despite its favorable features. Assessment of this potential antibacterial target therefore requires greater subtlety than is afforded by standard validation criteria.
MATERIALS AND METHODS
Bacterial strains and compounds.
S. aureus ATCC 29213 (methicillin-sensitive S. aureus [MSSA]), S. aureus ATCC 700699 (methicillin-resistant S. aureus [MRSA]), and Escherichia coli ATCC 25922 were obtained from the American Type Culture Collection, Manassas, VA. Pyridochromanone compounds 1 and 2 were synthesized at Achillion Pharmaceuticals. Adenosine 3′,5′-cyclic monophosphorothioate (Sp-cAMPs) was purchased from Sigma-Aldrich, St. Louis, MO.
Culture conditions.
S. aureus strains were grown with aeration at 35°C to 37°C in Mueller-Hinton II (MH II) broth (Becton, Dickinson; Sparks, MD).
Compound susceptibility assays.
MICs were determined by broth microdilution according to the CLSI approved guidelines (5).
Selection and characterization of resistant mutants.
S. aureus ATCC 29213 cultures were grown overnight under nonselective conditions in brain heart infusion (BHI) or MH II liquid medium. Spontaneous pyridochromanone-resistant mutants were recovered from these cultures by plating ∼1 × 109 organisms followed by overnight growth at 37°C on BHI or MH II agar containing 4 μg/ml compound 1. Recovered clones were assessed for compound susceptibility and ligA genotype. Genomic ligA DNA was amplified by PCR using the primers SaLig-N (ATATACCATGGCTGATTTATCGTCTCGTG; NcoI restriction site underlined) and SaLig-C (TTATATAAGCGGCCGCACTATTTAATTCATTTTGCTTATCTACA; NotI restriction site underlined). Genomic ligA products were purified by QIAquick PCR purification (Qiagen, Valencia, CA), and their coding sequences determined by automated DNA sequencing (W.M. Keck Foundation, Yale University, New Haven, CT, and SeqWright, Houston, TX).
LigA protein expression and purification.
ligA was amplified by PCR from S. aureus strain N315 with the primers SaLig-N and SaLig-C, and the product cloned into the NcoI and NotI sites of the pET21d vector (Novagen) to direct the expression of LigA with a C-terminal His tag. Missense mutants and the carboxy-terminally tagged LigA truncation (LigA:AD, the adenylation domain, comprising residues 1 to 315) were introduced by QuikChange site-directed mutagenesis according to the manufacturer's instructions (Stratagene, La Jolla, CA). All isoforms were expressed in Novagen E. coli BL21(DE3)pLysS (EMD Biosciences, Madison, WI), purified by HisTrap or HisGraviTrap chromatography (GE Healthcare, Piscataway, NJ), and deadenylated by incubation with excess nicotinamide mononucleotide (NMN) and MgCl2 followed by dialysis.
LigA adenylation assays.
A concentration of 10 nM LigA was incubated with 1 nM [32P]AMP-NAD+ in buffer A (10 mM HEPES, 25 mM KCl, 20 mM MgCl2, 1 mM dithiothreitol [DTT], 10% PEG 8000, pH 8.0). Reactions were stopped by the addition of EDTA after incubation periods ranging from 7.5 to 60 min, and [32P]AMP incorporation into LigA was assessed by SDS-PAGE or by a MultiScreen DE or IP filter binding assay (Millipore, Billerica, MA) similar to that of Miesel et al. (18), followed by scintillation counting in a Wallac MicroBeta reader (PerkinElmer, Waltham, MA). Reaction rates in the filter-binding assay were converted from counts per minute (cpm) to picomolar units using the empirically determined specific activity of [32P]NAD+ substrate.
DNA ligation assays.
Oligonucleotides 1 through 4 were synthesized by IDT (Coralville, IA) as defined by Chen et al. (2), with the duplex pairs 1-2 and 3-4 bearing complementary 4-bp overhangs. A concentration of 1 nM LigA was incubated in buffer A with 10 μM NAD+ and 1.25 ng/μl oligonucleotide duplex DNAs 1-2 and 3-4. Reaction mixtures were incubated for 10 to 12 min (wild-type LigA) or 40 min (mutant LigA) to optimize signal while keeping reactions within the linear range; reactions were terminated by the addition of EDTA. Ligation was assessed by electrophoresis through 15% polyacrylamide gels (Bio-Rad, Hercules, CA) in Tris-boric acid-EDTA buffer followed by SYBR gold staining (Invitrogen, Carlsbad, CA) and photographic quantitation (Alpha Innotech, San Leandro, CA).
Kinetic analyses.
