Abstract
LFF571 is a novel semisynthetic thiopeptide and potent inhibitor of Gram-positive bacteria. We report that the antibacterial activity of LFF571 against Clostridium difficile is due to inhibition of translation. Single-step mutants of C. difficile with reduced susceptibility to LFF571 were selected at frequencies of <4.5 × 10−11 to 1.2 × 10−9. Sequencing revealed a G260E substitution in the thiopeptide-binding pocket of elongation factor Tu. Importantly, this mutation did not confer cross-resistance to clinically used antimicrobials. These results support the development of LFF571 as a treatment for C. difficile infection.
TEXT
Clostridium difficile is a Gram-positive, anaerobic, spore-forming bacterium that is the leading cause of hospital-acquired antibiotic-associated diarrhea (22). Infection is potentially fatal and can involve complications such as severe colitis and toxic megacolon. The incidence of C. difficile infection (CDI) has risen dramatically over the past decade, and current estimates of 400,000 U.S. cases annually are almost quadruple the rates from the 1990s (1). This increase is associated with the emergence of hypervirulent strains, including B1/NAP1/027. CDIs are now more commonly found outside the hospital setting and in previously low-risk groups, such as children, pregnant women, and individuals with irritable bowel syndrome (1). Management of CDI involves treatment with the antibacterials metronidazole, vancomycin, and fidaxomicin. Unfortunately, recurrent disease occurs in 20 to 25% of patients receiving metronidazole and vancomycin (2, 11, 18). Less recurrence has been noted for fidaxomicin in clinical trials, although the risk was unchanged for patients infected with B1/NAP1/027 (8, 16, 17).
We have recently reported the discovery of LFF571 (14), a semisynthetic derivative of the natural metabolite GE2270 A (21). The structure of LFF571 is shown in Fig. 1. GE2270 A is a translation inhibitor that binds bacterial elongation factor Tu (EF-Tu) and blocks delivery of aminoacylated tRNA (aa-tRNA) to the ribosome (19). Like GE2270 A, LFF571 has antimicrobial activity against a range of Gram-positive bacteria, including C. difficile (4, 10, 14; S. Bushell, M. J. LaMarche, J. A. Leeds, and L. Whitehead, 18 June 2009, international patent application WO 2009/074605). LFF571 MIC90 values obtained in two independent studies were 0.25 and 0.5 μg/ml against C. difficile isolates (4, 12). Here, we investigate the mechanism of LFF571 and the frequency and mechanism of reduced susceptibility to this compound. An accompanying article by Trzasko et al. examines the efficacy of LFF571 in a Golden Syrian hamster model of CDI (21a).
Fig 1.
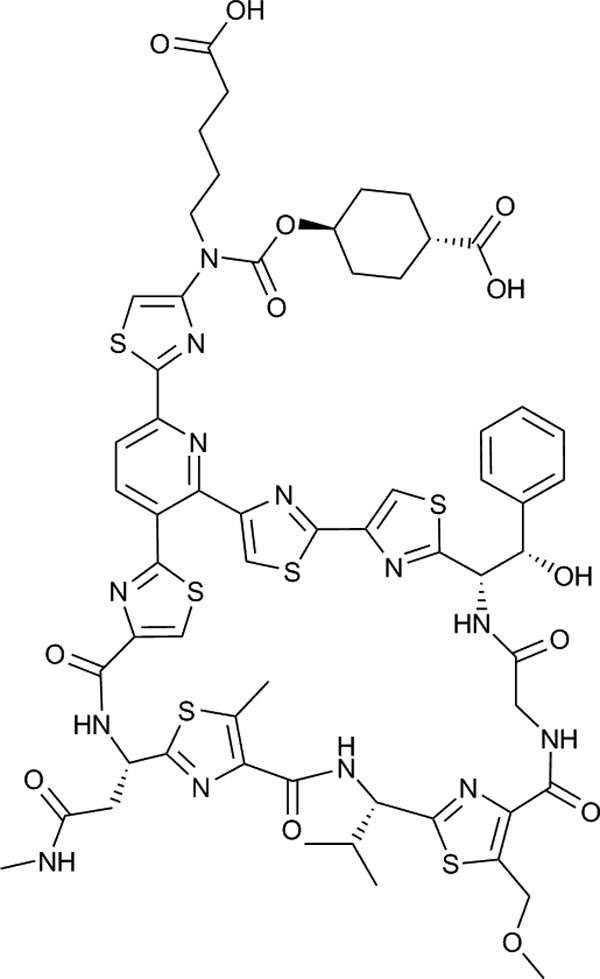
Chemical structure of LFF571. LFF571 is a semisynthetic thiopeptide derived from the natural product GE2270 A (21).
