Abstract
Hepatitis A virus (HAV) has previously been reported to bind to human red blood cells through interaction with glycophorin A. Residue K221 of VP1 and the surrounding VP3 residues are involved in such an interaction. This capsid region is specifically recognized by the monoclonal antibody H7C27. A monoclonal antibody-resistant mutant with the mutation G1217D has been isolated. In the present study, the G1217D mutant was characterized physically and biologically in comparison with the parental HM175 43c strain. The G1217D mutant is more sensitive to acid pH and binds more efficiently to human and rat erythrocytes than the parental 43c strain. In a rat model, it is eliminated from serum more rapidly and consequently reaches the liver with a certain delay compared to the parental 43c strain. In competition experiments performed in vivo in the rat model, the G1217D mutant was efficiently outcompeted by the parental 43c strain. Only in the presence of antibodies reacting specifically with the parental 43c strain could the G1217D mutant outcompete the parental 43c strain in serum, although the latter still showed a remarkable ability to reach the liver. Altogether, these results indicate that the G1217D mutation induces a low fitness phenotype which could explain the lack of natural antigenic variants of the glycophorin A binding site.
INTRODUCTION
Hepatitis A virus (HAV), classified as the type species of the genus Hepatovirus within the Picornaviridae family (16), is a medically important hepatotropic virus (14). The virion capsid is composed of the structural proteins VP1, VP2, VP3, and possibly VP4, encoded in the P1 region of the genome (14).
HAV has a limited number of antigenic sites. The immunodominant site, composed of closely clustered epitopes, is defined by two major groups of escape mutants that include mutations in residues 70, 71, and 74 of VP3 and residues 102, 171, and 176 of VP1 (25, 29). There is another apparently distinct antigenic site, represented by mutations around residue 221 of VP1, and an additional and still undefined third antigenic site for which no escape mutant has so far been isolated. A single serotype has been recognized up to now, and even descriptions of natural variants at these specific sites are scarce (8, 11, 27, 36), in spite of the quasispecies nature of HAV (34).
Recently, we described a molecular mechanism, i.e., codon usage constraints, that could contribute in part to this low antigenic variability (1). HAV presents a highly deoptimized codon usage with strategic locations of residues encoded by rare codons at or very close to the capsid epitopes. These residues encoded by rare codons are highly conserved among the different HAV strains (35), and their replacement is negatively selected even under specific immune pressure (1). The need to keep these residues encoded by rare codons probably relies on their role in controlling translation speed to guarantee proper capsid folding, i.e., fine-tuning translation kinetics or using the right combination of preferred and rare codons, through the induction of transient ribosome stalling due to the longer times required to obtain the proper tRNAs. In fact, under conditions of artificially induced cellular shutoff, with the consequent increase of tRNA pools, HAV changes its capsid codon usage, increasing the use of less abundant instead of the most abundant tRNAs, probably in an attempt to avoid the rapid translation associated with a higher tRNA availability (2).
Other mechanisms to explain the low antigenic variability of HAV may also rely on in vivo cycle constraints. Monoclonal antibody (MAb) escape mutants representing the above-mentioned epitopes show a completely different fitness pattern (1). While escape mutants of the H7C27 MAb, defined by mutations in the region around residue 221 of VP1, have a similar in vitro fitness to that of the parental virus, those of the K34C8 MAb, defined by mutations in residues 171 to 176 of the VP1 immunodominant site, have a significant lower in vitro fitness than the parental virus and are able to outcompete it only in the presence of antibodies to which they are resistant. In contrast, among the few antigenic variants isolated from patients, only representatives with variations of the immunodominant site are available (8, 11, 27, 36). This discrepancy may be explained by taking into consideration the biological constraints imposed by the virus enterohepatic cycle in the host. For the development of an infection cycle, HAV must overcome the challenges posed by the acid pH of the stomach and the action of intestinal proteases and detergents (particularly biliary salts) during the entry phase, the presence of decoy factors during the viremic phase, and again the action of proteases and detergents during the exit phase. The selective pressure of pH, proteases, and biliary salts calls for the need to shape a highly cohesive and stable capsid. However, the contribution of blood decoy factors to delineate such a capsid may seem less obvious. HAV replicates in hepatocytes and uses different membranes to enter and exit the polarized cell. As a consequence, the enterohepatic cycle is critical for the infection of new hepatocytes (23). In such a context, HAV needs to persist during the viremic phase. Erythrocyte glycoproteins may function as decoy receptors, attracting pathogens to the erythrocyte and keeping them away from target tissues (12). HAV interacts with glycophorin A of human erythrocytes, and the capsid region involved in this interaction is located around the putative pit area and coincides with the H7C27 MAb binding site (33). However, this interaction is optimal under acidic conditions and impaired at neutral physiological pH, suggesting that the actual HAV capsid conformation would allow escape from erythrocyte attachment. Avoiding blood clearance, i.e., the removal of viruses from the fluid compartment of blood, may constitute an advantage for a viremic infectious agent whose target organ is the liver, contributing to the final fitness outcome in vivo.
In the present work, the roles of all these physical and cycle constraints in the high level of antigenic stability of the glycophorin A binding site and their role in viral fitness are evaluated.
MATERIALS AND METHODS
Viruses and cells.
The cytopathogenic HM175 43c strain of HAV is a mutant (D3070A; replacement of D at position 70 of VP3 by A) resistant to a large number of MAbs against the immunodominant site (29) which was generated spontaneously during cell culture passage of the HM175 wild-type virus. The 43c strain is still sensitive to several MAbs against the immunodominant and glycophorin A binding sites (1, 29). The G1217D strain is a mutant (replacement of G at position 217 of VP1 by D) derived from the HM175 43c strain which was selected in the presence of the H7C27 MAb (1). The phenotype of resistance to the H7C27 MAb came from the G217D substitution in the VP1 protein. Residue 217 is very close to residue 221, which defines the H7C27 MAb (or glycophorin A) binding site (1). Both virus strains (43c parental strain and G1217D mutant) were used throughout this study. The 50% tissue culture infective dose (TCID50) was obtained for both viruses on FRhK-4 cell monolayers.
Quantification of resistance to acid pH, trypsin, and biliary salts.