Reactions were conducted in the absence or presence of the test compound. Kinetic parameters were calculated by nonlinear regression methods using Prism software (GraphPad, San Diego, CA). The parameters include initial velocity (Vo) for LigA adenylation reactions as derived from relative reaction rates, Km and kcat for DNA ligation at various NAD+ concentrations, IC50s for adenylation and ligation reactions in the presence of inhibitory compound concentration series, and the inhibition constant (Ki) for the ligation reaction catalyzed by the wild-type enzyme. The Ki for the ligation reaction catalyzed by the wild-type enzyme was also determined, using the Cheng-Prusoff equation Ki = IC50/(1 + [S]/Km), where [S] is the substrate concentration (3).
RESULTS
Inhibition of DNA ligation and LigA adenylation by pyridochromanones.
To explore the validity of LigA as an antibacterial target, we examined its susceptibility to two pyridochromanone inhibitors (Fig. 2). We confirmed that these compounds have antibacterial activities against MSSA and MRSA strains of S. aureus (MIC = 1 μg/ml for both strains) but not against E. coli (Table 1). We also confirmed that these compounds are potent in vitro inhibitors of DNA ligation catalyzed by S. aureus LigA (Table 1), and we determined that these compounds inhibit the DNA-independent autoadenylation activity of both full-length LigA and a truncated enzyme, LigA:AD, comprising the isolated adenylation domain (amino acids 1 to 315) (Table 1). This inhibition of LigA autoadenylation directly establishes the adenylation domain as the locus of pyridochromanone inhibition, consistent with previous suggestions and our finding (Table 1) that these compounds compete for NAD+ substrate binding.
Fig 2.
Pyridochromanone inhibitors. Compounds 1 and 2 correspond to compounds 2 and 3, respectively, of Brötz-Oesterhelt et al. (1).
Table 1.
Potency of pyridochromanones against bacterial test strains and S. aureus LigA enzymatic activities
| Compound | MIC (μg/ml) |
Ki (nM) of LigA DNA ligationa | IC50 (nM) of enzyme adenylation |
|||
|---|---|---|---|---|---|---|
| S. aureus ATCC 29213 | S. aureus ATCC 700699 | E. coli ATCC 25922 | LigA:ADb | LigA:FLc | ||
| Compound 1 | 1 | 1 | >64 | 4.0 | 36 | 28 |
| Compound 2 | 1 | 1 | >64 | NDd | 29 | ND |
| Ciprofloxacin | 0.25 | 64 | 0.016 | ND | ND | ND |
Ki for compound 1 against full-length LigA was determined by nonlinear regression analysis using a mixed competitive model. Inhibition was found in this analysis to be partly competitive (α = 3.2).
LigA:AD is truncated after residue 315. The adenylation domain is included, comprising domain Ia and the NTase region, but the downstream DNA-binding domains are omitted.
LigA:FL refers to the full-length LigA enzyme.
ND, not determined.
Pyridochromanone resistance mutations dispersed throughout ligA.
To characterize the mechanism of pyridochromanone action in S. aureus, 22 resistant derivatives of MSSA strain 29213 were collected from 13 independent cultures following growth on agar plates under selection by compound 1 at 4× MIC (Table 2). Resistant mutants were observed to arise at a frequency of 4 × 10−7. All recovered mutants showed high-level resistance, as MIC values for compound 1 were elevated >64-fold above that of the parental strain (Table 2). All resistant colonies appeared phenotypically normal, and three showed normal doubling times when measured in liquid culture (ACH-0342, ACH-0343, and ACH-0344; not shown).
Table 2.
S. aureus mutants with resistance to compound 1
| Strain | LigA substitution | MIC (μg/ml)c |
|---|---|---|
| Selection I (single culture)a | ||
| Parentd | Wild type | 1 |
| ACH-0275 | Arg326Gln | >64 |
| ACH-0276 | Ala549Val | >64 |
| ACH-0277 | His352Gln | >64 |
| ACH-0278 | Arg326Leu | >64 |
| ACH-0279 | His352Gln | >64 |
| ACH-0280 | Arg326Leu | >64 |
| ACH-0281 | Gly481Glu | >64 |
| ACH-0282 | His352Gln | >64 |
| ACH-0283 | His352Gln | >64 |
| ACH-0284 | His352Gln | >64 |
| Selection II (independent cultures)b | ||
| Parentd | Wild type | 1 |
| ACH-0341 | Gly545Val | >64 |
| ACH-0342 | Ala303Asp | >64 |
| ACH-0343 | Arg61Ile | >64 |
| ACH-0344 | Ala349Val | >64 |
| ACH-0345 | Leu351Phe | >64 |
| ACH-0346 | Ala349Val | >64 |
| ACH-0347 | Asp361Tyr | >64 |
| ACH-0348 | Ser206Tyr | >64 |
| ACH-0349 | Ala349Thr | >64 |
| ACH-0350 | Ala349Val | >64 |
| ACH-0351 | Ala349Val | >64 |
| ACH-0352 | Pro332Ser | >64 |
Selection I: 10 colonies resistant colonies were isolated from a single culture.
Selection II: 12 independent resistant colonies were obtained from 12 separate cultures.