The MICs of LFF571 against the five C. difficile strains used in this study are shown in Table 1. LFF571, which was synthesized at Novartis according to published methods (Bushell et al., international patent application WO 2009/074605), demonstrated potent antibacterial activity against all C. difficile strains tested, with MICs of ≤0.5 μg/ml.
Table 1.
MICs of LFF571 against C. difficile strains used in the studya
| Strain | Description | LFF571 MIC (μg/ml) |
|---|---|---|
| NB95002 | Clinical isolateb | 0.125 |
| NB95013 | ATCC 43255 | 0.5 |
| NB95026 | Clinical isolate (MOH838)c | 0.5 |
| NB95031 | Clinical isolate (MOH082, REA type AA)c | 0.5 |
| NB95047 | Clinical isolate (MOH108, REA type J)c | 0.25 |
MICs were determined by agar dilution methods according to Clinical and Laboratory Standards Institute (CLSI) guidelines (5). REA, restriction endonuclease analysis.
Novartis collection.
Kindly provided by Donald Low at Mount Sinai Hospital, Toronto, Canada.
To investigate LFF571's mechanism of action, we monitored translation and cell wall biosynthesis by the incorporation of [3H]leucine (Perkin Elmer, Billerica, MA) or N-[3H]acetylglucosamine (American Radiolabeled Chemicals, St. Louis, MO), respectively. Mid-exponential-phase cultures of C. difficile NB95026 in chemically defined medium (13) supplemented with 0.2% glucose, 0.5 μg/ml vitamin K1, and 5 μg/ml hemin were treated with radiolabeled precursor for 60 min at 37°C under anaerobic conditions (15). For LFF571, the 50% inhibitory concentration (IC50) for [3H]leucine incorporation (0.06 μg/ml) was similar to the antibacterial concentration under the same testing conditions (0.03 μg/ml) (Table 2). In contrast, no inhibition of cell wall synthesis was observed, even after treatment with >30× the antibacterial concentration. Similar results were seen for translational inhibitor tetracycline (Sigma-Aldrich, St. Louis, MO), while the peptidoglycan synthesis inhibitor vancomycin (US Pharmacopeia, Rockville, MD) (20) blocked incorporation of [3H]UDP-GlcNAc, but not [3H]leucine. These results indicate that the antibacterial activity of LFF571 is via inhibition of C. difficile protein synthesis.
Table 2.
Antibacterial and macromolecular inhibitory concentrations of LFF571
| Test agent | IC50 (μg/ml ± SD)a |
Antibacterial concn (μg/ml)b | |
|---|---|---|---|
| [3H]leucine | [3H]NAGc | ||
| LFF571 | 0.06 ± 0.004 (n = 5) | >1 (n = 5) | 0.03 |
| Tetracycline | 10.2 ± 4.3 (n = 5) | >32 (n = 5) | 8 |
| Vancomycin | >32 (n = 5) | 2.6 ± 1.9 (n = 6) | 4 |
Values represent means ± standard deviation (SD).
