Abstract
The primary binding sites for Bacillus stearothermophilus proteins B-L5 and B-L22 and the Escherichia coli proteins E-L5, E-L18 and E-L25 on B. stearothermophilus 5S RNA were determined by limited ribonuclease digestion of the corresponding 5S RNA-protein complexes. The results obtained in this study are in agreement with our previous experiments in which the binding sites of E. coli and B. stearothermophilus proteins were determined for E. coli 5S RNA and lead to the conclusion that the proteins interact with the most conserved regions of 5S RNA. A comparison of the results obtained in this study with those of other published experiments suggest that the proposed interaction of nucleotides 16-21 with those of 58-63 is facilitated by protein binding to 5S RNA.
Full text
PDF








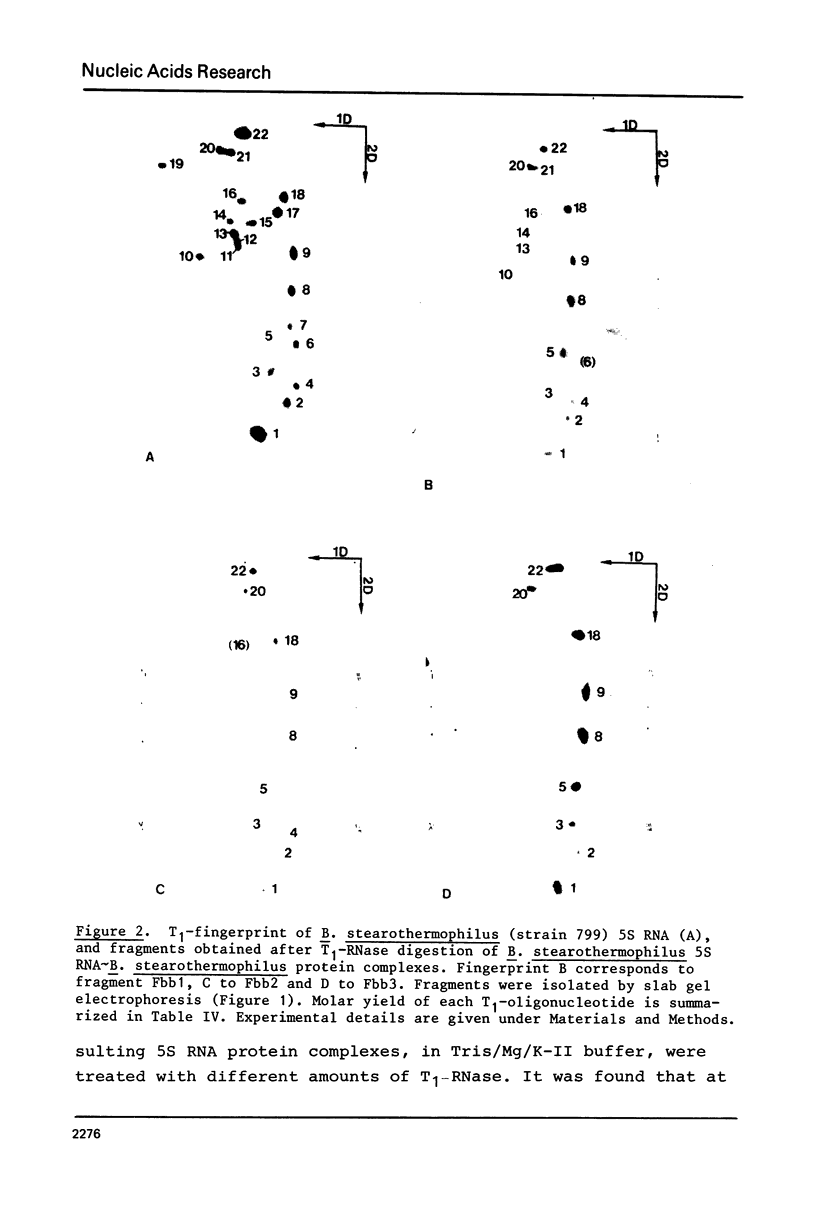
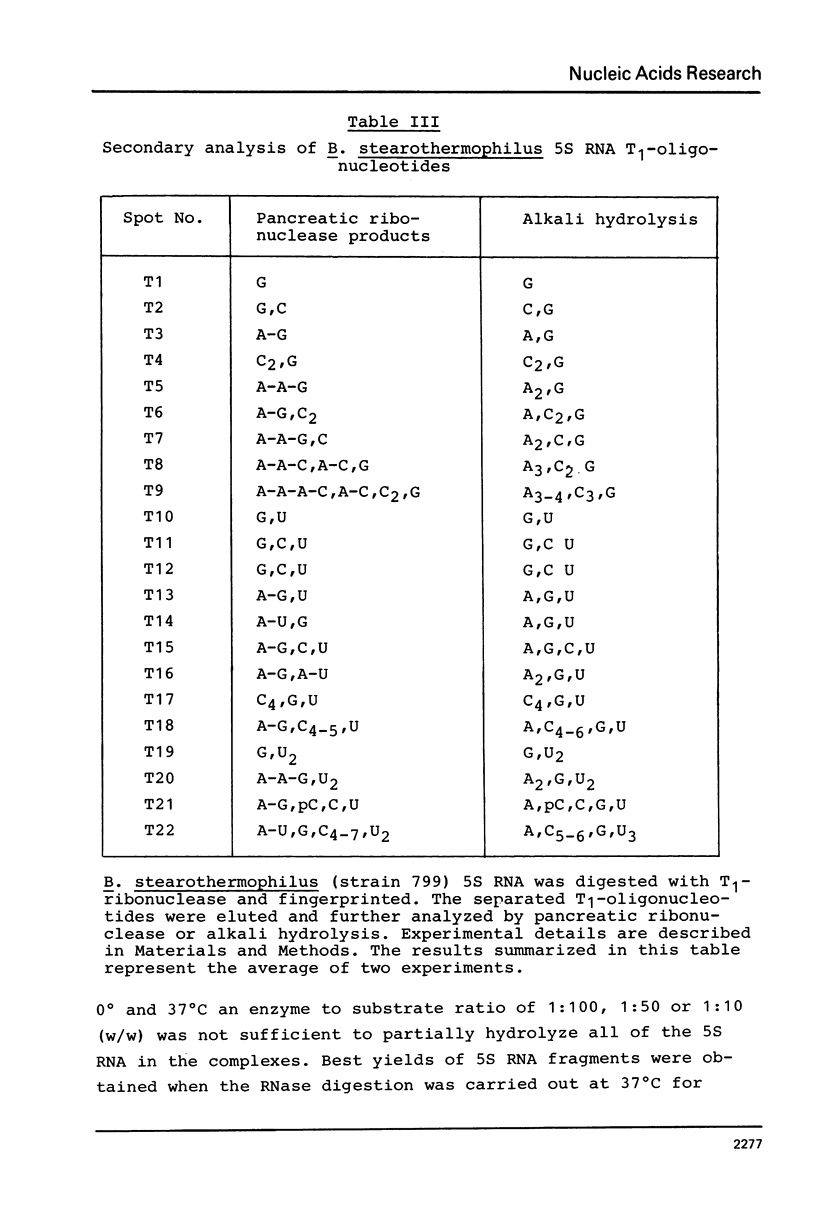

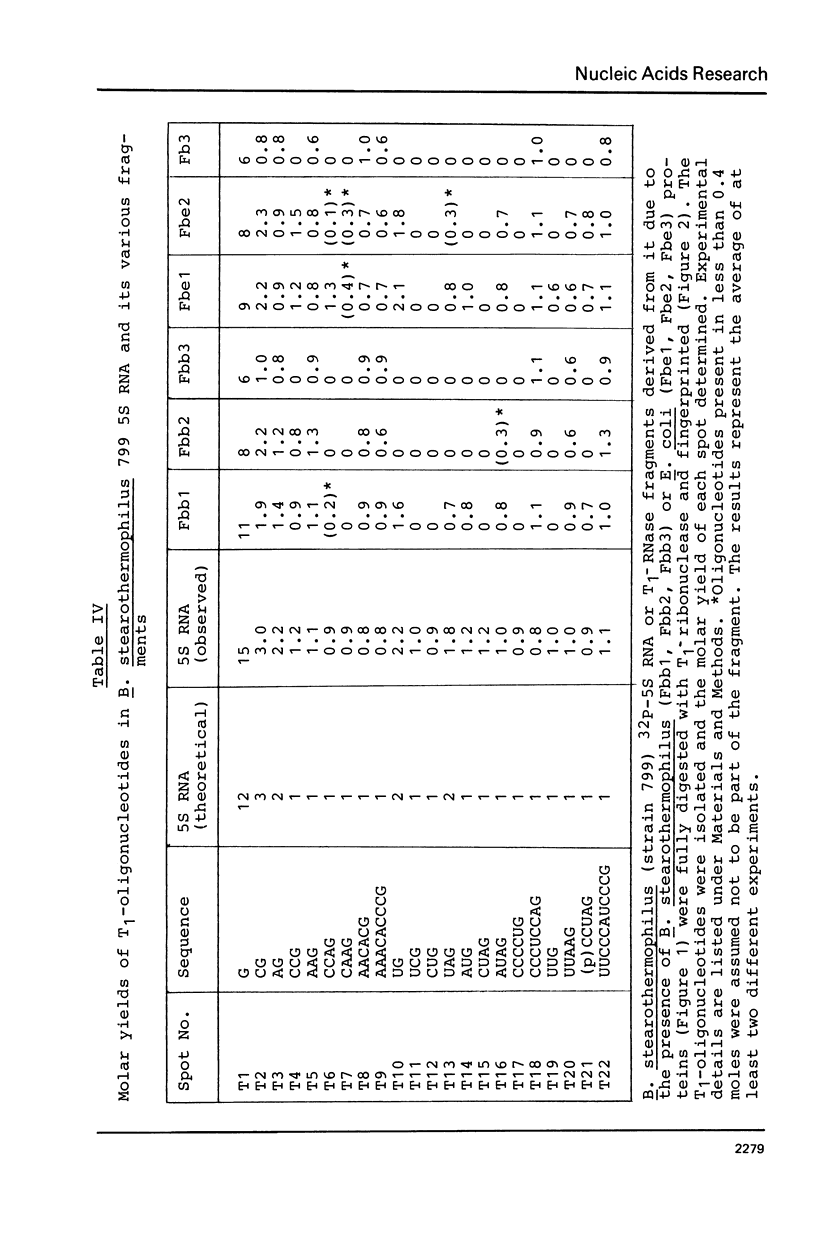

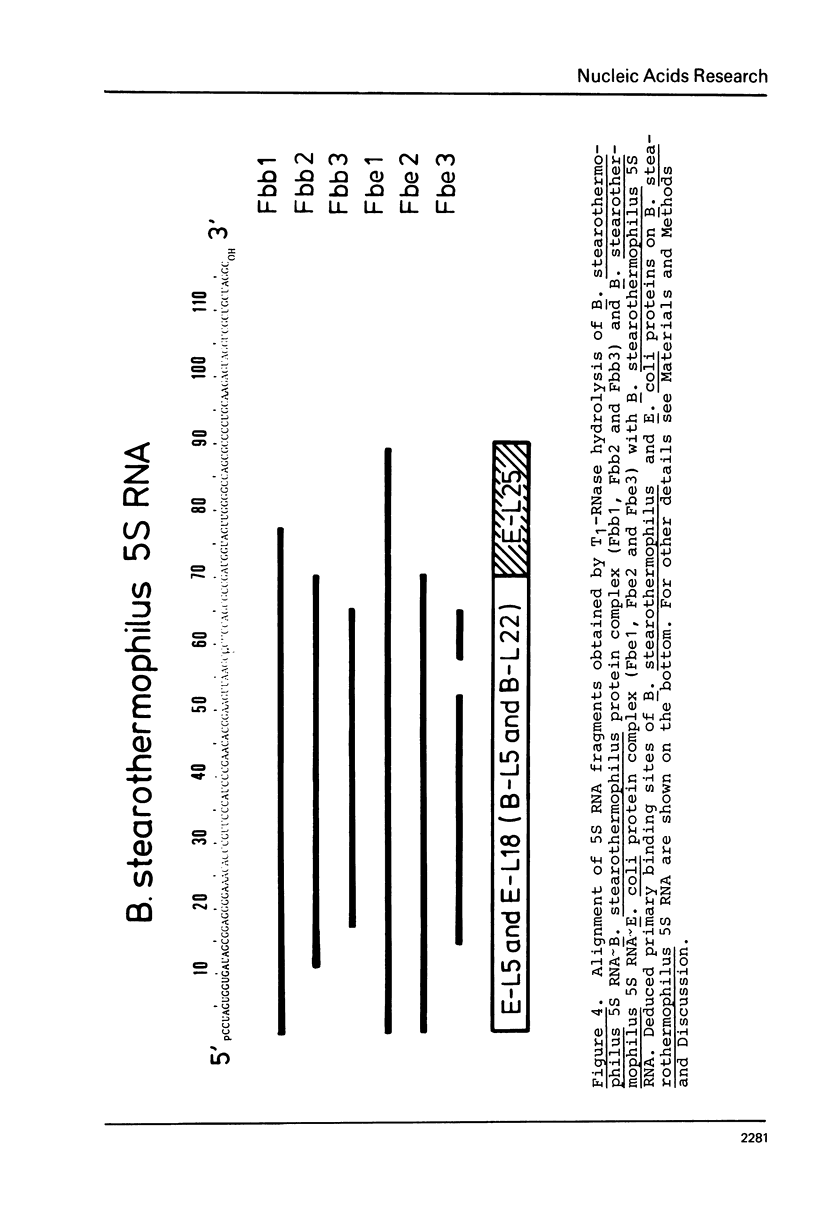