Antibacterial activity of compound 1 against parent or mutant as represented by MIC.
S. aureus ATCC 29213.
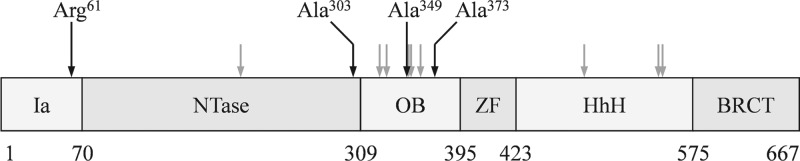
Missense point mutations were identified within the ligA gene in all 22 resistant colonies, establishing ligA as the immediate target of compound 1 activity (Table 2). Surprisingly, the mutations were dispersed throughout the ligA gene (Fig. 3). Only three were located within the adenylation domain, despite the demonstrated localized activity of compound 1 upon this domain. The remaining 19 were located in the DNA-binding OB fold and HhH domains, with the majority in the OB fold domain, like the previously reported resistance mutation Ala373 (1). The concentration of lesions in these downstream domains suggests a facile resistance pathway whereby mutations in the DNA-binding domains can overcome pyridochromanone inhibition in the adenylation domain, such as by altering the conformational landscape of the enzyme or its interaction with DNA, yet without compromising the essential functions of this enzyme in bacterial replication.
Fig 3.
Dispersed distribution of pyridochromanone resistance mutations. Modular architecture of the bacterial LigA enzyme is indicated: Ia, N-terminal NAD+-specific portion of the adenylation domain (S. aureus amino acids [aa] 1 to 69); NTase, nucleotidyl transferase portion of the adenylation domain (aa 70 to 309); OB, oligonucleotide-binding-fold domain (aa 310 to 397); ZF, zinc finger domain (398 to 424); HhH, helix-hairpin-helix domain (aa 425 to 578); BRCT, BRCA C-terminal domain (aa 580 to 667). ligA lesions identified in this study are indicated by arrows; the four mutations subjected to enzymological analysis in this study, comprising three identified here and Ala373 of Brötz-Oesterhelt et al. (1), are emphasized by black arrows and amino acid designations.
Kinetic properties defining three functional classes of LigA mutant.
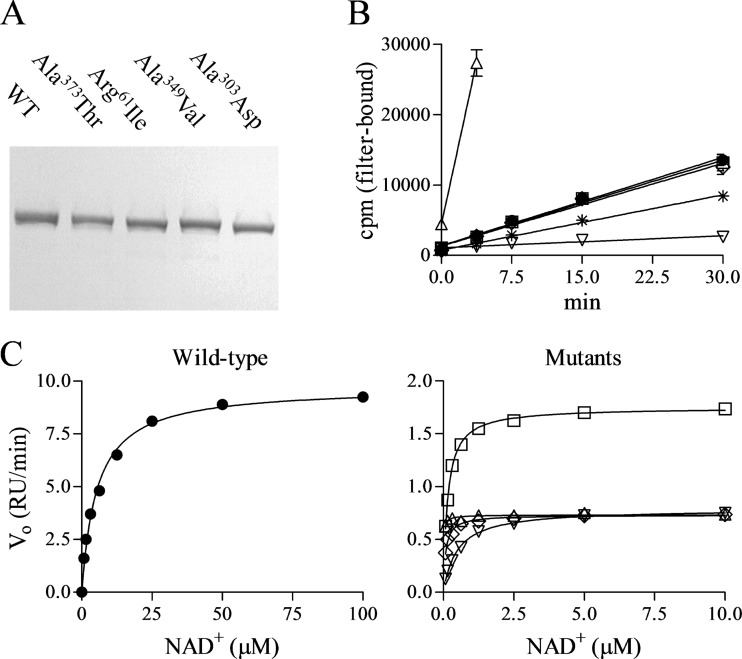
To dissect the mechanism underlying high-level pyridochromanone resistance, we engineered four of the observed resistance mutations individually into the full-length LigA enzyme (Fig. 4A). Arg61Ile and Ala303Asp were chosen to represent the relatively uncommon adenylation domain mutations; Ala349Val and Ala373Thr represented the more common OB fold domain mutations that probably confer pyridochromanone resistance from a distance. All four LigA mutant variants were expressed, purified, and subjected to enzymological study.
Fig 4.