Antibacterial concentrations were determined using the same medium and inoculum with which the macromolecular synthesis experiments were performed. Strain NB95026 was used for all assays.
NAG, N-aeetylglucosamine.
To characterize the frequency of selection of spontaneous mutants with reduced susceptibility to LFF571, C. difficile suspensions (109 to 1010 CFU/ml) were plated on brucella agar containing 0.5 to 1 μg/ml (1 to 4× MIC) antibiotic and incubated anaerobically for 48 to 72 h at 37°C. Resistance frequency was defined as the number of colonies selected divided by total CFU plated. Reduced susceptibility to LFF571 was observed at the following frequencies: 1.7 × 10−10 (NB95002 selected at 0.5 and 1 μg/ml LFF571), 1.2 × 10−9 and <6.2 × 10−10 (NB95013 at 0.5 and 1 μg/ml, respectively), and 3.0 × 10−11 and <3.0 × 10−11 (NB95026 at 0.5 and 1 μg/ml, respectively). We were unable to select colonies of NB95031 under the conditions tested (<4.5 × 10−11).
To understand the genetic basis for reduced susceptibility to LFF571, mutants were analyzed for changes in EF-Tu. C. difficile possesses two identical copies of the gene encoding EF-Tu, tufA and tufB (Table 3), which were amplified using primers to nonidentical flanking sequences: 5′-CTTACCATAAGCTTATTGTGAGCA-3′ (forward) and 5′-GAGGAGCATAACCCCTCTTT-3′ (reverse) for tufA and 5′-ATTCGATCACTATGAGCAAGTTC-3′ (forward) and 5′-TATATGCTTTAGCGCTACTTGTC-3′ (reverse) for tufB. All mutants exhibited tufB mutation G782A, resulting in amino acid substitution G260E; NB95013-JAL0759 harbored the G782A change in both tufA and tufB.
Table 3.
Genetic changes in C. difficile upon selection with LFF571a
| Strain | Nucleotide/amino acid change |
MIC (μg/ml)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| tufa | tufB | LFF571 | VAN | MET | FDX | TET | CLI | ERY | MOX | LNZ | |
| NB95009 | NCc | NC | 0.25 | 1 | 0.5 | 0.03 | 64 | 2 | 1 | 2 | 2 |
| NB95002 | NC | NC | 0.5 | 0.5 | 1 | 0.125 | 32 | 4 | 1 | 1 | 2 |
| NB95002–JAL0777 | NC | G782A/G260E | >128 | 0.5 | 1 | 0.03 | 64 | 4 | 0.5 | 0.5 | 2 |
| NB95002–JAL0783 | NC | G782A/G260E | >128 | 0.5 | 1 | 0.03 | 32 | 4 | 0.5 | 0.5 | 2 |
| NB95013 | NC | NC | 0.25 | 0.5 | 0.5 | 0.03 | 0.06 | 4 | 1 | 0.5 | 2 |
| NB95013–JAL0758 | NC | G782A/G260E | >128 | 0.5 | 0.5 | 0.03 | 0.06 | 4 | 1 | 0.5 | 2 |
| NB95013–JAL0759 | G782A/G260E | G782A/G260E | >128 | 1 | 0.5 | 0.03 | 0.06 | 4 | 1 | 0.5 | 2 |
| NB95026 | NC | NC | 0.25 | 0.5 | 0.5 | 0.06 | 0.06 | 2 | 1 | 32 | 2 |
| NB95026–JAL0792 | NC | G782A/G260E | >128 | 0.5 | 0.5 | 0.06 | 0.125 | 2 | 1 | 32 | 2 |
All strains were selected on 0.5 μg/ml LFF571, except NB95002-JAL0783, which was selected on 1 μg/ml.
MICs were determined by agar dilution methods according to Clinical and Laboratory Standards Institute (CLSI) guidelines (5). VAN, vancomycin; MET, metronidazole; FDX, fidaxomicin; TET, tetracycline; CLI, clindamycin; ERY, erythromycin; MOX, moxifloxacin; LNZ, linezolid.