Resistance treatment at acid pH consisted of virus incubations at pH 1, 2, and 3.5 for 1 and 3 h at 37°C. Resistance to 1% biliary salts and 0.25% trypsin was also tested for 4 and 8 h at 37°C. To quantify virus inactivation, a control test of nontreated viruses kept for the same length of time at 37°C was run in parallel. The log(Nt/N0) value, where Nt is the infectious viral titer after treatment and N0 is the infectious viral titer of the parallel nontreated virus control, was figured.
Purification of infectious (150S) particles.
Concentrated viral stocks were obtained as previously described (4). Briefly, at 5 to 6 days postinfection, cells from a T-175 flask were harvested by trypsin treatment, collected by centrifugation, resuspended in 500 μl of NT buffer (0.1 M NaCl, 10 mM Tris-HCl, 1% NP-40, pH 7.4), and incubated for 30 min at room temperature. These lysed cell suspensions were centrifuged at 1,700 × g for 5 min and the supernatants again centrifuged at 13,000 × g for 5 min. Viruses recovered in the supernatants were subjected to three sonication cycles of 30 s at 60 W in the presence of 0.4% SDS. Five hundred microliters of each concentrated viral stock was layered onto a 5 to 45% sucrose gradient in TNMg buffer (20 mM Tris-HCl, 10 mM NaCl, and 50 mM MgCl2, pH 6.7) and spun at 205,000 × g for 165 min (31). Fractions containing infectious virus (150S) were identified by both determination of the refraction index and a sandwich enzyme-linked immunosorbent assay (ELISA) consisting of HAV capture by a convalescent-phase serum followed by detection with MAb K2-4F2 (30). Positive fractions from six gradients were collected and dialyzed against water to remove sucrose. Finally, pooled samples were concentrated by ultracentrifugation at 229,600 × g for 4 h, and the pellet was recovered in a final volume of 500 μl.
Viral titers were quantified by real-time reverse transcription-PCR (RT-PCR) (7) and the TCID50.
Erythrocyte purification.
Human type O erythrocytes (provided by the blood bank of the Hospital Vall d'Hebron, Barcelona, Spain) and erythrocytes from adult female Wistar rats were purified on preformed self-generated gradients of Percoll (Pharmacia Biotech) following the manufacturer's specifications, as previously described (33). Briefly, a gradient was formed by spinning 10 ml of a 70% solution of Percoll in 150 mM NaCl at 2,000 × g for 15 min. Two milliliters of gradient material was removed from the bottom of the tube, and 2 ml of 50% heparinized blood in 150 mM NaCl was layered on top of the gradient. The sample was centrifuged for 5 min at 400 × g, the platelets on the top of the gradient were removed and replaced by saline, and the gradient was centrifuged again at 800 × g for 15 min. Cells from the erythrocyte-containing fraction were washed three times in Alserver's solution (2.05% [wt/vol] glucose, 0.8% sodium citrate, 0.055% citric acid, and 0.42% NaCl, pH 6.7), collected by centrifugation at 500 × g for 5 min, and resuspended in the same solution. Before hemagglutination assays, erythrocytes were again washed three times in PBS (0.137 M NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4) at pH 5.5 or 7.2.
Hemagglutination assays.
Hemagglutination assays were performed in 96-well U-bottom plates as previously described (33). Briefly, serial 2-fold dilutions of 150S particles were made in phosphate-buffered saline (PBS) at pH 5.5 or 7.2. Twenty microliters of diluted particles was dispensed in each well, and 20 μl of a suspension containing 106 erythrocytes at pH 5.5 or 7.2 was added, mixed, and incubated for 1 h at room temperature. Negative controls were also assayed by incubating 20 μl of PBS with the same volume of the erythrocyte suspension under the same conditions. The hemagglutination titer was expressed as the reciprocal of the greatest dilution of HAV that caused complete agglutination of the erythrocytes (endpoint dilution) in comparison with negative controls. The hemagglutination unit was defined as the number of particles present at the hemagglutination endpoint dilution able to hemagglutinate 106 erythrocytes.
In vivo injection of HAV strains into rats.
In order to test blood clearance of the 43c strain and the G1217D mutant, two groups of seven adult female Wistar rats (175 to 200 g) were inoculated with 1 ml of virus at 108 genome copies/ml via the tail vein. Blood (200 to 300 μl) was obtained through the saphenous vein at 1, 5, 15, 30, and 60 min postinoculation (p.i.) and 3, 6, 12, 24, and 48 h p.i. from rats under isoflurane inhalation anesthesia, and the titers of virus genome copies in both blood compartments (serum and blood cells) were determined. At 48 h p.i., animals were sacrificed by lethal injection, and the liver and spleen were removed for quantitative PCR (qPCR) determination of HAV genome copy numbers (7).
To test the influence of blood clearance on the biodistribution of both strains, two groups of nine rats were inoculated with 1 ml of virus at 108 genome copies/ml. On days 7, 14, and 21, three rats of each group were sacrificed and the virus in sera, livers, and spleens quantified. Fecal samples were obtained on days 3, 5, 7, 10, 12, 14, and 21 from available animals to monitor viral excretion. For the liver and spleen, both positive- and negative-strand RNA copy numbers were determined, while for serum and feces only the positive-strand RNA was titrated. A supplementary experiment was performed similarly, but using four groups of animals, two of which received the untreated parental and mutant strains as described above and two of which received the same strains inactivated through a UV-light (254 nm) irradiation treatment for 3 h. In this experiment, all samples were equally obtained and processed, with the exception of fecal samples, which were collected daily during the interval between days 14 and 21.
Additionally, clearance competition experiments were performed by inoculating four groups of seven rats with mixtures of both strains at a 1:1 (43c:G1217D mutant) ratio, as well as a 1:9 ratio, in the absence and presence of the H7C27 MAb at a concentration inducing 90% neutralization of strain 43c. The 1-ml inoculum that each rat received contained 108 genome copies/ml; in the 1:1 mixtures, there were 5 × 107 genome copies of each virus strain, and in the 1:9 mixtures, there were 1 × 107 genome copies of the 43c strain plus 9 × 107 genome copies of the G1217D mutant strain. Blood samples were taken following the same schedule as that described above. However, animals were sacrificed not at 48 h but at 21 days. After sacrifice, samples of the liver and spleen were also taken.
