Abstract
The structure of Mycobacterium tuberculosis peptidoglycan is atypical since it contains a majority of 3→3 cross-links synthesized by l,d-transpeptidases that replace 4→3 cross-links formed by the d,d-transpeptidase activity of classical penicillin-binding proteins. Carbapenems inactivate these l,d-transpeptidases, and meropenem combined with clavulanic acid is bactericidal against extensively drug-resistant M. tuberculosis. Here, we used mass spectrometry and stopped-flow fluorimetry to investigate the kinetics and mechanisms of inactivation of the prototypic M. tuberculosis l,d-transpeptidase LdtMt1 by carbapenems (meropenem, doripenem, imipenem, and ertapenem) and cephalosporins (cefotaxime, cephalothin, and ceftriaxone). Inactivation proceeded through noncovalent drug binding and acylation of the catalytic Cys of LdtMt1, which was eventually followed by hydrolysis of the resulting acylenzyme. Meropenem rapidly inhibited LdtMt1, with a binding rate constant of 0.08 μM−1 min−1. The enzyme was unable to recover from this initial binding step since the dissociation rate constant of the noncovalent complex was low (<0.1 min−1) in comparison to the acylation rate constant (3.1 min−1). The covalent adduct resulting from enzyme acylation was stable, with a hydrolysis rate constant of 1.0 × 10−3 min−1. Variations in the carbapenem side chains affected both the binding and acylation steps, ertapenem being the most efficient LdtMt1 inactivator. Cephalosporins also formed covalent adducts with LdtMt1, although the acylation reaction was 7- to 1,000-fold slower and led to elimination of one of the drug side chains. Comparison of kinetic constants for drug binding, acylation, and acylenzyme hydrolysis indicates that carbapenems and cephems can both be tailored to optimize peptidoglycan synthesis inhibition in M. tuberculosis.
INTRODUCTION
β-Lactams are usually not considered for treatment of tuberculosis (TB) since Mycobacterium tuberculosis produces a species-specific broad-spectrum class A β-lactamase (BlaC) (12). In spite of BlaC, carbapenems and the combination of amoxicillin and clavulanic acid have been reported to be bactericidal in vitro (4, 5, 7) and to reduce the burden of M. tuberculosis in the sputum of patients with pulmonary tuberculosis (4, 5). However, clinical assessments of the drugs for treatment of multidrug-resistant tuberculosis (MDR-TB) are limited to anecdotal cases involving combined therapy with second-line drugs (23, 24). The potential interest in β-lactams for the treatment of extensively drug-resistant tuberculosis (XDR-TB) has recently been renewed by detailed characterization of BlaC that showed that this β-lactamase is irreversibly inactivated by clavulanic acid and hydrolyzes carbapenems only at a low rate (12, 13). Combined with clavulanic acid, carbapenems are not only bactericidal against exponentially growing M. tuberculosis but are also active against nonreplicating forms of the bacilli (13). Furthermore, the combination was uniformly active against a collection of XDR strains (13).
The main target of meropenem in M. tuberculosis is unlikely to be the d,d-transpeptidase activity of classical penicillin-binding proteins (PBPs) since the peptidoglycan of this bacterium contains a high proportion (80%) of cross-links connecting residues at the third position of stem peptides (3→3 cross-links) (Fig. 1A) (15). These cross-links are formed by l,d-transpeptidases (Ldts) and replace the 4→3 cross-links synthesized by PBPs (Fig. 1B). Ldts and PBPs are structurally unrelated and contain active-site cysteine and serine residues, respectively (2, 19). The chromosome of M. tuberculosis strain H37Rv encodes five l,d-transpeptidases (Ldts) designated LdtMt1 to LdtMt5. Among these paralogues, LdtMt1 and LdtMt2 are both functional in an in vitro peptidoglycan cross-linking assay and inactivated by carbapenems (10, 15). LdtMt2 is essential for virulence in a mouse model of acute infection (10), whereas LdtMt1 is thought to play a critical role in peptidoglycan adaptation to the nonreplicative state of the bacilli (15).
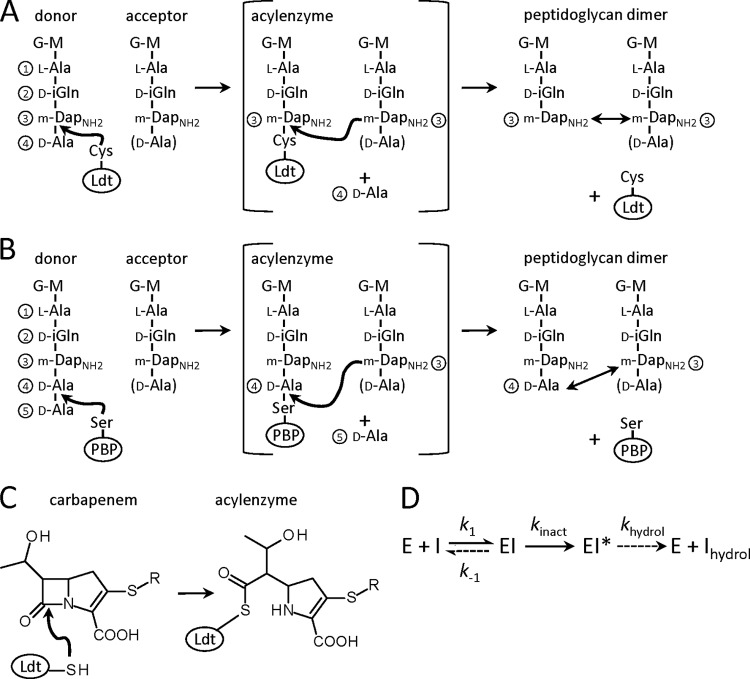
Fig 1.
