Abstract
A major limitation in the identification of novel antichlamydial compounds is the paucity of effective methods for large-scale compound screening. The immunofluorescence assay is the preferred approach for accurate quantification of the intracellular growth of Chlamydia. In this study, an immunofluorescence image-based method (termed image-based automated chlamydial identification and enumeration [iBAChIE]) was customized for fully automated quantification of Chlamydia infection using the freely available open-source image analysis software program CellProfiler and the complementary data exploration software program CellProfiler Analyst. The method yielded enumeration of different species and strains of Chlamydia highly comparably to the conventional manual methods while drastically reducing the analysis time. The inhibitory capability of established antichlamydial activity was also evaluated. Overall, these data support that iBAChIE is a highly effective tool for automated quantification of Chlamydia infection and assessment of antichlamydial activities of molecules. Furthermore, iBAChIE is expected to be amenable to high-throughput screening studies for inhibitory compounds and fluorescently labeled molecules to study host-pathogen interactions.
INTRODUCTION
Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infection (STI) in the United States, for which novel preventative and therapeutic compounds are needed (3). The vast majority (up to 75%) of C. trachomatis infections in women are asymptomatic (19), and if left untreated, these infections can lead to serious complications, including pelvic inflammatory disease (PID), ectopic pregnancy, and infertility (5, 6, 15). Moreover, genital C. trachomatis infection has been shown to associate with cervical cancer development and to contribute significantly to HIV transmission (8, 22). While current antibiotics are effective, resistance to the clinically preferred antibiotic (i.e., doxycycline) has already been observed in a Chlamydia species, albeit a nonhuman isolate (C. suis) (13; A. A. Andersen and D. G. Rogers, presented at the 9th International Symposium on Human Chlamydial Infection, International Chlamydia Symposium, San Francisco, CA, 1998). Prevention strategies are essential given the high rate of asymptomatic C. trachomatis infections; however, a vaccine or vaginal microbicide effective in preventing Chlamydia infection is currently unavailable for clinical use.
A major limitation in the development of antichlamydial compounds is the lack of a well-established method for quantifying Chlamydia infection that is adaptable to a high-throughput screening format. The paucity of high-throughput screening methods is heavily influenced by the obligate intracellular growth characteristics and the inability for axenic cultivation of Chlamydia. As a result, the direct immunofluorescent assay (IFA) is predominately relied on for accurate detection of Chlamydia infection. The established procedure depends on visual inspection of the intracellular vacuole formed by Chlamydia, termed an inclusion, typically within a relatively low number of fields (e.g., 3 to 10). The limitations of manual enumeration are evident; the process is labor intensive, field selection is prone to subjectivity, and results are based on a relatively restricted data set.
Freely available open-source computational image analysis software, CellProfiler (http://www.cellprofiler.org), has been shown to be an effective tool for quantifying visual information in a variety of biological images, particularly in large-scale imaging experiments (2). The complement software, CellProfiler Analyst, performs analysis of image-derived quantitative information as defined within a very large collection of image-feature measurements (i.e., a cytological profile) produced by CellProfiler. CellProfiler Analyst contains a supervised machine-learning system that is intuitive for development of image analysis algorithms via visual perception using a user-friendly interface (9, 10).
In this study, image-based automated chlamydial identification and enumeration (iBAChIE) was developed using CellProfiler and CellProfiler Analyst. To validate iBAChIE, the commonly utilized laboratory strain (LGV-2) and the more clinically relevant strain (serovar D) of C. trachomatis were analyzed. The accuracy and expediency of the automated method were compared to those of the conventional visual inspection in a 96-well format. For evaluating the efficacy and potential of iBAChIE to measure antichlamydial compounds, C. trachomatis infections were measured following treatment with four anti-Chlamydia molecules (tetracycline, polymyxin B, hydroxyethyl cellulose, and anti-C. trachomatis polyclonal antibody) that have contrasting mechanisms of inhibitory activity toward C. trachomatis (4, 7, 11, 12, 20, 23, 26).
MATERIALS AND METHODS
Bacteria and cell culture.