Experiments were conducted according to European Union regulations for animal experiments (Directive 86/609 CEE) under the supervision of the Ethical Committee for Animal Procedures (CEEA) of the University of Barcelona. Procedure 3656 (clearance kinetics of hepatitis A virus [HAV] in adult Wistar rats) was previously approved by the Environment Department of the Generalitat de Catalunya.
Determination of virus clearance from blood.
The number of positive-strand RNA genome copies per ml of serum at each sampling time was determined by qPCR, using a previously described standardized procedure which includes both RNA extraction and RT-PCR inhibition controls (7, 32). Blood samples were centrifuged at 3,000 × g for 10 min and RNA extracted from 150 μl of the serum supernatant and 150 μl of the PBS-resuspended cell pellet by use of a NucleoSpin RNA virus kit (Macherey-Nagel). Five microliters of each RNA suspension was used for one-step real-time RT-PCR.
At 2 weeks p.i., sequencing of a fragment of the VP3 coding region corresponding to amino acids 1 to 123 and a fragment of the VP1 coding region corresponding to amino acids 176 to 245 was performed as previously described (1) to monitor the evolution of the inoculated virus population.
In the clearance competition experiments, the follow-up of the proportion of each viral strain was done by using the mutation at position 217 of VP1, present only in the G1217D mutant strain (G → A mutation at nucleotide [nt] 2857 in HM175; GenBank accession number M14707), as a genetic marker allowing semiquantitative monitoring of the strains through determination of the proportional heights of the two peaks inferred from the chromatogram of the consensus sequence. The absolute number of genome copies per ml of serum was determined at 60 min p.i., and the relative titer of each strain was estimated through the percentage of each strain in the sequencing chromatogram.
Determination of genome copy numbers in liver and spleen.
The titer of positive-strand RNA genome copies per mg of tissue was determined by qPCR essentially as described above. Additionally, the number of negative-strand (antigenomic) RNA copies was also obtained, by a two-step real-time RT-PCR derived from the above-mentioned method for genome quantification, but using a primer annealing with the negative strand as the reverse primer for RT. Five hundred milligrams of minced tissue was digested in 500 μl of a solution containing 100 μg/ml of proteinase K for 2 h at 37°C with constant agitation. Tissue debris was removed by centrifugation at 3,000 × g for 5 min, and the supernatant was used for virus quantification. RNA was extracted from 150 μl of this supernatant by use of the kit described above, and 5 μl of this RNA suspension was used in the real-time RT-PCR assays.
In the clearance competition experiments, the follow-up of the proportion of each viral strain was done using the same genetic marker as that stated in the previous section. The absolute titer of genome copies per mg of liver was determined at 21 days p.i., and the relative titer of each strain was estimated through the percentage of each strain in the sequencing chromatogram.
Determination of genome copy numbers in feces.
Genome copy numbers per g of feces were determined by qPCR essentially as described above. A 10% suspension of feces in PBS was prepared, vigorously agitated, and centrifuged at 2,000 × g for 5 min. RNA was extracted from 150 μl of the supernatant obtained after centrifugation as described above, and 5 μl of this RNA suspension was used for one-step real-time RT-PCR.
Statistical analyses.
Statistical differences between parental 43c and G1217D mutant behaviors were assessed by using the Student t test. Additionally, statistical differences in percentages and RNA copy numbers of a given strain along time were also ascertained. RNA copy numbers are expressed as means ± standard errors.
RESULTS
Comparative stability of 43c and G1217D mutant strains of HAV to acid pH, biliary salts, and trypsin.
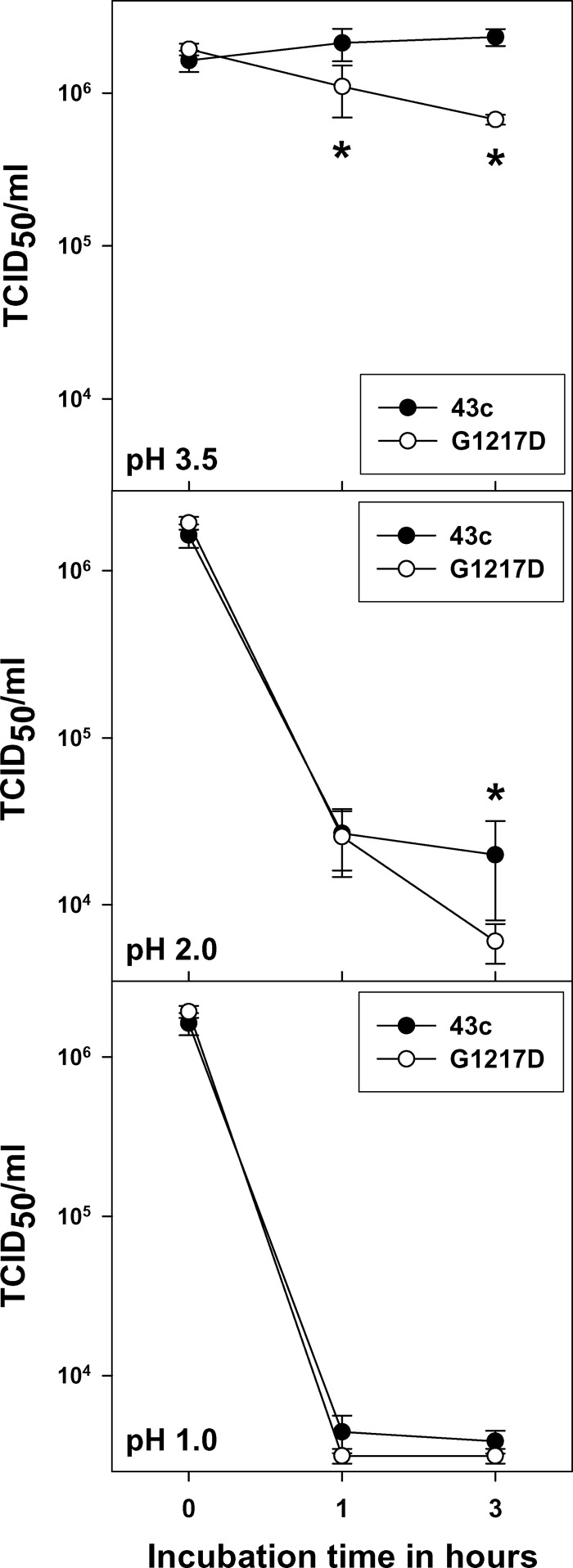
A comparative analysis of the stability of the parental 43c and G1217D mutant strains in the presence of acid pH, trypsin, and biliary salts was performed. Both strains were highly resistant to acid pH, although the G1217D mutant was somewhat more sensitive (P < 0.05) than the parental 43c strain (Fig. 1). After 3 h at pH 3.5, the G1217D mutant showed a reduction of 0.43 ± 0.07 log10, while the 43c strain was not affected. After 3 h at pH 2, the 43c strain showed a reduction of 1.94 ± 0.34 log10 and the G1217D mutant showed a reduction of 2.50 ± 0.22 log10, and after 3 h at pH 1, reductions of 2.40 ± 0.10 and 2.73 ± 0.12 log10, respectively, were observed. Resistance to both 1% biliary salts and 0.25% trypsin was also evaluated, and in both cases, reductions of <1 log10 were detected, with no statistical differences between strains.