Reaction catalyzed by peptidoglycan transpeptidases. (A) Formation of 3→3 cross-links by l,d-transpeptidases (Ldt). Cross-links are indicated by double arrows. G, N-acetylglucosamine; M, N-glycolyl-muramic acid; mDap, meso-diaminopimelic acid; mDapNH2, mDap with an amidated ε carboxyl group; D-iGln, d-iso-glutamine. (B) Formation of 4→3 peptidoglycan cross-links by classical d,d-transpeptidases belonging to the penicillin-binding protein (PBP) family. (C) Ldt acylation by carbapenems. (D) Full catalytic cycle. Binding of a β-lactam (I) to the enzyme (E) leads to formation of a noncovalent complex (EI). The chemical step of the reaction leads to formation of an acylenzyme (EI*), which is slowly hydrolyzed to produce free enzyme and hydrolyzed β-lactam (Ihydrol). Dashed arrows indicate slow reactions.
Mass spectrometry analyses have previously shown that carbapenems bind covalently to LdtMt1 and LdtMt2 (Fig. 1C) (10, 15), but the kinetics of this reaction have not been explored and the interaction of l,d-transpeptidases with drugs belonging to other β-lactam classes has not been investigated in detail. Here, we investigate the mechanism and kinetics of LdtMt1 inactivation by four carbapenems (meropenem, doripenem, imipenem, and ertapenem) and three cephalosporins (cefotaxime, cephalothin, and ceftriaxone) using a combination of mass spectrometry and stopped-flow fluorescence spectroscopy. We show that both classes of drugs form covalent adducts with LdtMt1, although enzyme acylation with cephalosporins is slower and leads to the elimination of one of their side chains. Comparisons of kinetic constants for drug binding, acylation, and hydrolysis of β-lactam–LdtMt1 adducts (Fig. 1D) indicate that the carbapenem and cephem rings can both be tailored for peptidoglycan synthesis inhibition in M. tuberculosis.
MATERIALS AND METHODS
Production and purification of LdtMt1.
We have previously reported the construction of a derivative of vector pET2818 (Ap) harboring the gene ldtMt1, previously designated Rv0116C (15). For this study, LdtMt1 production was improved by using a synthetic gene (GeneCust) with codon usage optimized for expression in Escherichia coli. The gene was cloned into vector pET28a (Km) (Novagen) to avoid contamination of LdtMt1 preparations by vector-encoded β-lactamase. E. coli BL21(DE3) was used for the production of recombinant LdtMt1 consisting of a polyhistidine tag followed by a tobacco etch virus (TEV) protease cleavage site (MHHHHHHENLYFQGHM) and residues 32 to 251 of the protein. Bacteria were grown at 37°C with vigorous shaking in 2 liters of brain heart infusion broth (Difco) containing kanamycin (50 μg/ml) to an optical density at 600 nm of 0.8. Isopropyl-β-d-thiogalactopyranoside was added (0.5 mM), and incubation was continued for 18 h at 16°C. LdtMt1 was purified from a clarified lysate by affinity chromatography on Ni2+-nitrilotriacetate-agarose resin (Qiagen GmbH) and by size exclusion chromatography (Superdex 75 HL26/60 column; Amersham Pharmacia Biotech) in 100 mM sodium-phosphate buffer (pH 6.4) containing 300 mM NaCl. LdtMt1 was concentrated by ultrafiltration (Amicon Ultra-4 centrifugal filter devices; Millipore) to a final concentration of 1.5 mg/ml and stored at −65°C in the same buffer.
Mass spectrometry analyses.
The formation of drug-enzyme adducts was tested by incubating LdtMt1 (20 μM) with β-lactams (100 μM) at 37°C in 100 mM sodium-phosphate buffer (pH 6.0) for 1 h. Experiments were also performed with a 2-fold excess of LdtMt1 (20 μM) over carbapenems (10 μM) in order to simultaneously observe the acylenzyme and the native enzyme in the same injection. Five microliters of acetonitrile and 1 μl of 1% formic acid were extemporaneously added, and the reaction mixture was injected directly into the mass spectrometer (Qstar Pulsar I; Applied Biosystem) at a flow rate of 0.05 ml/min (acetonitrile, 50%, water, 49.5%, and formic acid, 0.5%; per volume). Spectra were acquired in the positive mode as previously described (21).
Kinetics of LdtMt1 acylation by carbapenems.