C. trachomatis lymphogranuloma venereum (LGV) serovar L2/434/Bu elementary bodies (EBs) were purified from infected L929 cells using a 30% Renografin density gradient (21) and stored in sucrose phosphate glutamate (SPG) at −80°C until use. C. trachomatis serovar D/UW-3/Cx was isolated from infected HeLa 229 cells and C. muridarum strain Nigg was purified from infected L929 cells as described previously (21). L929 cells and HeLa 229 cells were routinely cultured in RPMI 1640 tissue culture medium (Mediatech, Inc., Manassas, VA) supplemented with 5% fetal bovine serum (FBS) (Thermo Fisher Scientific, Liverpool, NY) and 10 μg/ml gentamicin (MP Biomedicals, Santa Ana, CA) at 37°C in a humidified atmosphere of 5% CO2. Cells were seeded in 96-well plates (Bioexpress, Kaysville, UT) at a density of 7 × 104 cells/ml, 200 μl/well, and incubated overnight prior to infection. In order to minimize the edge effect (well-to-well variations in the number of cells), plated cells were incubated for 1 h at room temperature prior to incubation at 37°C in an atmosphere of 5% CO2, as previously described (14).
Microbicide and antibiotic preparation.
Polymyxin B sulfate (Enzo Life Science, New York, NY) and tetracycline hydrochloride (USB Corporation, Cleveland, OH) were obtained in powder form. Stock solution of polymyxin B sulfate was prepared in Hanks buffered salt solution (HBSS) (Mediatech, Inc., Holly Hill, FL) to a concentration of 10 mM. Tetracycline was dissolved in sterile water to a stock concentration of 1 mg/ml. Hydroxyethyl cellulose (HEC) (90 kDa; Sigma-Aldrich, St. Louis, MO) was prepared as a stock solution at a concentration of 12% (wt/vol) in water. The HEC gel (polymeric liquid) was adjusted to pH 4.5 using 10 M NaOH and stored at 4°C. At the time of the assay, a 12% HEC gel was diluted to a final concentration of 2.8% (wt/vol) in phosphate-buffered saline (PBS). The mixture was shaken for 1 h at 37°C to achieve a uniform solution, centrifuged for 1 min at 300 × g to remove any bubbles, adjusted to pH 7.0, and stored at 4°C until use.
EB dilution assay.
Purified C. trachomatis L2 EBs were serially diluted 2-fold in HBSS and transferred to ∼70% confluent L929 cell monolayers (90 μl/well) in a 96-well plate. Ninety microliters of HBSS alone was added to mock-infected control cells. Cells were incubated for 2 h at room temperature. Following the incubation period, chlamydial inocula were removed and cells were washed once with 200 μl of HBSS. Two hundred microliters of fresh medium (RPMI–5% fetal bovine serum–10 μg per ml gentamicin) was added to each well, and cells were incubated at 37°C in an atmosphere of 5% CO2 for 24 h.
Antichlamydial compound inhibitory assay.
C. trachomatis L2 EBs were diluted in HBSS. Polymyxin B and hydroxyethyl cellulose were serially diluted 2-fold in HBSS containing EBs to the concentration range of 1 mM to 15.625 μM and 2.8% to 0.175%, respectively. Compound-EB mixtures were incubated for 60 min at room temperature. Compound-treated EB inocula were mixed using a multichannel pipette (LabNet Excel, Edison, NJ) at the start and end of the incubation period to ensure uniformity of the inocula. The mixtures containing the compound and EBs were then transferred to ∼70% confluent L929 cell monolayers in a 96-well plate and incubated at room temperature for 2 h. Following the incubation period, the inocula were removed and cells were washed once with 200 μl of HBSS. Two hundred microliters of fresh medium (RPMI–5% fetal bovine serum–10 μg per ml gentamicin) was added to each well. For tetracycline treatment, fresh medium containing an appropriate concentration (0.1 to 0.7 μg/ml) of tetracycline was added following 2-h inoculation period. Cells were incubated at 37°C in an atmosphere of 5% CO2 for 24 h.
C. trachomatis serovar D and C. muridarum infection.
Purified C. trachomatis serovar D or C. muridarum was serially diluted 4-fold in HBSS and transferred to ∼70% confluent HeLa 229 or L929 cell monolayers (90 μl/well) in a 96-well plate, respectively. Ninety μl of HBSS alone was added to mock-infected control cells. The plate was centrifuged at 900 × g for 1 h at room temperature (21). Following the incubation period, chlamydial inocula were removed and cells were washed once with 200 μl of HBSS. Two hundred microliters of fresh medium (RPMI–5% fetal bovine serum–10 μg per ml gentamicin) was added to each well, and cells were incubated at 37°C in an atmosphere of 5% CO2 for 48 h for serovar D and for 24 h for C. muridarum before they were analyzed.