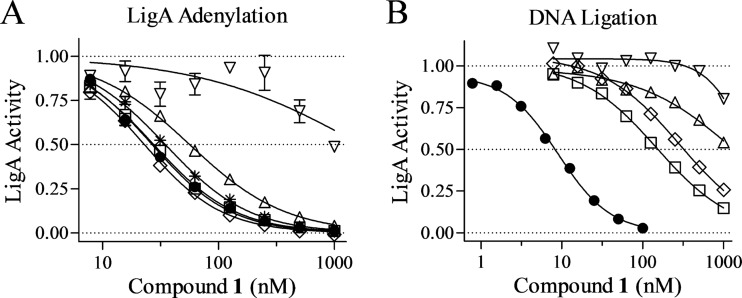
Enzymological properties of mutant LigA. (A) Purified wild-type and mutant LigA visualized by SDS-PAGE and Coomassie staining. (B) Adenylation time course with 10 nM LigA and 1 nM [32P]NAD+. Means ± standard errors of the means are shown for the duplicate wells of a single representative experiment. (C) DNA ligation reactions catalyzed by 1 nM LigA at various NAD+ concentrations. Data points indicate reaction rates from a single representative experiment; curves indicate Michaelis-Menten curves as fitted by nonlinear regression analysis. Data for wild-type and mutants are plotted in separate panels because of their differing NAD+ concentration scales. RU, relative units. Symbols: ●, LigA; *, LigA:AD (panel B only); △, LigA(Arg61Ile); ▽, LigA(Ala303Asp); ◊, LigA(Ala349Val); □, LigA(Ala373Thr).
First, we examined the NAD+-dependent but DNA-independent LigA adenylation step that preactivates the enzyme (Fig. 4B and Table 3). LigA is consumed as a substrate during this single-turnover reaction, which therefore is not amenable to classic enzymological analysis. We reduced the NAD+ concentration to 1 nM, far below its expected Km, to slow this reaction sufficiently for measurement of adenylation rates. The two adenylation domain mutations had opposite effects on the adenylation rate, as the Arg61Ile lesion effected a 15-fold increase in rate whereas the Ala303Asp lesion conferred a 7-fold decrease. In contrast, the two OB fold mutations did not affect the adenylation rate, consistent with their locations apart from the adenylation domain.
Table 3.
Activities of wild-type and mutant LigA isoforms
| LigA isoform | Adenylation Vo (pM · min−1)a | DNA ligationb |
||
|---|---|---|---|---|
| kcat (normalized)c | Km (μM NAD+) | kcat/Km (normalized)d | ||
| LigA | 1.9 ± 0.10 | 1.0 | 5.4 ± 0.26 | 1.0 |
| LigA:AD | 1.3 ± 0.023 | NAe | NA | NA |
| LigA(Arg61Ile) | 28 ± 1.9 | 0.077 ± 0.0043 | <0.078 | >5.6 |
| LigA(Ala303Asp) | 0.29 ± 0.015 | 0.075 ± 0.0035 | 0.59 ± 0.042 | 0.73 |
| LigA(Ala349Val) | 1.9 ± 0.0036 | 0.070 ± 0.0021 | 0.092 ± 0.0082 | 4.4 |
| LigA(Ala373Thr) | 1.9 ± 0.0070 | 0.17 ± 0.012 | 0.13 ± 0.014 | 7.1 |
Vo is the adenylation rate, as depicted in Fig. 4B. Data are means ± standard errors of the means (SEM) from 2 experiments.
Kinetic parameters for the complete DNA ligation reaction, as depicted in Fig. 4C. Data are means ± SEM from 4 (wild type) or 2 (mutants) experiments.
kcat was determined for each isoform in relative units (relative fluorescence intensity of product generated per unit time and unit enzyme) and then normalized to the results for wild-type LigA.
kcat/Km for each isoform was normalized to that of wild-type LigA in each experiment as described above in footnote c.
NA, not applicable.
Next, we examined the Michaelis-Menten kinetics of the full DNA ligation reaction using an oligonucleotide substrate and a range of NAD+ concentrations (Fig. 4C and Table 4). All four LigA mutations caused 6.0-fold to 14-fold reductions in the maximal ligation rate, as represented by kcat, but also 9.1-fold to >69-fold reductions in the NAD+ Km. Therefore, the DNA ligation reaction is not much affected at a low NAD+ concentration, as evidenced by the relatively modest impacts of the lesions on catalytic efficiency (kcat/Km), but is significantly compromised at higher NAD+ concentrations.
Table 4.
Inhibition of wild-type and mutant LigA adenylation and DNA ligation activities
| LigA isoform | LigA adenylation with compound 1 (IC50 [nM])a | DNA ligation |
|||
|---|---|---|---|---|---|
| Compound 1 |
Sp-cAMPs |
||||
| IC50 (nM)a | Ki (nM)b | IC50 (μM) | Ki (μM)b | ||
| LigA | 28 ± 1.1 | 9.0 ± 0.48 | 3.1 | 1.5 ± 0.03 | 0.51 |
| LigA:AD | 36 ± 1.5 | NAc | NA | NA | NA |
| LigA(Arg61Ile) | 58 ± 1.3 | >1,000 | NDd | >100 | ND |
| LigA(Ala303Asp) | >1,000 | >1,000 | >56 | >100 | >5.2 |
| LigA(Ala349Val) | 25 ± 1.7 | 320 ± 27 | 2.7 | >100 | 0.91 |
| LigA(Ala373Thr) | 28 ± 0.66 | 160 ± 7.1 | 2.2 | >100 | 1.3 |
Data are IC50s of inhibitor in LigA adenylation and DNA ligation reactions, as depicted in Fig. 5. Data are means ± SEM from 2 or 3 experiments.