Fluorescence kinetic data were acquired with a stopped-flow apparatus (RX-2000, Applied Biophysics) coupled to a spectrofluorimeter (Cary Eclipse; Varian) in 100 mM sodium phosphate (pH 6.0) at 10°C. Trp residues were excited at 224 nm with a slit of 10 nm and an optical path length of 2 mm. Fluorescence emission was determined at 335 nm with a slit of 10 nm and an optical path length of 10 mm. The detector voltage was set to 550 V. For kinetics simulations (26), variations in the concentrations of the three forms of the enzyme (free enzyme, E; noncovalent complex of enzyme and β-lactam inhibitor [I], EI; and acylenzyme, EI*) (Fig. 1D) over time were defined for free enzyme (d[E]/dt = k−1[EI] − k1[E][I], which is equal to d[I]/dt), for the noncovalent complex (d[EI]/dt = k1[E][I] − kinact[EI] − k−1[EI], where kinact is the rate constant of the acylation step), and for the acylenzyme (d[EI*]/dt = kinact[EI]). Using the initial conditions ([E] = [Etotal] and [I] = [Itotal] at time = 0), the concentrations of the three forms of the enzyme were iteratively computed for sequential time increments of 0.005 min using Excel software (Microsoft), as previously described (26) (Fig. 2A). Fluorescence intensity (F) was calculated using the relative fluorescence intensities (a, b, and c, as indicated in the following equation) of the three forms of the enzyme (F = a[E] + b[EI] + c[EI*]), and normalized fluorescence was expressed as F/a[Etotal] (Fig. 2B and C). In order to fit simulations with experimental data, three parallel plots with three β-lactam concentrations were generated. The catalytic constants (k1, k−1, and kinact) and the relative fluorescence intensity of EI were adjusted to simultaneously obtain the best superposition between simulations and experimental data in the three plots, as previously described (26).
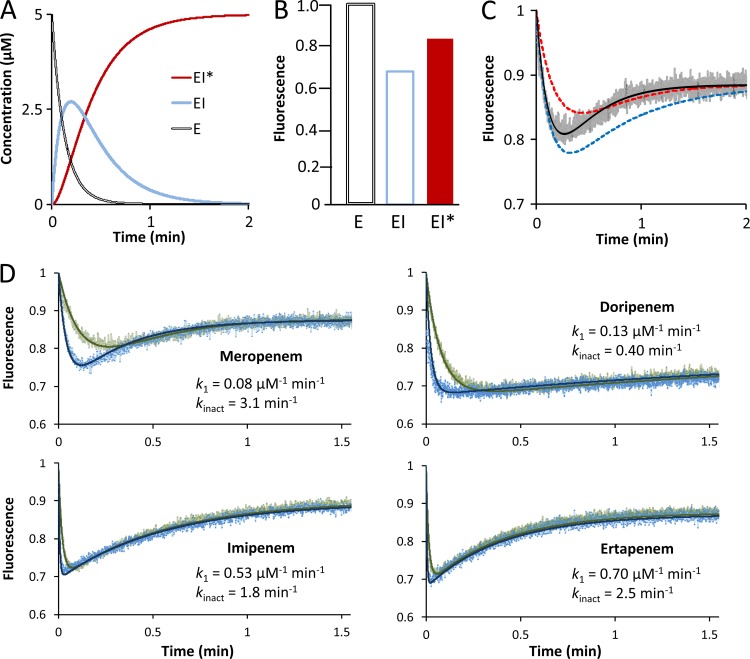
Fig 2.
Fluorescence kinetics of LdtMt1 inactivation by carbapenems. (A) Simulation of variations in the concentrations of the three enzyme forms. The initial concentrations of enzyme (E = Etotal at time zero) and inhibitor (I = Itotal at time zero) were 5 and 100 μM, respectively. The values 0.08 μM−1 min−1, 0.1 min−1, and 3.1 min−1 were attributed to catalytic constants k1, k−1, and kinact, respectively. (B) Relative fluorescence intensities of the three enzyme forms. (C) Simulation of fluorescence kinetics. The black curve was computed with the kinetic constants and fluorescence intensities used for the data shown in panels A and B. The red and blue curves show the impact of 2-fold decreases in k1, and kinact, respectively. The gray curve corresponds to experimental data. (D) Determination of kinetic constants for meropenem, doripenem, imipenem, and ertapenem. The values of k1 and kinact were determined by fitting simulations (solid lines) to experimental data for drug concentrations of 100 μM (green) and 300 μM (blue).
Kinetics of LdtMt1 acylation by cephems.
Fluorescence data were acquired as described above for carbapenems. Regression analyses of fluorescence kinetics were performed with the equation [EI* ] =[Etotal](1 − e−kobst), in which [Etotal] is the total enzyme concentration, kobs is the observed constant for acylation, and t is time (26). For determination of the kinact over Kapp (inhibitor concentration that leads to half-maximal acylation velocity) ratio, the values of kobs were plotted as a function of cephem concentration [I] and regression analysis was performed using the equation kobs = kinact[I]/Kapp + [I], in which kinact is the first-order constant for acylenzyme formation and Kapp a constant (21). In order to compare the efficacy of LdtMt1 inactivation by carbapenems and cephems, the kinact/Kapp ratio was deduced from simulations for the former drugs by using the experimentally determined catalytic constants k1, k−1, and kinact (see above).
Kinetics of acylenzyme hydrolysis.