Neutralization assay.
Guinea pig polyclonal serum against Chlamydia L2 EBs was purchased from Abcam (Cambridge, MA). Rabbit polyclonal anti-RpoA antibody was affinity purified using the AminoLink Plus immobilization kit (Thermo Scientific, Rockford, IL). The protein concentration of purified antibody was determined to be 1.01 mg/ml by Bradford assay (Bio-Rad Laboratories, Hercules, CA). Sera and purified antibody were serially diluted 5-fold in HBSS. Prior to infection, purified C. trachomatis L2 EBs were added to each serum or purified antibody sample and incubated for 30 min at 37°C to allow interaction between EBs and the antibody to take place (24). HBSS alone was used as a negative control. Following the incubation period, 90 μl of each sample containing antibody-EB mixture was added to a monolayer of L929 cells in a 96-well plate and incubated at room temperature for 2 h. After the incubation period, inocula were removed and cells were washed once with HBSS. Two hundred microliters of fresh medium (RPMI–5% fetal bovine serum–10 μg per ml gentamicin) was added to each well, and cells were incubated at 37°C for 24 h.
Immunofluorescence assay (IFA).
At 24 or 48 h postinfection (hpi) for serovar L2 or D, respectively, infected cells were fixed with 100% methanol (200 μl/well) for 10 min at room temperature and washed once with PBS. Cells were stained using the MicroTrack C. trachomatis culture confirmation test (Syva Co., Palo Alto, CA) with dilution to 1:40 in PBS (50 μl/well) for 60 min in the dark, followed by a 5-min staining with 1 μM 4′,6-diamidino-2-phenylindole (DAPI) in PBS (50 μl/well). DAPI was removed, and 1 ml of PBS was added. Plates were stored in the dark at 4°C until imaging.
Image acquisition.
Images were automatically captured with a BD Pathway BioImager 855 microscope (Becton, Dickinson and Company, Franklin Lakes, NJ) with a 20×, 0.4-numerical-aperture objective, fully equipped for multicolor capture, and an Orca ER camera (Hamamatsu Photonics, Bridgewater, NJ). The system was controlled by Attovision collection software (Becton, Dickinson and Company, Franklin Lakes, NJ), with automated infrared and image autofocus capture for 15 fields per well for a 96-well plate. Multiple plate capture was enabled by a Twister II robotic arm (Caliper Lifesciences Inc., Hopkinton, MA), which was integrated with the Attovision software.
Image analysis using CellProfiler and CellProfiler Analyst.
Images acquired by automated microscopy were loaded into the open-source software program CellProfiler (Broad Institute, Cambridge, MA), followed by segmentation of images and identification of objects. The process had four main steps: segmentation of nuclei, identification of whole cells, segmentation of chlamydial inclusions, and tabulation of measurements (see Fig. S1 in the supplemental material). The first task, performed by CellProfiler, was identification of the objects. Using CellProfiler modules, the nuclei, referred to as primary objects within CellProfiler, were identified first from the DNA-stained images. Once the nuclei had been identified, the subsequent module used the whole-cell-stained images and previously identified nuclei to identify the cell boundaries (whole cell), which were referred to as secondary objects. The area of the identified nuclei was subtracted from the whole cells to identify the cytoplasm, referred to as tertiary objects. For identification of chlamydial inclusions, CellProfiler expanded each area of nuclei by 15 pixels and used this expanded region to search for inclusions. Inclusions were then identified and segmented. The second task performed by CellProfiler was generating a cytological profile of each cell. CellProfiler measured a large number of cellular and subcellular features once all objects (nuclei, cells, cytoplasm, and identified chlamydial inclusions) had been identified. These quantitative measurements consist of apparent cellular and subcellular features, as well as an extensive amount of less-evident details, such as intensity of the stain, size, shape, and pixel correlation between objects. Once these measurements were taken, the cytological profile was created for each cell to be utilized by CellProfiler Analyst (Broad Institute, Cambridge, MA) and exported to a spreadsheet. Both nuclei and chlamydial inclusions were related to the identified cells so that each inclusion was assigned to only one nucleus. For enumeration of inclusion-forming units (IFU), the last module within the pipeline (ExportToDatabase) was modified to allow for classification of individual inclusion rather than cells by CellProfiler. To enumerate infected cells, the cytological profiles were loaded into CellProfiler Analyst. Random images, each containing a single cell, were presented to initiate the training process. Approximately 20 images from the positive (infected) and negative (uninfected) controls were manually classified by dragging and dropping the images into the appropriate bins within the CellProfiler Analyst interface (see Fig. S2a in the supplemental material). Based on the cytological profiles of these classified cells, CellProfiler Analyst identified the parameters necessary to accurately distinguish between uninfected and infected cells by generating a set of 50 rules (i.e., parameters). The software then presented a new set of cells that had been classified as positive or negative based on the rules generated. Any cells classified inaccurately were manually sorted into the appropriate bin to continue the training process. To assess the accuracy of the training, images from selected wells were opened and individual cells were automatically marked as uninfected or infected by CellProfiler Analyst based on the established rules (see Fig. S2b in the supplemental material). Any cell that had been classified incorrectly was manually reassigned to the appropriate bin. After the correction, a set of refined rules was generated and used to score every cell in every image. These steps were repeated until the system was able to accurately classify greater than 95% of cells into the correct category. Upon completion of scoring, a spreadsheet containing the numbers of positive and negative cells for each well was generated and exported.