NC, no change.
We investigated whether reduced susceptibility to LFF571 resulted in cross-resistance to other antibiotics. Fidaxomicin was prepared at Novartis by fermentation of Catellatospora sp. strain Bp3323-81; its activity against the Clinical and Laboratory Standards Institute (CLSI) quality control strain of C. difficile (ATCC 700057) was within the acceptable range (6). Linezolid (from Pfizer as Zyvox) and moxifloxacin (from Bayer as Avelox) were extracted and purified at Novartis. Tetracycline, ampicillin, clindamycin, and erythromycin were purchased from Sigma-Aldrich; vancomycin and metronidazole were obtained from US Pharmacopeia. As expected, an increase in the LFF571 MIC was observed for the selected mutants (>128 μg/ml versus 0.25 to 0.5 μg/ml for parental strains). Strains with reduced susceptibility to LFF571 continued to be sensitive to structurally and mechanistically unrelated antibiotics, including fidaxomicin, vancomycin, and metronidazole (Table 3). Sensitivity to the protein synthesis inhibitor tetracycline was also unchanged within the acceptable 2-fold range of the assay, and the decreased susceptibility of NB95026 to moxifloxacin was not affected by the tufB mutation.
In this study, we characterized the mechanisms of action and loss of susceptibility to LFF571. We hypothesized that the mechanism of action of LFF571 against C. difficile would parallel that of related semisynthetic monoacidic derivatives of GE2270 A against S. aureus (15). Indeed, LFF571 specifically blocked C. difficile protein synthesis. Furthermore, selection on inhibitory concentrations of LFF571 resulted in a substitution at the C. difficile residue analogous to G257 in E. coli EF-Tu. Substitutions at E. coli G257 render the protein resistant to the inhibitory activity of GE2270 A when measured in an in vitro translation assay (23). Along with our previous observation that LFF571 interacts directly with E. coli EF-Tu in vitro (9), these data support the hypothesis that LFF571 inhibits C. difficile translation by binding EF-Tu. The frequency of selection of single-step mutants with reduced susceptibility to LFF571 was low (≤1.2 × 10−9); for comparison, Critchley et al. (7) reported frequencies of C. difficile with reduced susceptibility to the metRS inhibitor REP3123 of ≤1 × 10−8. Our observation that only one amino acid substitution is selected in C. difficile in vitro likely explains the lower frequency of resistant colonies obtained for LFF571 compared to those of agents that select multiple, independent substitutions, including REP3123 (7). Overall, the excellent potency of LFF571, low frequency of susceptibility loss in vitro, and absence of cross-resistance to standard-of-care antibiotics support the development of LFF571 for treatment of CDI in humans.
ACKNOWLEDGMENT
We acknowledge Jade Bojkovic and Stacey TiamFook for technical assistance and Catherine Jones for editing the manuscript.
Footnotes
Published ahead of print 29 May 2012
REFERENCES
- 1. Ananthakrishnan AN. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat. Rev. Gastroenterol. Hepatol. 8:17–26 [DOI] [PubMed] [Google Scholar]
- 2. Bauer MP, Kuijper EJ, van Dissel JT. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): treatment guidance document for Clostridium difficile infection (CDI). Clin. Microbiol. Infect. 15:1067–1079 [DOI] [PubMed] [Google Scholar]
- 3. Reference deleted.