To determine the hydrolysis rates of acylenzymes (khydrol) (Fig. 1D), β-lactams (100 μM) were incubated with increasing concentrations of LdtMt1 (0, 2.5, 5, and 10 μM) in sodium phosphate buffer (100 mM; pH 6.0). Rupture of the β-lactam ring was determined at 20°C by measuring the absorbance decrease at 265 nm for cephems and at 299 nm for carbapenems on a Cary 100 spectrophotometer (Cary 100 Bio; Varian). The hydrolysis velocities (V) were plotted as a function of LdtMt1 concentration, and enzyme turnover (khydrol) was deduced from the slope.
RESULTS
Inactivation of LdtMt1 by carbapenems.
LdtMt1 was incubated with meropenem, doripenem, imipenem, and ertapenem (Fig. 3A), and the formation of covalent adducts was determined by electrospray mass spectrometry (Fig. 3B). The masses of adducts were equal to the mass of the protein plus that of the antibiotic. Mass conservation upon acylation indicates that acylenzymes were generated by opening of the β-lactam ring and formation of a thioester bond (Fig. 1C) (21).
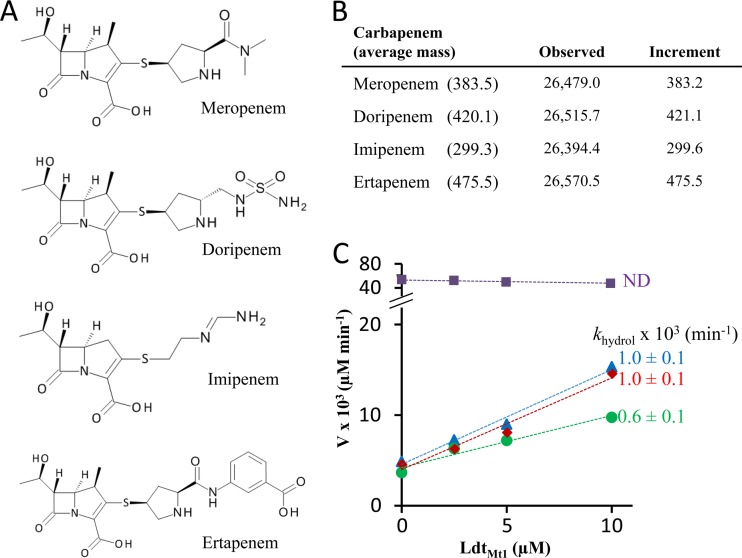
Fig 3.
LdtMt1 inactivation by carbapenems. (A) Structures of carbapenems. (B) Masses (atomic mass units) of covalent adducts resulting from acylation of LdtMt1 by carbapenems. Increments were obtained by subtracting the observed mass of the native enzyme from that of acylenzymes. (C) Determination of the rate constant for acylenzyme hydrolysis (khydrol) for meropenem (blue), doripenem (green), imipenem (purple), and ertapenem (red) (values are fit ± standard deviation [SD]). Rates of hydrolysis were deduced from the slopes. V, hydrolysis velocity; ND, not detected.
Since kinetic analyses of l,d-transpeptidase inactivation have not been performed for mycobacterial enzymes, we applied a recently developed fluorescence-based approach (26) to determine the catalytic constants of the reaction (see Materials and Methods). Briefly, concentration variations were simulated for the three enzyme forms (Fig. 2A) according to the reaction scheme depicted in Fig. 1D. Since free enzyme (E), the noncovalent complex (EI), and the acylenzyme display different fluorescence intensities (Fig. 2B), the fluorescence kinetics are biphasic (Fig. 2C). In the first phase, fluorescence intensity decreases because EI is rapidly formed to the detriment of free enzyme and the fluorescence intensity of EI is lower than that of E. In the second phase, fluorescence intensity increases since EI* is formed to the detriment of EI (acylation step) and the fluorescence intensity of EI* is greater than that of EI. As shown in Fig. 2C, the fluorescence kinetics is exquisitely sensitive to variations in k1 and kinact, the second- and first-order rate constants for formation of complex EI and enzyme acylation, respectively. Fitting simulations to experimental data therefore provided estimates of the catalytic constants (Fig. 2D).
For the four carbapenems, the kinetic constants were determined by simultaneously fitting simulations to experimental data at three drug concentrations, two of which are shown in Fig. 2D. The constant k−1 could be set to zero or to any arbitrary value smaller than 0.1 min−1. This indicates that k−1 is much smaller than kinact and, therefore, that the noncovalent complex is committed to acylenzyme formation, as the rate constant for EI dissociation is much lower than that of acylation. Acylenzyme hydrolysis was not taken into account in this analysis since the reaction occurs at a different time scale (3 versus 1,000 min; see below). As shown in Fig. 2D, ertapenem and then imipenem were the most efficient drugs for in vitro LdtMt1 inactivation (high k1 and kinact). The meropenem side chain moderately impaired drug binding (lower k1), whereas that of doripenem had an additional unfavorable impact on the acylation step (lower kinact). Together, these data indicate that all carbapenems inactivate LdtMt1 by acylation, although differences in antibiotic side chains modulate drug binding and acylation rates.