Fig 1.
Comparative stability of HAV parental 43c and G1217D mutant strains under different acid conditions. Statistical analyses to compare the behaviors of both strains were performed for each pH condition and at each sampling time. Asterisks indicate statistically significant differences (P < 0.05) between the 43c and G1217D mutant strains at a given sampling time.
Differential hemagglutination pattern between 43c and G1217D mutant strains of HAV.
The hemagglutination capacities of the 43c and G1217D mutant strains are shown in Table 1. The hemagglutination unit of human erythrocytes was significantly higher (P < 0.05) for the 43c parental strain than for the G1217D mutant strain at both acid and neutral pHs, with an average difference of 1 log10. To ascertain whether this higher erythrocyte binding capacity of the G1217D mutant strain, which is associated with the G217D substitution of the VP1 protein, could be related to an increased clearance from blood, the Wistar rat was used as an in vivo model. Previously, the capacity of both viral strains to hemagglutinate rat erythrocytes was tested at physiological pH. In this scenario, the hemagglutination unit of the 43c strain was also significantly higher (P < 0.05) than that of the G1217D mutant, again with an average difference of 1 log10. These differences in hemagglutination capacity were not due to differences in the total virus/infectious virus ratios of both strains, whose values are not statistically significantly different (P < 0.05) (76 ± 59 and 49 ± 34 for the parental and mutant strains, respectively).
Table 1.
Hemagglutination of parental 43c strain and H7C27 escape mutant (G1217D) viruses
| Strain | No. of viruses able to hemagglutinate 106 erythrocytes (hemagglutination unit)a |
||
|---|---|---|---|
| Human O erythrocytes |
Wistar rat erythrocytes at pH 7.2 | ||
| pH 5.5 | pH 7.2 | ||
| 43c | 2.5 × 106 ± 2.1 × 106a | 4.7 × 106 ± 3.0 × 106a | 3.6 × 106 ± 1.4 × 106a |
| G1217D mutant | 1.1 × 105 ± 0.8 × 105b | 1.3 × 105 ± 1.2 × 105b | 4.4 × 105 ± 3.4 × 105b |
Different superscript letters indicate statistically significant differences (P < 0.05) between the 43c parental strain and the G1217D mutant.
Differential blood clearance pattern and biodistribution of 43c and G1217D mutant strains of HAV.
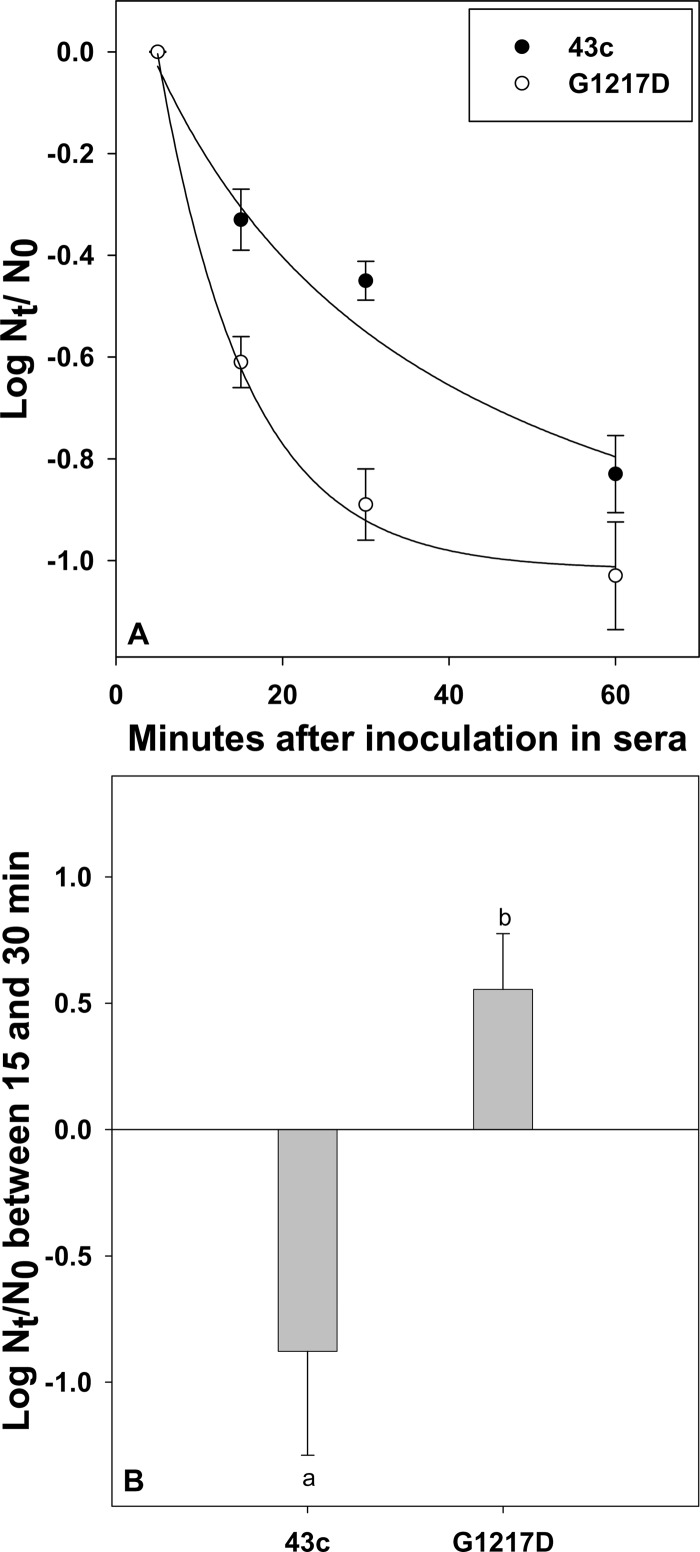
The differential blood clearance of the 43c parental and G1217D mutant strains was ascertained by quantifying the viral genome half-life in serum after virus inoculation into Wistar female adult rats. After an initial abrupt drop in genome copies observed with both strains at 5 min p.i., due to virus distribution in the whole blood volume as well as nonspecific trapping in blood cells, subsequent virus decay in serum was more rapid for the G1217D mutant than for the 43c parental