RESULTS
Evaluation of iBAChIE in a 96-well format and comparison to manual enumeration.
To evaluate the suitability of iBAChIE for large-scale application, image acquisition and quantification of Chlamydia infection were performed in a 96-well-plate format. High sensitivity, reproducibility, and precision of the method were demonstrated by quantification of infection levels among host cells infected with 2-fold serial dilutions of C. trachomatis EBs (see Fig. S3 in the supplemental material). In order to assess the accuracy and efficiency of iBAChIE relative to the conventional manual inspection, manual enumeration was performed on three fields randomly selected from the immunofluorescence images acquired for the automated method (see Table S1 in the supplemental material). Comparison of the number of host cells and inclusion-positive cells, as well as the infection level reported by each method, indicated that the automated method is capable of performing highly reproducible analysis among a large set of samples with high sensitivity (a dynamic range of infections) and precision (low standard deviations).
Validation of iBAChIE with known chemical inhibitors.
The ability of iBAChIE to accurately assess the inhibitory activities of compounds against Chlamydia infection was demonstrated using three previously known antichlamydial compounds that inhibit different stages of the developmental cycle: tetracycline, hydroxyethyl cellulose (HEC), and polymyxin B (PMB). Tetracycline is a broad-spectrum antibiotic, for which Chlamydia is known to be exceptionally sensitive. Tetracycline interferes with the metabolic process of Chlamydia by targeting translational machinery and inhibiting protein biosynthesis (4). Complete inhibition of inclusion formation by tetracycline at a concentration of 0.51 μg/ml has been previously reported (26). Infection of L929 cells with C. trachomatis EBs along with compound treatment was performed in a 96-well plate and analyzed 24 hpi following automated image acquisition. iBAChIE revealed the antichlamydial effect of tetracycline in a dose-dependent manner with complete inhibition of inclusion formation at 0.5 μg/ml, an inhibitory concentration consistent with the previous report (26) (Fig. 1a).
Fig 1.
Inhibitory properties of known inhibitors for C. trachomatis serovar L2 infection. Inhibitory activities of three known antichlamydial compounds, tetracycline, 90-kDa HEC, or PMB, were determined using iBAChIE. Percent inhibition was calculated relative to the wells without treatment at 24 hpi. (a) Tetracycline treatment. L929 cells were infected with C. trachomatis, and 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, or 0.7 μg/ml of tetracycline was added along with growth medium. Error bars indicate standard deviations of the means for triplicate wells. (b) Ninety-kilodalton-HEC treatment. L929 cells were infected with C. trachomatis preincubated with decreasing concentrations of 5.6 to 0.175% of HEC for an hour prior to infection. Error bars represent the standard deviations of the means for triplicate wells. (c) PMB treatment. L929 cells were infected with C. trachomatis preincubated with decreasing concentrations of 1,000 to 15.625 μM PMB for an hour prior to infection. Error bars represent the standard deviations of the means for triplicate wells.
HEC is a common polysaccharide excipient used in vaginally delivered microbicides for STIs. Although excipients are typically used as delivery vehicles and are expected to be pharmacologically inactive, a 90-kDa HEC has been shown to have an inhibitory effect on C. trachomatis infection, purportedly as a competitive inhibitor for adhesion (20). When EBs were treated with HEC, the inhibitory effect of HEC was directly proportional to its concentration in the range of 0.175% to 0.7% and reached complete inhibition at 1.4% (Fig. 1B). This proportionality between the concentration and the inhibitory effect supports a previously suggested mechanism for HEC inhibition, that HEC may act as a competitive inhibitor by competing with the host cell for the recognition site on the chlamydial surface (20).