The stability of acylenzymes was assayed by incubating a fixed concentration of carbapenem with increasing concentrations of LdtMt1 for 1,000 min. The decreasing absorbance was used to determine hydrolysis velocities, which were plotted as a function of LdtMt1 concentration (Fig. 3C). The intercept at [LdtMt1] = 0 corresponds to the rate of spontaneous hydrolysis of the drug. The slope corresponds to the enzyme turnover number for the full catalytic cycle depicted in Fig. 1D. This number provides a direct estimate of the rate constant for acylenzyme hydrolysis (khydrol) since hydrolysis is the kinetically limiting step of the catalytic cycle (see above). Acylenzymes formed with meropenem, doripenem, and ertapenem were hydrolyzed at similar low rates (khydrol = 0.6 to 1.0 × 10−3 min−1). Hydrolysis of the imipenem-LdtMt1 adduct was not detected. Thus, LdtMt1 formed stable acylenzymes with all four carbapenems.
Mechanism of LdtMt1 inactivation by cephalosporins.
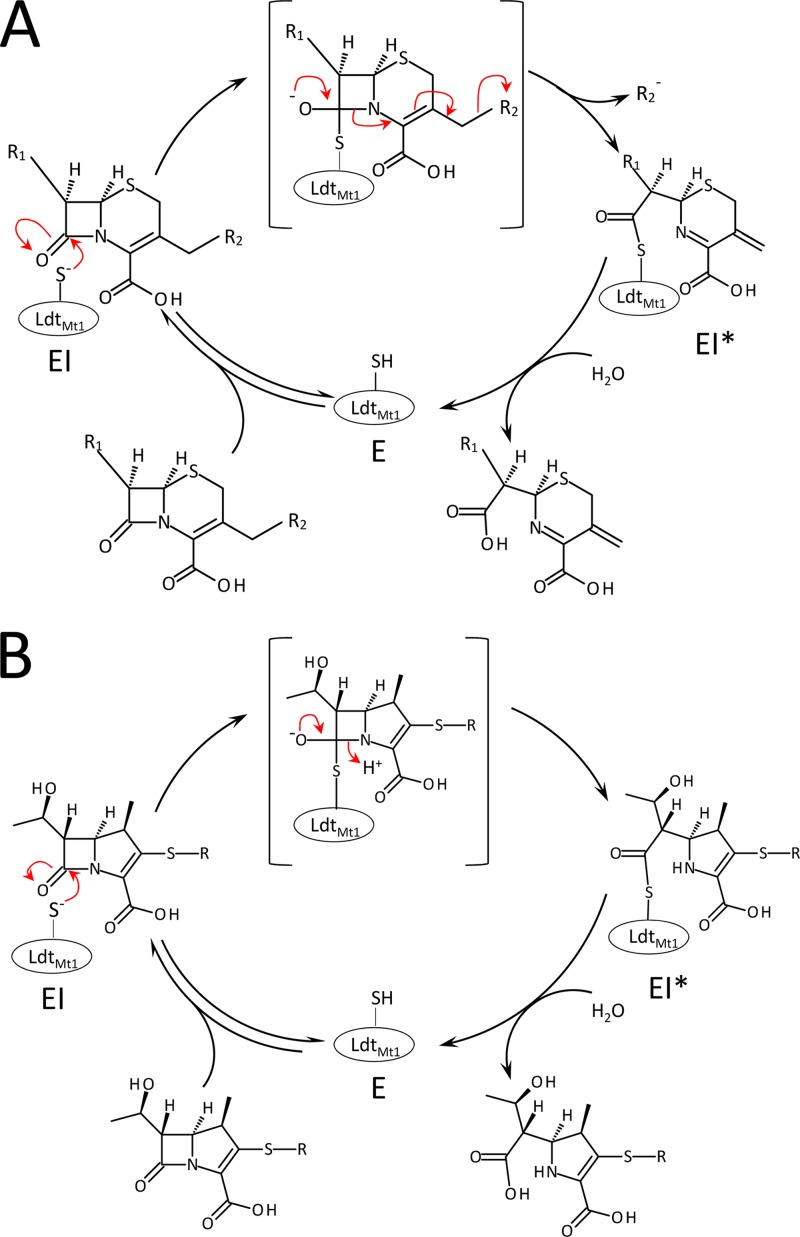
As found for carbapenems, LdtMt1 formed a covalent adduct with cefotaxime (Fig. 4A). However, the mass increase observed upon acylation was smaller than the mass of the antibiotic, indicating that a fragment of the drug was lost (Fig. 4B). In order to map the fragment lost upon acylation of LdtMt1 by cefotaxime, two additional cephalosporins, cephalothin and ceftriaxone (Fig. 4A), were tested. These two drugs were chosen to provide two pairs of compounds differing only in the R1 (cephalothin and cefotaxime) or R2 (cefotaxime and ceftriaxone) side chain. Incubation of LdtMt1 with ceftriaxone and cefotaxime led to adducts with the same mass (Fig. 4B), indicating that R2 was lost upon acylation. In agreement, adducts obtained with cephalothin and cefotaxime revealed loss of the acetate group of the R2 side chain, which is common to the two drugs. Based on these results, we propose that nucleophilic attack of the β-lactam carbonyl of cephalosporins by the active-site Cys residue of LdtMt1 is followed by the elimination of a portion of the R2 side chains, according to the reaction scheme depicted in Fig. 5A.