strain, with a significantly (P < 0.05) more pronounced slope of the curve during the first 30 min p.i. (Fig. 2A). Accordingly, virus particles of the G1217D mutant moved specifically from the serum compartment to the erythrocyte compartment, showing an increase of cell-bound viruses of 0.56 ± 0.22 log10 from 15 to 30 min p.i. (Fig. 2B). In contrast, those particles of the 43c parental strain that were nonspecifically trapped in the blood cells at 15 min p.i. were further released, with a decrease of bound viruses of −0.88 ± 0.41 log10 (Fig. 2B) from 15 to 30 min p.i. This difference, i.e., an increase and a decrease of the bound viruses of the G1217D mutant and 43c parental strains, respectively, was statistically significant (P < 0.05). Overall, the estimated half-lives of the G1217D mutant and 43c parental strains in serum were 8 and 17 min, respectively.
Fig 2.
Comparative kinetics of clearance from blood of HAV parental and G1217D mutant strains. Rats were injected intravenously with 1 ml of virus at 108 genome copies/ml via the tail vein. Blood (200 to 300 μl) was obtained through the saphenous vein at 1, 5, 15, 30, and 60 min p.i., and virus genome copy numbers in both blood compartments (serum and blood cells) were determined. (A) Genome copy number decay in serum, expressed as the log10(Nt/N0) value, where Nt is the genome copy number at 15, 30, or 60 min p.i. and N0 is the genome copy number at 5 min p.i. Curves were fitted by nonlinear regression with SigmaPlot (SPSS, Chicago, IL). (B) Genome copy number variation in blood cells between 15 and 30 min p.i., expressed as the log10(Nt/N0), where Nt is the genome copy number at 30 min p.i. and N0 is the genome copy number at 15 min p.i. Different letters indicate statistically significant differences (P < 0.05).
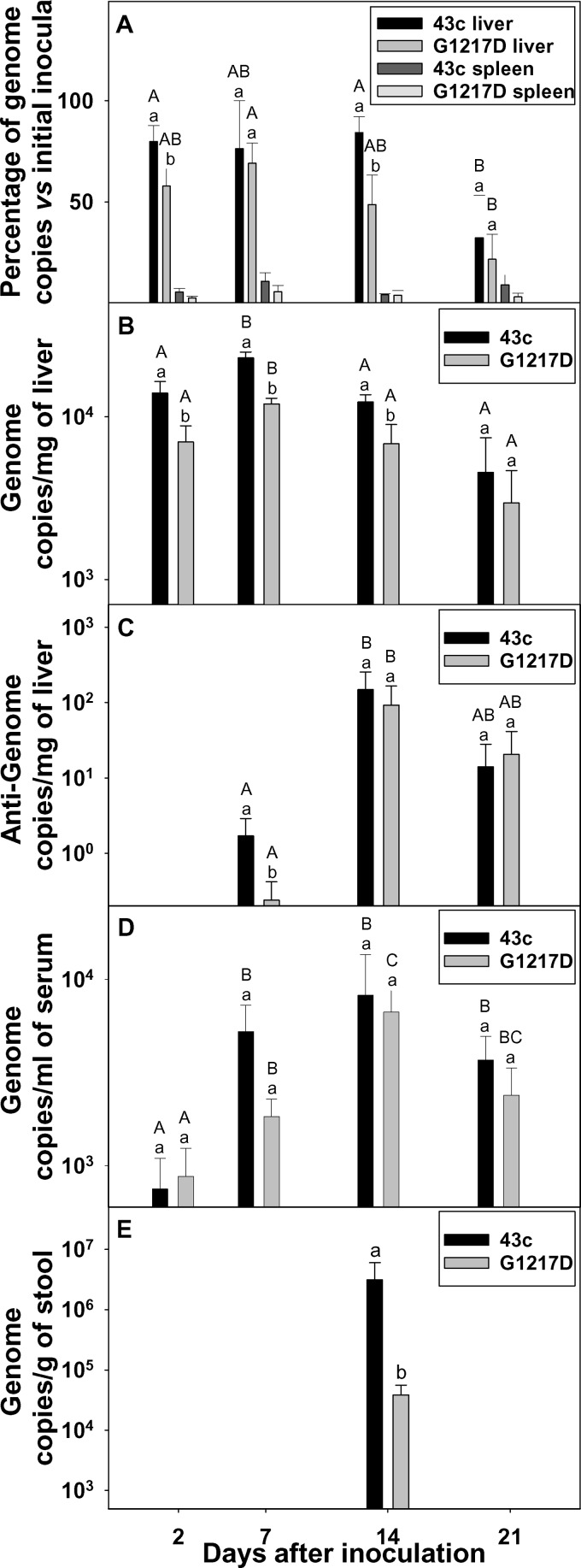
To address whether blood clearance efficiency plays any role in pathogenesis, the tissue distribution of the 43c and G1217D mutant strains was studied (Fig. 3). At 48 h p.i., both strains were present in the liver, but at significantly (P < 0.05) different levels, with averages of around 77% and 59% of the total inoculated genomes for the 43c and G1217D mutant strains, respectively (Fig. 3A). Only around 5% of the inoculated viruses were detected in the spleen (Fig. 3A) throughout the study period, with no differences between strains. After 1 week p.i., the percentage of inoculated 43c in the liver remained unchanged, while that of the G1217D mutant increased to 69%. At 14 days p.i., these percentages were again significantly different: 84% and 49% for the 43c and G1217D mutant strains, respectively. Thereafter, the decline was clear for both strains, with only 32% and 22% of the initial inocula at 21 days p.i. for the 43c and G1217D mutant strains, respectively.