PMB is an antibiotic that typically acts by disrupting the outer membrane of Gram-negative bacteria through interaction with the lipid A portion of lipopolysaccharide (17, 27). PMB is active against nearly all species of Gram-negative bacteria, including C. trachomatis, and has been used as a positive inhibition control for antichlamydial activity in numerous studies (11, 12, 23). In contrast to HEC treatment, when EBs were treated with PMB, the concentration of the inhibitor did not show a proportional effect on inclusion formation, suggesting a mode of inhibition different from that of HEC. In addition, complete inhibition of inclusion formation was not achieved even at the highest concentration of PMB, reaching maximum inhibition of approximately 70% at 500 μM. Interestingly, previous studies show dose-dependent antichlamydial activity by PMB that plateaued without reaching complete inhibition (7, 16).
Efficacy of iBAChIE for measuring neutralizing antibodies.
Given the current lack of an effective vaccine against Chlamydia, identifying and evaluating neutralizing antibodies would be extremely advantageous. iBAChIE was used to examine inhibitory properties of polyclonal sera for chlamydial EBs. Affinity-purified polyclonal antibody against C. trachomatis RpoA (bacterial cytosol RNA polymerase α subunit) was used as a negative control. L929 cell monolayers were inoculated with C. trachomatis EBs that had been preincubated with anti-Chlamydia EB sera or affinity-purified polyclonal anti-RpoA antibody, and infection levels were analyzed 24 hpi. At the highest dose (1:100 dilution), anti-Chlamydia sera inhibited chlamydial infection by 98.7% relative to the infection level of the control wells, where EBs were not pretreated with antibody (Fig. 2). Anti-RpoA antibody treatment showed insignificant neutralization activity, demonstrating that the neutralization activity of anti-Chlamydia sera was not due to a nonspecific reaction. When the amount of anti-Chlamydia sera used was reduced by 5-fold (1:500 dilution), the inhibitory effect was reduced to 44.1%. No inhibitory effect was observed at the lowest concentration (1:2,500). Together, these results indicate that this automated method may be used to accurately identify neutralizing antibody and protective concentrations for Chlamydia infection in cell culture.
Fig 2.
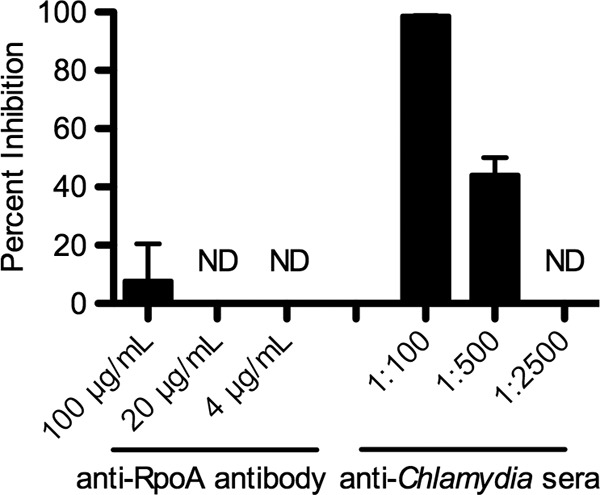
Inhibitory properties of anti-Chlamydia sera for infectivity of C. trachomatis L2 EBs. The ability of anti-Chlamydia sera to neutralize the infectivity of C. trachomatis L2 EBs was evaluated using iBAChIE. L929 cells were infected with C. trachomatis EBs preincubated with various concentrations of anti-RpoA antibody or anti-Chlamydia sera for 30 min prior to infection. Percent inhibition was calculated relative to the wells without treatment following 24 h of incubation. ND, no inhibition was detected.
Assessment of iBAChIE with C. trachomatis (serovar D) and the mouse Chlamydia species C. muridarum.
Due to the relative ease of cultivation and shorter developmental cycle, LGV serovar L2 is a commonly used C. trachomatis prototype strain in chlamydial research. However, trachoma serovars D, E, and F are the most prevalent urogenital strains of C. trachomatis worldwide and therefore of more clinical relevance (18). C. muridarum is a natural pathogen of mice and is commonly used as a model for infection of the human genital tract caused by C. trachomatis.