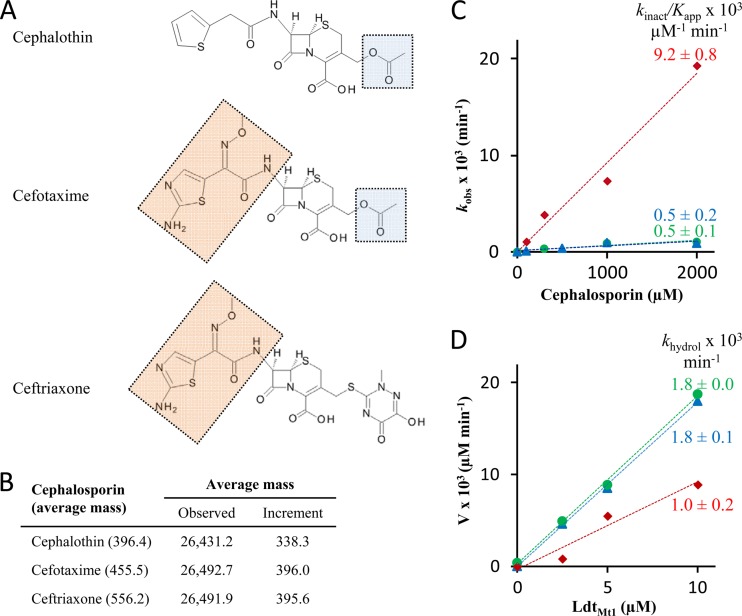
Fig 4.
LdtMt1 inactivation by cephalosporins. (A) Structure of cephalosporins with common R1 (wheat) and R2 (blue) side chains highlighted. (B) Average mass (atomic mass units) of acylenzymes. (C) Kinetics of LdtMt1 inactivation by cephalothin (blue), cefotaxime (green), and ceftriaxone (red) (values are fit ± SD). The slope provides an estimate of the ratio of kinact over Kapp. (D) Kinetics of acylenzyme hydrolysis. The slope provides an estimate of the rate constant khydrol for hydrolysis of acylenzymes formed with cephalothin (blue), cefotaxime (green), and ceftriaxone (red) (values are fit ± SD).
Fig 5.
Reactions of LdtMt1 with cephalosporins (A) and carbapenems (B).
Kinetics of LdtMt1 inactivation by cephalosporins and acylenzyme stability.
The kinetics of LdtMt1 inactivation by cephalosporins revealed monophasic fluorescence decreases that could be fitted to exponential decays (data not shown). The resulting rate constants (kobs) were determined for three drug concentrations. The rate constant kobs increased linearly with the drug concentration for the three cephalosporins (Fig. 4C). The slope (kinact /Kapp), which provides an estimate of the overall efficacy of LdtMt1 inactivation, was 18-fold higher for ceftriaxone than for the other two cephalosporins (Table 1). The acylenzymes were stable, with hydrolysis rates (khydrol) in the order of 1.0 × 10−3 min−1 (Fig. 4D).
Table 1.
Efficacy of LdtMt1 inactivation by β-lactams
| β-Lactam | Kinetic constant |
|
|---|---|---|
| kinact/Kapp × 103 μM−1 min−1 | khydrol [(fit ± SD) × 103 min−1] | |
| Carbapenems | ||
| Meropenem | 67 | 1.0 ± 0.1 |
| Doripenem | 160 | 0.6 ± 0.1 |
| Imipenem | 400 | NDa |
| Ertapenem | 500 | 1.0 ± 0.1 |
| Cephems | ||
| Cephalothin | 0.5 | 1.8 ± 0.1 |
| Cefotaxime | 0.5 | 1.8 ± 0.0 |
| Ceftriaxone | 9.2 | 1.0 ± 0.2 |
ND, not detected.
DISCUSSION
Recent increases in the incidence of XDR-TB reinforce the critical need for new drugs (29). l,d-Transpeptidases are promising targets for the development of novel antitubercular β-lactams since the essential cross-linking step of peptidoglycan synthesis is predominantly catalyzed by these enzymes in M. tuberculosis (20). Inhibitors of these targets in general and of β-lactams in particular have many assets. l,d-Transpeptidases polymerize peptidoglycan in the periplasm and therefore are not protected by the permeability barrier of the cytoplasmic membrane (22, 27). In addition, intensive drug development in the past 70 years has proven that the β-lactam ring is highly versatile for obtaining safe antibiotics with various antibacterial spectra and pharmacokinetic properties. To date, β-lactam research has not yet been applied to the development of antitubercular drugs since M. tuberculosis is intrinsically resistant to this family of compounds due to the production of the broad-spectrum β-lactamase BlaC. This bottleneck has recently been overcome by the discovery that clavulanic acid acts as an irreversible inhibitor of BlaC (12, 13). Thus, the development of l,d-transpeptidase inhibitors belonging to the β-lactam family is a novel and promising approach to obtain drugs for XDR-TB treatment.