Fig 3.
Comparative biodistribution, replication, and elimination of HAV parental and G1217D mutant strains. (A) Percentages of initial virus inocula in the liver and spleen at different times p.i., measured via genome copies (positive-strand RNA). (B) Genome copy numbers (positive-strand RNA) in the liver at different times p.i. (C) Antigenome copy numbers (negative-strand RNA) in the liver at different times p.i. (D) Genome copy numbers (positive-strand RNA) in serum at different times p.i. (E) Genome copy numbers (positive-strand RNA) in stool at different times p.i. Different letters indicate statistically significant differences (P < 0.05). Lowercase letters indicate comparison between both strains at a given sampling time. Capital letters indicate comparison at different sampling times for a given strain.
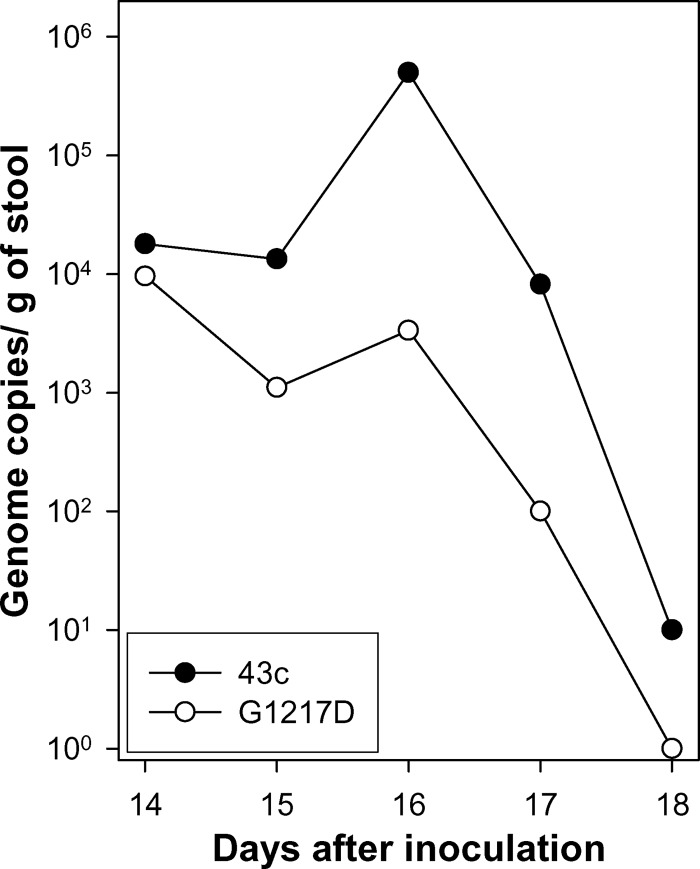
HAV may reach the liver in at least two different compartments: (i) erythrocyte bound and able to be phagocytosed by Kupffer cells of the reticuloendothelial system in the liver and (ii) free and able to infect both Kupffer cells and hepatocytes. Thus, the total number of genomes in the liver must be the result of the number of viruses reaching the organ (by means of any of the aforementioned ways), their increase due to replication (mainly after a period of weeks), and their elimination. The obtained data indicated a delay of the G1217D mutant strain in reaching the liver as well as a more rapid elimination from it than that of the 43c parental strain, which could be related to its higher level of erythrocyte binding. To confirm this hypothesis, replication of both strains in the liver was also studied through the increase of the positive-strand and, particularly, negative-strand RNA titers. At 1 week p.i., titers of positive-strand RNA were significantly (P < 0.05) different: 23,000 ± 2,000 copies/mg of liver and 12,000 ± 1,000 copies/mg of liver for the 43c and G1217D mutant strains, respectively. Titers of negative-strand RNA were also significantly (P < 0.05) different: 1.70 ± 1.20 copies/mg of liver and 0.24 ± 0.18 copy/mg of liver for the 43c and G1217D mutant strains, respectively (Fig. 3B and C). A similar trend was observed at 2 weeks p.i., with significantly (P < 0.05) higher titers of positive-strand RNA for 43c (12,333 ± 1,333 copies/mg of liver and 6,833 ± 2,123 copies/mg of liver for the 43c and G1217D mutant strains, respectively), while only slightly different levels of the negative-strand RNA (150 ± 106 copies/mg of liver and 93 ± 74 copies/mg of liver for the 43c and G1217D mutant strains, respectively) were observed. The increase in negative-strand RNA copy numbers between days 7 and 14 p.i. clearly indicated replication. However, the replication level was low, and consequently, the expected increase in viremia as well as fecal excretion, although present, was also moderate. Regarding excretion in stools, HAV genomes were detected in only 30% of the inoculated rats and only at 2 weeks p.i., with titers of 3.1 × 106 ± 2.9 × 106 genome copies/g and 3.9 × 104 ± 1.8 × 104 genome copies/g for the 43c and G1217D mutant strains, respectively. This significant (P < 0.05), 2-log difference in numbers of genome copies in stool reflects a more effective outcome of the 43c parental strain, likely due to a more effective way of reaching the target organ rather than more effective replication. A second experiment was performed in which fecal samples were collected daily during the interval between days 14 and 21 p.i. (Table 2 and Fig. 4). The total titers of genomic and antigenomic RNAs in the liver for the parental strain at 7 days p.i. were significantly higher than those for the G1217D mutant (Table 2). Virus excretion was detected in 60% and 33% of rats receiving the 43c parental and G1217D mutant strains, respectively. Viruses were detected from day 14 through day 17, with the highest titers at day 16, and the total number of genome copies in feces during this window was significantly higher for animals receiving the parental strain than for those receiving the G1217D mutant strain (Table 2 and Fig. 4).