- 4. Citron DM, Tyrrell KL, Merriam CV, Goldstein EJ. 2012. Comparative in vitro activities of LFF571 against Clostridium difficile and 630 other intestinal strains of aerobic and anaerobic bacteria. Antimicrob. Agents Chemother. 56:2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2010. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 7th ed. Approved standard M11-7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S21. Twenty-first informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Critchley IA, et al. 2009. Spectrum of activity and mode of action of REP3123, a new antibiotic to treat Clostridium difficile infections. J. Antimicrob. Chemother. 63:954–963 [DOI] [PubMed] [Google Scholar]
- 8. Crook D, et al. 2010. Randomized clinical trial in Clostridium difficile infection confirms equivalent cure rate and lower recurrence rate of fidaxomicin vs vancomycin, abstr P-LB2401. Abstr. 20th Eur. Congr. Clin. Microbiol. Infect. Dis., Vienna, Austria [Google Scholar]
- 9. Deng G, et al. 2011. Investigation of mode of binding of elongation factor tu inhibitor LFF571, abstr F1-1859. Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL [Google Scholar]
- 10. Dzink-Fox J, et al. 2011. Antimicrobial activity of the novel elongation factor Tu Inhibitor, LFF571, abstr F1-1346. Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL [Google Scholar]
- 11. Gerding DN, Muto CA, Owens RC., Jr 2008. Treatment of Clostridium difficile infection. Clin. Infect. Dis. 46(Suppl 1):S32–S42 [DOI] [PubMed] [Google Scholar]
- 12. Hecht D, Osmolski J, Gerding D. 2012. Activity of LFF571 against 103 clinical isolates of C. difficile, abstr P1440. Abstr. 22nd Eur. Society of Clinical Microbiology and Infectious Diseases, London, United Kingdom [Google Scholar]
- 13. Karasawa T, Ikoma S, Yamakawa K, Nakamura S. 1995. A defined growth medium for Clostridium difficile. Microbiology 141:371–375 [DOI] [PubMed] [Google Scholar]
- 14. LaMarche MJ, et al. 2012. Discovery of LFF571: an investigational agent for Clostridium difficile infection. J. Med. Chem. 55:2376–2387 [DOI] [PubMed] [Google Scholar]
- 15. Leeds JA, et al. 2011. In vitro and in vivo activities of novel, semisynthetic thiopeptide inhibitors of bacterial elongation factor Tu. Antimicrob. Agents Chemother. 55:5277–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Louie TJ, et al. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 364:422–431 [DOI] [PubMed] [Google Scholar]
- 17. Miller M, Mullane KM, Weiss K. 2009. OPT-80 versus vancomycin in Clostridium difficile infection: results of randomized clinical trial. Gastroenterology 136(Suppl 1):A-115 doi:10.1016/S0016-5085(09)60516-3 [Google Scholar]
- 18. Nelson RL. 18 July 2007. Antibiotic treatment for Clostridium difficile-associated diarrhea in adults. Cochrane Database Syst. Rev. 2007:CD004610. [DOI] [PubMed] [Google Scholar]
- 19. Parmeggiani A, et al. 2006. Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu. Biochemistry 45:6846–6857 [DOI] [PubMed] [Google Scholar]
- 20. Perkins HR, Nieto M. 1974. The chemical basis for the action of the vancomycin group of antibiotics. Ann. N. Y. Acad. Sci. 235:348–363 [DOI] [PubMed] [Google Scholar]
- 21. Selva E, et al. 1991. Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J. Antibiot. (Tokyo) 44:693–701 [DOI] [PubMed] [Google Scholar]
- 21a. Trzasko A, Leeds JA, Praestgaard J, LaMarche MJ, McKenney D. 2012. Efficacy of LFF571 in a hamster model of Clostridium difficile infection. Antimicrob. Agents Chemother. 56:4459–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Viswanathan VK, Mallozzi MJ, Vedantam G. 2010. Clostridium difficile infection: an overview of the disease and its pathogenesis, epidemiology and interventions. Gut Microbes 1:234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuurmond AM, et al. 2000. GE2270A-resistant mutations in elongation factor Tu allow productive aminoacyl-tRNA binding to EF-Tu.GTP.GE2270A complexes. J. Mol. Biol. 304:995–1005 [DOI] [PubMed] [Google Scholar]