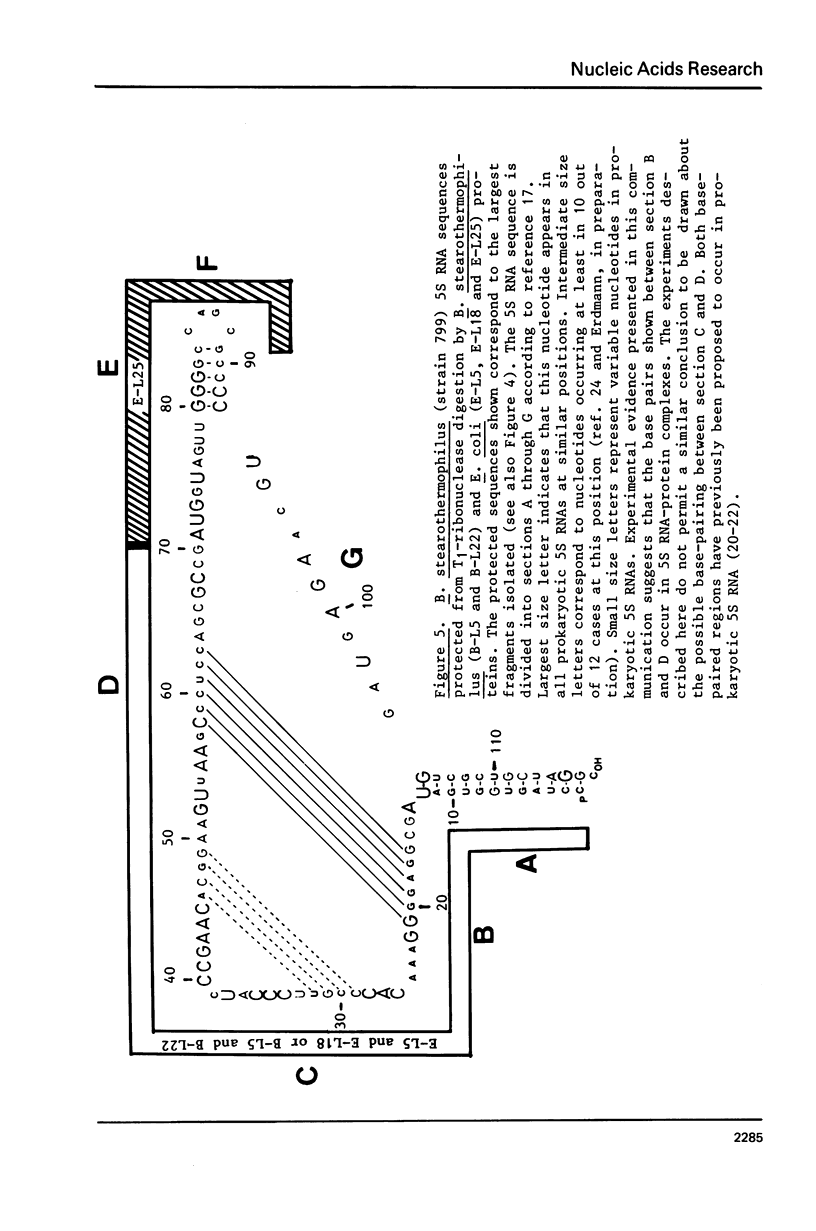



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brownlee G. G., Sanger F., Barrell B. G. The sequence of 5 s ribosomal ribonucleic acid. J Mol Biol. 1968 Jun 28;34(3):379–412. doi: 10.1016/0022-2836(68)90168-x. [DOI] [PubMed] [Google Scholar]
- Cronenberger J. H., Erdmann V. A. Stimulation of polypeptide polymerization by blocking of free sulphydryl groups in Escherichia coli ribosomal proteins. J Mol Biol. 1975 Jun 15;95(1):125–137. doi: 10.1016/0022-2836(75)90340-x. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1978 Jan;5(1):r1–r13. [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Doberer H. G. Structure and function of 5S RNA: the role of the 3' terminus in 5S RNA function. Mol Gen Genet. 1972;114(2):89–94. doi: 10.1007/BF00332779. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Fahnestock S., Higo K., Nomura M. Role of 5S RNA in the functions of 50S ribosomal subunits. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2932–2936. doi: 10.1073/pnas.68.12.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Sprinzl M., Pongs O. The involvement of 5S RNA in the binding of tRNA to ribosomes. Biochem Biophys Res Commun. 1973 Oct 1;54(3):942–948. doi: 10.1016/0006-291x(73)90785-7. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Fox J. W., Wong K. P. Changes in the conformation and stability of 5 S RNA upon the binding of ribosomal proteins. J Biol Chem. 1978 Jan 10;253(1):18–20. [PubMed] [Google Scholar]
- Gray P. N., Bellemare G., Monier R., Garrett R. A., Stöffler G. Identification of the nucleotide sequences involved in the interaction between Escherichia coli 5 RNA and specific 50 S subunit proteins. J Mol Biol. 1973 Jun 15;77(1):133–152. doi: 10.1016/0022-2836(73)90367-7. [DOI] [PubMed] [Google Scholar]
- Hindennach I., Kaltschmidt E., Wittmann H. G. Ribosomal proteins. Isolation of proteins from 50S ribosomal subunits of Escherichia coli. Eur J Biochem. 1971 Nov 11;23(1):12–16. doi: 10.1111/j.1432-1033.1971.tb01585.x. [DOI] [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Horne J. R., Erdmann V. A. Isolation and characterization of 5S RNA-protein complexes from Bacillus stearothermophilus and Escherichia coli ribosomes. Mol Gen Genet. 1972;119(4):337–344. doi: 10.1007/BF00272091. [DOI] [PubMed] [Google Scholar]
- Marotta C. A., Varricchio F., Smith I., Weissman S. M. The primary structure of Bacillus subtilis and Bacillus stearothermophilus 5 S ribonucleic acids. J Biol Chem. 1976 May 25;251(10):3122–3127. [PubMed] [Google Scholar]
- Nierhaus K. H., Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Structure and function of 5S ribosomal ribonucleic acid from Torulopsis utilis. II. Partial digestion with ribonucleases and derivation of the complete sequence. J Biochem. 1974 Nov;76(5):935–947. [PubMed] [Google Scholar]
- Nomura M., Erdmann V. A. Reconstitution of 50S ribosomal subunits from dissociated molecular components. Nature. 1970 Nov 21;228(5273):744–748. doi: 10.1038/228744a0. [DOI] [PubMed] [Google Scholar]
- Nomura M., Traub P., Bechmann H. Hybrid 30S ribosomal particles reconstituted from components of different bacterial origins. Nature. 1968 Aug 24;219(5156):793–799. doi: 10.1038/219793b0. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Traub P., Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci U S A. 1968 Mar;59(3):777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Erdmann V. A. Activities of B. stearothermophilus 50 S ribosomes reconstituted with prokaryotic and eukaryotic 5 S RNA. FEBS Lett. 1973 Jul 15;33(3):315–319. doi: 10.1016/0014-5793(73)80219-4. [DOI] [PubMed] [Google Scholar]
- Wrede P., Pongs O., Erdmann V. A. Binding oligonucleotides to Escherichia coli and Bacillus stearothermophilus 5 S RNA. J Mol Biol. 1978 Mar 25;120(1):83–96. doi: 10.1016/0022-2836(78)90296-6. [DOI] [PubMed] [Google Scholar]