Table 2.
Total log10 copy numbers of the 43c parental and G1217D mutant strains, inoculated untreated or after UV treatment
| Strain, treatment | Total log10 copy numbera |
||
|---|---|---|---|
| Genomic RNA in liver at 7 days p.i. | Antigenomic RNA in liver at 7 days p.i. | Genomic RNA in feces at 14 to 17 days p.i. | |
| 43c, untreated | 7.79 ± 0.12a | 4.03 ± 0.02a | 5.78 ± 0.51a |
| 43c, UV treatedb | Undetectable | Undetectable | Undetectable |
| G1217D mutant, untreated | 6.31 ± 0.63b | 2.36 ± 0.09b | 4.56 ± 0.38b |
| G1217D mutant, UV treatedb | Undetectable | Undetectable | Undetectable |
Superscript letters indicate statistically significant differences (P < 0.05).
Detection at all sampling times, not simply at 7 days p.i. (data shown), gave negative results.
Fig 4.
Kinetics of virus excretion in feces. Rats were inoculated with 108 genome copies of either the 43c parental HAV strain or the G1217D mutant, and feces was collected daily in the window period from day 14 to day 21. The excretion pattern of a single animal per group, which represents the average observed pattern, is shown.
Inoculation of 108 genome copies of UV-inactivated strains never resulted in virus detection in the serum, liver, or feces at any sampling time, thus confirming that HAV RNA detection in the different body compartments and feces after inoculation of live strains indeed reflected virus replication, rather than turnover, in the Wistar rat model. However, the replication capacity was limited and very low. A further confirmation of replication was the generation during the course of infection in rats of mutations not present in the consensus sequence of the original population used for inoculation. Eighty percent of viruses isolated at 2 weeks p.i. from sera of rats inoculated with both the 43c parental and G217D mutant strains showed replacements at position 12 of VP3. The replacement detected in the parental strain-derived isolates was A3012G. Among the G217D mutant-derived isolates, two replacements were detected, namely, A3012G in one isolate and A3102V in two other isolates.
In vivo competitive biodistribution of 43c and G1217D mutant strains of HAV.
To confirm the differential efficiency of both strains in reaching the liver, competitive biodistribution experiments were performed. The parental 43c strain clearly outcompeted the G1217D mutant strain in serum in the absence of specific antibodies (Fig. 5A and C). When both strains were inoculated at equal proportions (1:1), only 5 min was necessary to detect a significant (P < 0.05) increase, from 50% ± 0% to 74% ± 10%, for the 43c strain and a decrease, from 50% ± 0% to 26% ± 10%, for the G1217D mutant strain (Fig. 5A). The larger proportion of the 43c strain in serum correlated with an almost complete dominance (94% ± 8%) of the same strain in the liver at 21 days p.i. (Fig. 5A). When the 43c and G1217D mutant strains were inoculated at a 1:9 proportion (Fig. 5C), the 43c strain again increased, from 10% ± 0% to 70% ± 10%, and the G1217D mutant decreased, from 90% ± 0% to 30% ± 9%, although 15 min was necessary to detect such changes in sera (Fig. 5C). Accordingly, in the liver, a proportion of 58% ± 25% strain 43c was observed at 21 days p.i. (Fig. 5C), in spite of its very low inoculated proportion (10% ± 0%). Only when specific antibodies against the glycophorin A binding site were present (Fig. 5B and D) did the G1217D mutant strain (resistant to these antibodies) outcompete the 43c parental strain (sensitive to these antibodies) in serum, with an increase from 50% ± 0% to 70% ± 5% at 5 min when the starting ratio was 1:1 (43c:G1217D mutant) (Fig. 5B) and an increase from 90% ± 0% to 95% ± 2% (Fig. 5D) when the starting ratio was 1:9 (43c:G1217D mutant). However, even under these conditions of immune clearance of the 43c parental strain, the clear dominance of the latter strain in reaching the liver was evident, since in the first case its proportion was 55% ± 20% (Fig. 5B) and in the second case it was 30% ± 7% (Fig. 5D).
Fig 5.
Growth competition experiments. The parental 43c strain of HAV and the G1217D mutant were inoculated into rats in the absence (A and C) or presence (B and D) of the H7C27 MAb, to which the G1217D mutant is resistant. The 43c/G1217D mutant ratios tested were 1:1 (A and B) and 1:9 (C and D). The mean proportion of each virus in serum was determined at different times p.i. (at 21 days p.i. for the liver).
The absolute genome copy numbers per ml of serum and per mg of liver were determined at 60 min and 21 days p.i., respectively, for all rats in each experiment, and the relative numbers of copies of each strain were inferred through the percentages of each strain. The data presented in Table 3 show again that despite the same number of genomes inoculated in each experiment, when most of them were of the mutant type (43c/G1217D mutant ratio of 1:9) the total number of genome copies/ml of serum at 60 min p.i. was statistically lower (P < 0.05) than when equal proportions of the parental and mutant types (1:1) were inoculated. Additionally, only in the latter case did the H7C27 MAb have a statistically significant (P < 0.05) effect on the total number of viruses in serum at 60 min p.i., due to its specific neutralization of the parental strain. However, this different effect of the H7C27 MAb depending on the initial ratio of both strains was not observed in the liver, likely due to the fact that the phenotype of resistance to this MAb shown by the G1217D mutant strain is masked by its higher rate of clearance from blood.
Table 3.