Data are Ki values for DNA ligation reactions, as calculated according to the Cheng-Prusoff equation: Ki = IC50/(1 + [S]/Km), where [S] = 10 μM NAD+ and Km is taken from Table 3.
NA, not applicable.
ND, not determined: Ki could not be calculated from available data.
These LigA mutations therefore define three functional classes according to their kinetic properties, in which DNA-independent preactivation of enzyme (LigA adenylation) is accelerated, hindered, or unaffected, and yet the parameters of the complete reaction cycle (DNA ligation) are significantly compromised in all cases. Each class likely represents a distinct mechanism by which LigA is altered conformationally or energetically to produce enzyme that retains sufficient activity for viability yet has acquired resistance to active-site inhibition.
Resistance of mutant LigA enzymatic activities to inhibition.
Finally, we examined the impacts of the three classes of resistance mutations on LigA susceptibility to compound 1 inhibition in the enzyme reaction assays (Fig. 5A and Table 4). In the LigA adenylation reaction, the Arg61Ile lesion caused minimal elevation in IC50 (2.1-fold) whereas Ala303Asp increased the IC50 by >36-fold. The OB fold domain mutations Ala349Val and Ala373Thr, as expected, had no effect on the inhibition of the adenylation reaction by compound 1. We conclude that only the Ala303Asp lesion significantly affects the affinity between LigA and the pyridochromanone inhibitor.
Fig 5.
Resistance of LigA mutants to pyridochromanone inhibition. (A) Inhibition by compound 1 of adenylation reactions conducted with 10 nM LigA and 1 nM [32P]NAD+. (B) Inhibition by compound 1 of DNA ligation reactions catalyzed by 1 nM LigA with 10 μM NAD+. (A and B) Data points indicate normalized reaction rates from a single representative experiment; error bars in panel A represent standard error for duplicate wells. Curves indicate sigmoidal inhibition curves as fitted by nonlinear regression. Symbols: ●, LigA; *, LigA:AD (panel A only); △, LigA(Arg61Ile); ▽, LigA(Ala303Asp); ◊, LigA(Ala349Val); □, LigA(Ala373Thr).
In the DNA ligation reaction, in contrast, all four LigA lesions conferred significant resistance to pyridochromanone inhibition (Fig. 5B and Table 4). The IC50s for compound 1 were elevated by >110-fold for the two adenylation domain mutants and by 18-fold and 35-fold for the two OB fold domain mutants, accounting for the antibacterial resistance phenotypes of the corresponding mutant staphylococci. Mutant LigA was similarly resistant to the ATP analog Sp-cAMPs, which competes for access to the active site of the adenylation domain (Table 4) (18). The elevated IC50s against the two OB fold LigA mutants in the DNA ligation assay were not accompanied by corresponding increases in Ki values. Together, these findings support the view that the majority of LigA mutations confer resistance without direct changes in inhibitor binding at the active site, instead altering active-site accessibility by affecting the downstream steps of the three-step DNA ligation reaction.
DISCUSSION
We have reexamined the potential of NAD+-dependent DNA ligase (LigA) as an antibacterial target in S. aureus. We recovered 22 mutants with ligA coding lesions that were highly resistant to a known pyridochromanone inhibitor yet showed no deficits in viability or growth. Three mutants whose lesions were within the adenylation domain were recovered; the remaining 19 had lesions located within the DNA-binding OB fold (16 of 22) and HhH (3 of 22) DNA-binding domains. Although the possibility of distal lesions was anticipated because of the previously identified resistance mutation (1), their predominance was unexpected, as we have established the adenylation domain as the focus of pyridochromanone action. Our enzymological analysis of four representative mutant LigA isoforms has identified three mechanistic classes of resistance. We suggest that the observed tolerance of staphylococci to resistance mutations, despite the altered enzymology of DNA ligation, presents a general resistance pathway that renders LigA a questionable antibacterial target despite its favorable features.