HeLa 229 cells or L929 cells were infected with 4-fold serial dilutions of serovar D or C. muridarum, respectively. HeLa 229 cells were used to demonstrate that various eukaryotic cells may be used as host cell lines for the automated enumeration. Due to the longer developmental cycle of serovar D than of L2, serovar D infections were quantified 48 hpi and C. muridarum was quantified 24 hpi. iBAChIE measured the infection level, ranging from 81.9% to 14.0% for serovar D and 95.7% to 4.8% for C. muridarum in a dose-dependent manner (Fig. 3). These results support that accurate enumeration of chlamydial infection by the automated method is highly adaptable to a wide range of serovars and species of Chlamydia despite the differences in the duration of developmental cycle, and eukaryotic host cell lines.
Fig 3.

Automated enumeration of C. trachomatis serovar D and C. muridarum infection. Percent infectivity for a 4-fold dilution series of C. trachomatis serovar D (a) or C. muridarum (b) was determined by the automated method. Infections were carried out in triplicate wells for each dilution. Percent infectivity was calculated based on the numbers of infected and uninfected cells identified by CellProfiler Analyst. Error bars indicate standard deviations of the means for triplicate wells.
DISCUSSION
When the two methods were contrasted, a major advantage iBAChIE exhibited over manual enumeration was the ability to rapidly analyze large data sets. Analysis of a 96-well plate by the automated method, accommodating enumeration of 1,440 images consisting of approximately 130,000 cells on average per plate, was completed within 2 h following automated image acquisition. During these 2 h, the average hands-on time was less than 30 min, with the rest spent on computer processing. Notably, analysis of multiple 96-well plates did not require additional time compared to a single plate analysis. This is due to the fact that regardless of the number of samples, only one training session is required to establish a differential rule set that can be applied across the entire experiment. Considering that manual enumeration required up to 2 min per field of view, equivalent analysis would take up to 48 h by the manual method. Thus, the automated method achieved substantial time savings in sample analysis.
iBAChIE is equally effective in analysis of small-scale (up to hundreds of images) or large-scale screens containing thousands of images. While analysis of hundreds of images by CellProfiler can be completed on a single computer in a few hours, the experiments described in this study consisted of thousands of images, and therefore more computational processing capabilities were needed to achieve rapid analysis (25). This challenge was addressed by utilizing a dedicated “cluster” of processors (16 quad-core 2.5 GHz processors) for image processing. With addition of an extra module to the end of a pipeline, CellProfiler automatically divided images into small batches and created the files containing batches of images, which were then submitted as individual jobs to be processed separately by the cluster, significantly minimizing the time required for data processing (1).
In summary, an automated method (iBAChIE) utilizing CellProfiler and CellProfiler Analyst has been developed and validated for rapid and accurate analysis of Chlamydia infection. The ability of iBAChIE for evaluation of various inhibitory molecules, along with simultaneous and rapid analysis of large-scale experiments (i.e., multiple 96-well plates), supports that this method could readily be applied to high-throughput screening of small-molecule libraries for antichlamydial agent and vaccine development. In addition to the potential screening capabilities, iBAChIE offers a high-throughput tool for a wide variety of chlamydial research that currently relies on manual analysis of fluorescence microscopy images by using fluorescently labeled chlamydial proteins or host subcellular markers. The combined use of CellProfiler and CellProfiler Analyst may find applications in the research of microorganisms where no well-established high-throughput quantification is currently available and offer a powerful tool in a wide range of studies, including drug discovery.
Supplementary Material
ACKNOWLEDGMENTS
We are extremely grateful for the technical efforts of Dru Walstrom, Wesley Mason, Charles Henry, and the University of Kansas Information and Telecommunication Technology Center. We acknowledge Taryn Cansler for her early contributions to this project. We also thank Lindsay Sammons and Vinidhra Sridharan for critical review of the manuscript.
This study was funded largely by an NIH Microbicide Innovation Program IV grant (no. AI082697).