In spite of the potential of β-lactams for the development of novel treatments for XDR-TB, the interaction of the drugs with M. tuberculosis l,d-transpeptidases has previously been assessed only qualitatively (10, 15). Here, we use stopped-flow fluorescence spectrophotometry to explore the inactivation kinetics of LdtMt1, the prototypic enzyme of M. tuberculosis (15). We show that meropenem, known to be uniformly bactericidal against XDR strains (13), rapidly binds to LdtMt1 with a second-order rate constant (k1) of 0.08 μM−1 min−1 (Fig. 2D). The enzyme cannot recover from this initial binding step since the dissociation rate constant of the noncovalent complex is low (k−1 < 0.1 min−1) in comparison to the acylation rate constant (kinact = 3.1 min−1) (Fig. 2D). In addition, the hydrolysis rate constant of the acylenzyme (khydrol = 1.0 × 10−3 min−1) is low, indicating that only 6% of the activity can potentially be recovered per hour in the total absence of free drug (Fig. 3C). The side chain of carbapenems affected the velocity of both drug binding (k1) and acylation (kinact), ertapenem being the most efficient drug with respect to in vitro LdtMt1 inactivation (Fig. 2). This indicates that the side chain of carbapenems is amenable to modifications in order to optimize interaction with the target, minimize hydrolysis by β-lactamase BlaC, and improve pharmacokinetic properties. Drug optimization should also take into account the collateral damage to resident flora and risks of selection of resistance in other pathogens, in particular in members of the family Enterobacteriaceae (3, 6). Of note, mycobacteria (15, 16, 22) and Clostridium difficile (25) are exceptions among human pathogens in having a large proportion of their peptidoglycan being cross-linked by l,d-transpeptidases. Thus, β-lactam optimization may lead to narrow-spectrum drugs with minimal interaction with classical d,d-transpeptidases.
To date, carbapenems were thought to be the only β-lactam class able to inactivate l,d-transpeptidases (10, 15, 16, 21). Analysis of LdtMt1 revealed that members of this enzyme family may also be inactivated by cephems (Fig. 4). The drugs inactivated LdtMt1 7- to 1,000-fold less efficiently than carbapenems (Table 1). Carbapenems and cephems also differed in the fact that the latter antibiotics lost a portion of their R2 side chains upon acylation (Fig. 4B), according to the reaction scheme depicted in Fig. 5A. Acylenzymes formed with carbapenems contained the entire drug molecules (Fig. 3B) since their side chains did not contain an appropriately located leaving group (Fig. 5B).
The elimination reaction observed with cephems may be a consequence of β-lactam ring rupture upon acylation since the reaction has also been reported for cephalosporin hydrolysis by β-lactamases (11), as well as in alkaline conditions (1). The efficiency of the acylation reaction by three cephems was compared based on determination of the kinact over Kapp ratio (Fig. 4 and Table 1). Replacement of the R2 side chain of cefotaxime by that of ceftriaxone led to an 18-fold increase in the velocity of the acylation reaction. In contrast, the nature of the R1 side chain had limited impact on the velocity of the acylation reaction and on acylenzyme stability. Together, these results indicate that the R2 side chain of cephems can be tailored to optimize l,d-transpeptidase inactivation and peptidoglycan synthesis inhibition in M. tuberculosis.
The recent report of in vitro bactericidal activity of meropenem against extensively drug-resistant M. tuberculosis (13) has stimulated two in vivo studies, which have specifically assessed the efficacy of carbapenems in combination with clavulanic acid (9, 28). In a chronic mouse model, significant reductions of bacterial burden in the lungs and spleens were obtained with meropenem, alone or in combination with clavulanic acid, despite a short half-life for meropenem (9). In combination with clavulanic acid, all carbapenems (imipenem, meropenem, doripenem, and ertapenem) significantly reduced the bacterial burden in M. tuberculosis-infected macrophages, with imipenem and meropenem having the largest effect (9). In a preventive mouse model, imipenem, meropenem, and ertapenem were inactive in reducing pulmonary lesions or splenomegaly (28). Bacterial counts were only reduced by the imipenem-clavulanate combination, and mortality was only reduced by imipenem and meropenem in combination with clavulanic acid. Ertapenem was inactive, and this correlated with a 4-fold higher MIC in comparison to those of imipenem and meropenem. In contrast, we show here that LdtMt1 is inactivated by ertapenem more rapidly than by imipenem and meropenem. Multiple factors may account for this discrepancy. Both in vivo studies (9, 28) concluded that the shortness of the half-lives of carbapenems in mice is a major limitation to evaluation of drug efficacy. In addition, the targets of carbapenems are likely to include a combination of l,d-transpeptidases and, perhaps, of additional d,d-transpeptidases or d,d-carboxypeptidases (14), with partially redundant functions (15, 18, 22). All l,d-transpeptidases that have been tested to date were found to be inactivated by carbapenems and to form stable covalent adducts with the drugs (8, 15, 17, 21, 26). However, the side chains of the drugs modulate the kinetic constants for the inactivation reaction (8), and these effects may vary among l,d-transpeptidases. As previously discussed for the classical targets of β-lactams (30), the existence of multiple l,d-transpeptidases in M. tuberculosis (10, 15) should be considered an asset since this limits the possibility for the emergence of resistance by target modification. The design of specific transpeptidase inhibitors for tuberculosis treatment will require the identification of the combinations of enzymes that are essential for the replication or survival of the bacilli in various environments.