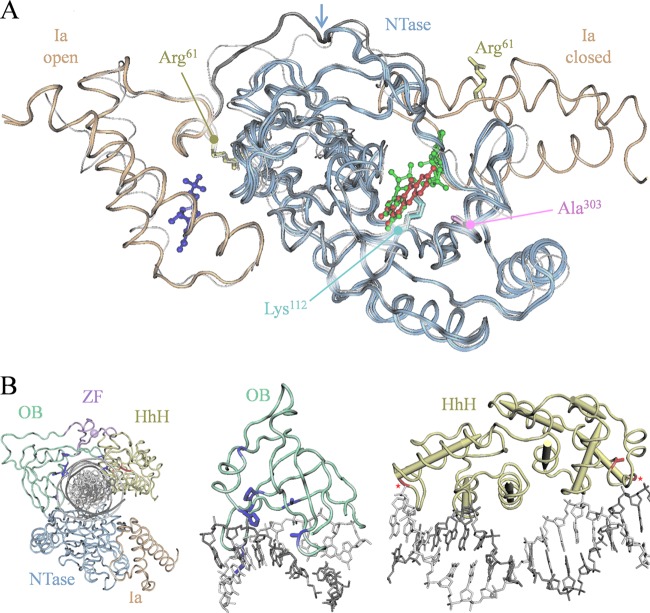
The adenylation domain mutations Arg61Ile and Ala303Asp exert opposing effects on the rate of DNA-independent LigA adenylation yet cause comparable reductions in the catalytic rate for DNA ligation. These effects can be illuminated by examination of the available crystal structures of LigA adenylation domains from several pathogenic bacteria, some featuring bound ligands, including substrate, product, and/or various inhibitors (Fig. 6A) (8, 12, 19, 27; also unpublished data of C. Pinko, A. Borchardt, V. Nikulin, and Y. Su). Most structures show an “open” orientation of domain Ia relative to the NTase domain, with two well-separated (>20 Å) ligand-binding sites at the domain Ia surface and the NTase active site. In contrast, the binding of NAD+ substrate directs the large-scale reorientation of domain Ia about a hinge region to enclose NAD+ within the active site, now in contact with both ligand-binding sites, and defines a “closed” orientation (8).
Fig 6.
Structural consideration of the three classes of resistance mutation. (A) Overlaid LigA adenylation domain structures showing aligned open and closed orientations of domain Ia (gold) relative to the NTase (blue). The overlay comprises three Enterococcus faecalis structures with bound ligands, including two in open conformation (Protein Data Bank accesion number 1TA8 with bound NMN, 3AB with bound NMN, and pyridochromanone compound 1) and one in closed conformation (1TAE, bound NAD+), and one S. aureus structure in the open conformation (3JSL apoenzyme, white strand) (8, 12; also unpublished data of C. Pinko et al.). Side chains are shown for the adenylation domain resistance loci Arg61 (yellow) and Ala303 (magenta) and for the catalytic amino acid Lys112 (cyan). Ligands: NMN bound to domain Ia in open conformation (indigo); compound 1 bound to active site in open conformation (red); and NAD+ bound to active site in closed conformation (green). Arrow, pivot point at conformational hinge for domain Ia reorientation. (B) Modular structure of E. coli LigA bound to nicked adenylated DNA (2OWO) (20) with resistance loci indicated for the OB fold and HhH domains. Left, complete structure; center, OB fold with proximate DNA base pairs; right, HhH domain with proximate DNA base pairs. In OB fold domain (green), resistance loci are shown with side chains (indigo); in HhH domain (yellow), resistance loci are shown with side chains or, for glycine residues, marked by asterisks (red). All numbering refers to the S. aureus LigA sequence.
The Arg61Ile mutation alters a well-conserved residue near this hinge region (Fig. 6A). This proximity suggests that the mutation might elicit the observed changes in LigA kinetics by favoring the closed orientation upon NAD+ binding, such as from the loss of interactions seen in the open orientation between Arg61 and NTase surface moieties, including the backbone carbonyl of Ile152. An accelerated LigA closing would increase the LigA adenylation rate as observed, and a disfavored return to the open conformation could also cause the observed retardation of DNA-dependent steps, such as DNA ligation and/or release. The latter could also increase active-site occupancy by the adenylate product and, thus, confer reduced susceptibility to pyridochromanones and other competitive inhibitors.
The Ala303Asp mutation, in contrast, alters a well-conserved residue underlying the catalytic region, abutting the residues Leu82 and Val281 that contact NAD+ substrate (Fig. 6A) (8). This mutation was observed to lower the catalytic rates of LigA adenylation and DNA ligation without lowering the catalytic efficiency of ligation, suggesting that the alteration diminishes active-site function without affecting the initial NAD+ binding. Additionally, the proximity of Ala303 to bound pyridochromanone compound 1 (5.3 Å) (unpublished data of C. Pinko, et al. [doi:10.2210/pdb3bac/pdb.]; 3BA8, 3AB9, 3BAA, 3BAB, and 3BAC) suggests that mutation could directly alter the inhibitor-binding interface to generate the observed high-level pyridochromanone resistance.
The high-frequency lesions within the DNA-binding OB fold domain define a third mechanistic class of resistance mutation, as the test mutations Ala349Val and Ala373Thr do not alter the kinetics of LigA adenylation yet still cause significant decreases in Km and kcat values for DNA ligation. These results highlight the importance of the DNA-binding domains in coordinating the adenylyl transfer steps with the DNA-dependent steps of ligation.