Footnotes
Published ahead of print 21 May 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Carpenter AE. 2009. Extracting rich information from images. Methods Mol. Biol. 486:193–211 [DOI] [PubMed] [Google Scholar]
- 2. Carpenter AE, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 doi:10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. CDC 2011. Chlamydia—CDC fact sheet. CDC, Atlanta, GA [Google Scholar]
- 4. Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232–260; second page, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chow JM, et al. 1990. The association between Chlamydia trachomatis and ectopic pregnancy. A matched-pair, case-control study. JAMA 263:3164–3167 [PubMed] [Google Scholar]
- 6. Cohen CR, Brunham RC. 1999. Pathogenesis of Chlamydia induced pelvic inflammatory disease. Sex. Transm. Infect. 75:21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fadel S, Eley A. 2008. Is lipopolysaccharide a factor in infectivity of Chlamydia trachomatis? J. Med. Microbiol. 57:261–266 [DOI] [PubMed] [Google Scholar]
- 8. Fleming DT, Wasserheit JN. 1999. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex. Transm. Infect. 75:3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones TR, et al. 2009. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc. Natl. Acad. Sci. U. S. A. 106:1826–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones TR, et al. 2008. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9:482 doi:10.1186/1471-2105-9-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lampe MF, Ballweber LM, Isaacs CE, Patton DL, Stamm WE. 1998. Killing of Chlamydia trachomatis by novel antimicrobial lipids adapted from compounds in human breast milk. Antimicrob. Agents Chemother. 42:1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lampe MF, Ballweber LM, Stamm WE. 1998. Susceptibility of Chlamydia trachomatis to chlorhexidine gluconate gel. Antimicrob. Agents Chemother. 42:1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lenart J, Andersen AA, Rockey DD. 2001. Growth and development of tetracycline-resistant Chlamydia suis. Antimicrob. Agents Chemother. 45:2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lundholt BK, Scudder KM, Pagliaro L. 2003. A simple technique for reducing edge effect in cell-based assays. J. Biomol. Screen. 8:566–570 [DOI] [PubMed] [Google Scholar]
- 15. Mardh PA. 2004. Tubal factor infertility, with special regard to chlamydial salpingitis. Curr. Opin. Infect. Dis. 17:49–52 [DOI] [PubMed] [Google Scholar]
- 16. Matsumoto A, Higashi N, Tamura A. 1973. Electron microscope observations on the effects of polymixin B sulfate on cell walls of Chlamydia psittaci. J. Bacteriol. 113:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morrison DC, Jacobs DM. 1976. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13:813–818 [DOI] [PubMed] [Google Scholar]
- 18. Papadogeorgakis H, et al. 2010. Chlamydia trachomatis serovar distribution and Neisseria gonorrhoeae coinfection in male patients with urethritis in Greece. J. Clin. Microbiol. 48:2231–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rahm VA, Gnarpe H, Odlind V. 1988. Chlamydia trachomatis among sexually active teenage girls. Lack of correlation between chlamydial infection, history of the patient and clinical signs of infection. Br. J. Obstet. Gynaecol. 95:916–919 [DOI] [PubMed] [Google Scholar]
- 20. Sater AAA, Ojcius DM, Meyer MP. 2008. Susceptibility of Chlamydia trachomatis to the excipient hydroxyethyl cellulose: pH and concentration dependence of antimicrobial activity. Antimicrob. Agents Chemother. 52:2660–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scidmore MA. 2005. Cultivation and laboratory maintenance of Chlamydia trachomatis. Curr. Protoc. Microbiol. Chapter 11:Unit 11A.1 [DOI] [PubMed] [Google Scholar]
- 22. Simonetti AC, Melo JH, de Souza PR, Bruneska D, de Lima Filho JL. 2009. Immunological's host profile for HPV and Chlamydia trachomatis, a cervical cancer cofactor. Microbes Infect. 11:435–442 [DOI] [PubMed] [Google Scholar]
- 23. Skinner MC, Stamm WE, Lampe ML. 2009. Chlamydia trachomatis laboratory strains versus recent clinical isolates: implications for routine microbicide testing. Antimicrob. Agents Chemother. 53:1482–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soriani M, et al. 2010. Exploiting antigenic diversity for vaccine design: the chlamydia ArtJ paradigm. J. Biol. Chem. 285:30126–30138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vokes MS, Carpenter AE. 2008. Using CellProfiler for automatic identification and measurement of biological objects in images. Curr. Protoc. Mol. Biol. Chapter 14:Unit 14.17 [DOI] [PubMed] [Google Scholar]
- 26. Walsh M, Kappus EW, Quinn TC. 1987. In vitro evaluation of CP-62,993, erythromycin, clindamycin, and tetracycline against Chlamydia trachomatis. Antimicrob. Agents Chemother. 31:811–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60:1206–1215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