ACKNOWLEDGMENTS
The research leading to these results has received funding from the European Union's Seventh Framework Program (FP7/2007-2013) under grant agreement no. 261378. V.D. was supported by a Poste d'Accueil, Institut National de la Santé et de la Recherche Médicale.
Footnotes
Published ahead of print 21 May 2012
REFERENCES
- 1. Akaho E, Nakayama H. 2003. An innovative classification of, and a new structure-activity-relationship approach to degradation kinetics of cephalosporins: an attempt to enhance the therapeutic activity. J. Antibiot. (Tokyo) 56:379–391 [DOI] [PubMed] [Google Scholar]
- 2. Biarrotte-Sorin S, et al. 2006. Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J. Mol. Biol. 359:533–538 [DOI] [PubMed] [Google Scholar]
- 3. Bush K, Fisher JF. 2011. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65:455–478 [DOI] [PubMed] [Google Scholar]
- 4. Chambers HF, Kocagoz T, Sipit T, Turner J, Hopewell PC. 1998. Activity of amoxicillin/clavulanate in patients with tuberculosis. Clin. Infect. Dis. 26:874–877 [DOI] [PubMed] [Google Scholar]
- 5. Chambers HF, Turner J, Schecter GF, Kawamura M, Hopewell PC. 2005. Imipenem for treatment of tuberculosis in mice and humans. Antimicrob. Agents Chemother. 49:2816–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 7. Cynamon MH, Palmer GS. 1983. In vitro activity of amoxicillin in combination with clavulanic acid against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 24:429–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dubée V, et al. 2012. Kinetic analysis of Enterococcus faecium l,d-transpeptidase inactivation by carbapenems. Antimicrob. Agents Chemother. 56:3409–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. England K, et al. 2012. Meropenem-clavulanic acid shows activity against Mycobacterium tuberculosis in vivo. Antimicrob. Agents Chemother. 56:3384–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gupta R, et al. 2010. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 16:466–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamilton-Miller JM, Newton GG, Abraham EP. 1970. Products of aminolysis and enzymatic hydrolysis of the cephalosporins. Biochem. J. 116:371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hugonnet JE, Blanchard JS. 2007. Irreversible inhibition of the Mycobacterium tuberculosis beta-lactamase by clavulanate. Biochemistry 46:11998–12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, III, Blanchard JS. 2009. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323:1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lavollay M, et al. 2009. The beta-lactam-sensitive d,d-carboxypeptidase activity of Pbp4 controls the l,d and d,d transpeptidation pathways in Corynebacterium jeikeium. Mol. Microbiol. 74:650–661 [DOI] [PubMed] [Google Scholar]
- 15. Lavollay M, et al. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J. Bacteriol. 190:4360–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lavollay M, et al. 2011. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by l,d-transpeptidases. J. Bacteriol. 193:778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lecoq L, et al. 26 January 2012. Backbone and side-chain (1)H, (15)N and (13)C assignment of apo- and imipenem-acylated l,d-transpeptidase from Bacillus subtilis. Biomol. NMR Assign. doi:10.1007/s12104-012-9358-1 [DOI] [PubMed]
- 18. Magnet S, et al. 2007. Identification of the l,d-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J. Bacteriol. 189:3927–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mainardi JL, et al. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J. Biol. Chem. 280:38146–38152 [DOI] [PubMed] [Google Scholar]
- 20. Mainardi JL, Hugonnet JE, Gutmann L, Arthur M. 2011. Fighting resistant tuberculosis with old compounds: the carbapenem paradigm. Clin. Microbiol. Infect. 17:1755–1756 [DOI] [PubMed] [Google Scholar]
- 21. Mainardi JL, et al. 2007. Unexpected inhibition of peptidoglycan ld-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J. Biol. Chem. 282:30414–30422 [DOI] [PubMed] [Google Scholar]
- 22. Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 32:386–408 [DOI] [PubMed] [Google Scholar]
- 23. Mitnick CD, et al. 2008. Comprehensive treatment of extensively drug-resistant tuberculosis. N. Engl. J. Med. 359:563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nadler JP, Berger J, Nord JA, Cofsky R, Saxena M. 1991. Amoxicillin-clavulanic acid for treating drug-resistant Mycobacterium tuberculosis. Chest 99:1025–1026 [DOI] [PubMed] [Google Scholar]
- 25. Peltier J, et al. 2011. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J. Biol. Chem. 286:29053–29062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Triboulet S, et al. 2011. Inactivation kinetics of a new target of beta-lactam antibiotics. J. Biol. Chem. 286:22777–22784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10:123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Veziris N, Truffot C, Mainardi JL, Jarlier V. 2011. Activity of carbapenems combined with clavulanate against murine tuberculosis. Antimicrob. Agents Chemother. 55:2597–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. World Health Organization 2011. Global tuberculosis control 2011. Document WHO/HTM/TB/2011.16. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/index.html [Google Scholar]
- 30. Zapun A, Contreras-Martel C, Vernet T. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361–385 [DOI] [PubMed] [Google Scholar]