The reported crystal structure of E. coli LigA bound to nicked adenylated DNA shows the OB fold and HhH domains in numerous contacts with the enveloped DNA duplex (Fig. 6B) (20). The OB fold domain contacts DNA at several of the residues identified as resistance loci (numbering from S. aureus): Arg326 forms a hydrogen bond with the DNA backbone, Ala349 and Ala373 directly abut the DNA backbone, His352 extends into the DNA minor groove, and Pro332 and Leu351 are adjacent to contact sites and probably influence the structure of the binding surface. The HhH domain also forms extensive contacts with bound DNA via four hexapeptide loop-helix motifs, each including an internal glycine that initiates the helix and creates an oxyanion hole that accepts a backbone phosphate. The resistance loci Gly481 and Gly585 define two such loop-helix glycines, and Ala549 resides within a loop-helix helix and probably also contributes to DNA binding.
The proximity of these lesions to DNA contact sites suggests that they reduce the catalytic rates for DNA ligation by lowering the affinity for DNA. We propose that these lowered catalytic rates can entirely explain the observed decreases in Km values and, most significantly, the high-level resistance to inhibition. First, by stalling the DNA ligation reaction, these lesions could stabilize the adenylated LigA intermediate and thus lower substrate Km values as observed. Second, this increased active-site occupancy would also necessarily confer resistance to pyridochromanone compounds and other competitive inhibitors. In this view, mutations in the distal domains can elicit high-level resistance as a secondary consequence of altered reaction kinetics.
The ligA gene is essential in every bacterial species examined to date, including S. aureus (13, 31). In this study, however, we recovered a series of highly pyridochromanone-resistant ligA mutants of S. aureus with no obvious growth defects despite dramatic deficits in DNA ligation kinetics. These results indicate that a significant portion of LigA function is dispensable for viability and that its modular composition facilitates ready diminishment by distal mutations that confer inhibitor resistance while leaving sufficient residual activity for viability. These results challenge the validity of LigA as a suitable target for antibacterial therapy and emphasize the need for subtler considerations of its biological requirement in staphylococci.
Classical genetic studies had previously indicated that E. coli can tolerate significant losses of LigA activity, as mutants with the conditional lethal mutation lig(Ts7) and the viable mutation lig-4 show 10- to 20-fold reductions in Okazaki fragment joining even under permissive growth conditions (10, 16). A complementation study in Mycobacteria has also suggested that significant LigA depletion has little effect on growth and viability (14). Our resistance study in S. aureus has confirmed that LigA has a large margin for variation with which to escape inhibitor action. The apparent excess of LigA activity in wild-type cells might be due to its multiple roles: low concentrations are sufficient for essential replicative functions, and yet higher concentrations are available for repair functions.
The difficulty in leveraging target enzymes into viable antibacterial therapies has been much discussed recently (21, 22, 24). Further study will be required to determine the true utility of LigA as a target for standard inhibitors or whether rare inhibitors might be required that kill via a gain-of-function “poison” mechanism analogous to the DNA breakage induced by quinolones (6). In summation, our study highlights the importance of a more thorough analysis of LigA target validity for antibacterial drug discovery.
ACKNOWLEDGMENTS
We acknowledge Akihiro Hashimoto, Godwin Pais, and Barton Bradbury for synthetic chemistry efforts and Christy Thoma and Jijun Cheng for technical support.
Footnotes
Published ahead of print 14 May 2012
REFERENCES
- 1. Brötz-Oesterhelt H, et al. 2003. Specific and potent inhibition of NAD+-dependent DNA ligase by pyridochromanones. J. Biol. Chem. 278:39435–39442 [DOI] [PubMed] [Google Scholar]
- 2. Chen XC, et al. 2002. Development of a fluorescence resonance energy transfer assay for measuring the activity of Streptococcus pneumoniae DNA ligase, an enzyme essential for DNA replication, repair, and recombination. Anal. Biochem. 309:232–240 [DOI] [PubMed] [Google Scholar]
- 3. Cheng Y, Prusoff WH. 1973. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22:3099–3108 [DOI] [PubMed] [Google Scholar]
- 4. Ciarrocchi G, MacPhee DG, Deady LW, Tilley L. 1999. Specific inhibition of the eubacterial DNA ligase by arylamino compounds. Antimicrob. Agents Chemother. 43:2766–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed, M07-A8, vol 29. CLSI, Wayne, PA [Google Scholar]
- 6. Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dwivedi N, et al. 2008. NAD+-dependent DNA ligase: a novel target waiting for the right inhibitor. Med. Res. Rev. 28:545–568 [DOI] [PubMed] [Google Scholar]
- 8. Gajiwala KS, Pinko C. 2004. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure 12:1449–1459 [DOI] [PubMed] [Google Scholar]
- 9. Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. 2004. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 279:20594–20606 [DOI] [PubMed] [Google Scholar]
- 10. Gottesman MM, Hicks ML, Gellert M. 1973. Genetics and function of DNA ligase in Escherichia coli. J. Mol. Biol. 77:531–547 [DOI] [PubMed] [Google Scholar]
- 11. Gul S, et al. 2004. Staphylococcus aureus DNA ligase: characterization of its kinetics of catalysis and development of a high-throughput screening compatible chemiluminescent hybridization protection assay. Biochem. J. 383:551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han S, Chang JS, Griffor M. 2009. Structure of the adenylation domain of NAD+-dependent DNA ligase from Staphylococcus aureus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaczmarek FS, et al. 2001. Cloning and functional characterization of an NAD+-Dependent DNA ligase from Staphylococcus aureus. J. Bacteriol. 183:3016–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Korycka-Machala M, et al. 2007. Evaluation of NAD+-dependent DNA ligase of mycobacteria as a potential target for antibiotics. Antimicrob. Agents Chemother. 51:2888–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lavesa-Curto M, et al. 2004. Characterization of a temperature-sensitive DNA ligase from Escherichia coli. Microbiology 150:4171–4180 [DOI] [PubMed] [Google Scholar]
- 16. Lehman IR. 1974. DNA ligase: structure, mechanism, and function. Science 186:790–797 [DOI] [PubMed] [Google Scholar]
- 17. Meier TI, et al. 2008. Identification and characterization of an inhibitor specific to bacterial NAD+-dependent DNA ligases. FEBS J. 275:5258–5271 [DOI] [PubMed] [Google Scholar]
- 18. Miesel L, et al. 2007. A high-throughput assay for the adenylation reaction of bacterial DNA ligase. Anal. Biochem. 366:9–17 [DOI] [PubMed] [Google Scholar]
- 19. Mills SD, et al. 2011. Novel bacterial NAD+-dependent DNA ligase inhibitors with broad-spectrum activity and antibacterial efficacy in vivo. Antimicrob. Agents Chemother. 55:1088–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nandakumar J, Nair PA, Shuman S. 2007. Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol. Cell 26:257–271 [DOI] [PubMed] [Google Scholar]
- 21. Parsons JB, Rock CO. 2011. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opin. Microbiol. 14:544–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40 [DOI] [PubMed] [Google Scholar]
- 23. Shuman S. 2009. DNA ligases: progress and prospects. J. Biol. Chem. 284:17365–17369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silver LL. 2011. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24:71–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sriskanda V, Schwer B, Ho CK, Shuman S. 1999. Mutational analysis of Escherichia coli DNA ligase identifies amino acids required for nick-ligation in vitro and for in vivo complementation of the growth of yeast cells deleted for CDC9 and LIG4. Nucleic Acids Res. 27:3953–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sriskanda V, Shuman S. 2002. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+. J. Biol. Chem. 277:9695–9700 [DOI] [PubMed] [Google Scholar]
- 27. Srivastava SK, et al. 2005. Mycobacterium tuberculosis NAD+-dependent DNA ligase is selectively inhibited by glycosylamines compared with human DNA ligase I. Nucleic Acids Res. 33:7090–7101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Srivastava SK, Tripathi RT, Ramachandran R. 2005. NAD+-dependent DNA ligase (Rv0314c) from Mycobacterium tuberculosis: crystal structure of the adenylylation domain and identification of novel inhibitors. J. Biol. Chem. 280:30273–30281 [DOI] [PubMed] [Google Scholar]
- 29. Srivastava SK, et al. 2007. NAD+-dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis: novel structure-function relationship and identification of a specific inhibitor. Proteins 69:97–111 [DOI] [PubMed] [Google Scholar]
- 30. Stokes SS, et al. 2011. Discovery of bacterial NAD+-dependent DNA ligase inhibitors: optimization of antibacterial activity. Bioorg. Med. Chem. Lett. 21:4556–4560 [DOI] [PubMed] [Google Scholar]
- 31. Streker K, et al. 2008. In vitro and in vivo validation of ligA and tarI as essential targets in Staphylococcus aureus. Antimicrob. Agents Chemother. 52:4470–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. 2006. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 106:687–699 [DOI] [PubMed] [Google Scholar]
- 33. Wang LK, Nair PA, Shuman S. 2008. Structure-guided mutational analysis of the OB, HhH, and BRCT domains of Escherichia coli DNA ligase. J. Biol. Chem. 283:23343–23352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang LK, Zhu H, Shuman S. 2009. Structure-guided mutational analysis of the nucleotidyltransferase domain of Escherichia coli DNA ligase (LigA). J. Biol. Chem. 284:8486–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu H, Shuman S. 2005. Structure-guided mutational analysis of the nucleotidyltransferase domain of Escherichia coli NAD+-dependent DNA ligase (LigA). J. Biol. Chem. 280:12137–12144 [DOI] [PubMed] [Google Scholar]