Log10 genome copy numbers per ml of sera at 60 min and per g of liver at 21 days after inoculation of different mixtures of the parental 43c and G1217D mutant strainsa
| Ratio of strain 43c to G1217D mutant | Serum datab |
Liver datab |
||||
|---|---|---|---|---|---|---|
| Absolute titer | Relative titer of 43c | Relative titer of G1217D mutant | Absolute titer | Relative titer of 43c | Relative titer of G1217D mutant | |
| 1:1 | 5.40 ± 0.10a | 5.30 ± 0.10 | 4.60 ± 0.10 | 3.20 ± 0.50a | 3.10 ± 0.50 | 2.7 ± 0.10 |
| 1:1 (with H7C27 MAb) | 4.50 ± 0.10b | 3.50 ± 0.10 | 4.50 ± 0.10 | 2.10 ± 0.20b | 1.90 ± 0.20 | 1.70 ± 0.30 |
| 1:9 | 4.80 ± 0.20b | 4.70 ± 0.20 | 4.10 ± 0.20 | 3.10 ± 0.30a | 3.00 ± 0.40 | 2.60 ± 0.30 |
| 1:9 (with H7C27 MAb) | 4.40 ± 0.10b | 3.30 ± 0.10 | 4.40 ± 0.10 | 2.30 ± 0.20b | 1.70 ± 0.20 | 2.10 ± 0.20 |
The different mixtures were inoculated into rats in the absence or presence of the H7C27 MAb, to which the G1217D mutant strain is resistant. Each rat received a 1-ml inoculum containing 8 log10 genome copies/ml; in the 1:1 mixtures, there were 7.70 log10 genome copies of each virus, and in the 1:9 mixtures, there were 7 log10 genome copies of the 43c strain plus 7.95 log10 genome copies of the G1217D mutant strain.
Data are expressed as means ± standard errors. Different letters indicate statistically significant differences (P < 0.05).
DISCUSSION
From the antigenicity point of view, HAV is a highly stable virus. In fact, only one serotype has been described so far. However, this antigenic stability is not the result of low mutation frequencies in the capsid region, whose values (1 × 10−3 to 1 × 10−4 substitution per nucleotide) are in the range of those for other picornaviruses (34), but of important structural and biological constraints (1, 2). Regarding the latter point, it is intriguing that some mutants with variations in the immunodominant site and very low in vitro fitness compared to the parental virus have been isolated naturally from patients, while natural mutants with variations in the glycophorin A binding site and similar in vitro fitness to that of the parental strain have never been described. This is probably related to the incomplete picture offered by in vitro assays to evaluate the actual in vivo fitness of a virus in its host, as reported very recently (39). Additionally, it has been documented that viral pathogenesis is determined in part by the spread of the virus in the organism to reach the target tissues (28, 38). In this sense, stability at acid pH, stability in the presence of biliary salts and trypsin, and blood clearance may be used as additional measures to predict the in vivo fitness outcomes of different HAV strains. While the first two factors may be tested easily in cell culture, determination of blood clearance kinetics requires an animal model. In the present study, Wistar rats were used as a suitable model, since our purpose was to study not virus replication but virus biodistribution. Nevertheless, we indeed detected virus replication, although at an extremely low level and not associated with clinical manifestations as previously described for guinea pigs (15). Indications of HAV replication in rats were, on the one hand, the detection of the negative-strand RNA in the liver and, on the other hand, the generation of a mutation at residue 12 of VP3 (A3012G; A3012V) during the infection cycle in most of the animals. However, the definite confirmation of replication came from the fact that UV-light treatment of the same virus inocula (parental and mutant strains) rendered neither genomic RNA in the serum, liver, and feces nor antigenomic RNA in the liver. This replication potential, although limited, opened the possibility of comparing the in vivo fitness of both viral strains in the rat model, which, although not perfect, enabled us to gather information that provides a better overview of actual fitness. Overall, the results presented here show that the 43c parental strain has a higher fitness than the G1217D mutant in the rat model and that a small difference in the initial infectious virus input in the liver has a large effect on the final outcome. Only one mutation (G217D mutation in the VP1 protein) differentiates both viruses, with this mutation altering the hemagglutination capacity and leading to increased red blood cell binding of the G1217D mutant, thus indicating that the lower fitness could be due to a higher rate of clearance from serum. In fact, the G1217D mutant showed faster elimination from serum and a slower transit to the liver, while the parental 43c strain had the completely opposite behavior: slower removal from serum and faster transit to the liver. All of these observations indicate a relationship between the amount of “free” viruses and their biodistribution in the liver. This phenomenon may be explained by the greater accumulation of the G1217D mutant in erythrocytes, which later is phagocytosed by Kupffer cells during the elimination of senescent and/or altered erythrocytes (18). The seminal role of delayed blood clearance in the ability to reach the target organs has also been documented for some other viruses, such as mengovirus, Sindbis virus, and adeno-associated parvovirus, among others (6, 5, 19). For both mengovirus and Sindbis virus, highly virulent strains were associated with delayed blood clearances, with half-lives of around 30 min. In contrast, attenuated strains showed shorter half-lives of around 5 min. A similar pattern occurs in HAV, with half-lives of 17 and 8 min for the parental and G1217D mutant strains, respectively.
These results were further confirmed in competition experiments between both HAV strains, which proved the outcompetition capacity of the parental strain over the G1217D mutant in serum and its higher level of delivery to the liver. Even in the presence of antibodies able to specifically neutralize 90% of the inoculated parental virus, its proportion in the infected liver was highly remarkable. Since no differences in replication capacity between both strains have ever been observed either in vitro (data not shown) or in the in vivo model used, with negative-strand RNA copy numbers equivalent in both strains at 14 days p.i., the higher infectivity potential of the parental virus must rely, at least in part, on its smaller erythrocyte-bound fraction and hence its larger “free” virus fraction. Nevertheless, HAV replication in Wistar rats is extremely low compared to replication in chimpanzees (21). The kinetics of HAV replication in both models are quite similar, but the magnitude of replication is much lower in rats, mainly bearing in mind the ratio between the inoculated dose (108 and 107 genomes in rats and chimpanzees, respectively) and animal weight. Genome copy numbers in the liver peaked during the first and the first and third weeks after inoculation in rats and chimpanzees, respectively, while in stool the peak was between the second and third weeks in both models. However, the genome copy numbers in the liver were 3 log lower, on average, in rats than in chimpanzees, and those in feces were between 2 and 3 log lower. This low replication capacity is likely due to an inefficient use of the putative receptor(s) present in rat hepatocytes. Since the inoculated viruses were produced in cell culture, viral particles were not IgA bound, and thus the use of the proposed asialoglycoprotein receptor (ASGPR) (10), which binds and internalizes IgA molecules, must be precluded. On the other hand, the occurrence of an ortholog of the hepatitis A virus cellular receptor 1 (HAVCR1/TIM1) (17) in rats has been proven (24), and provided that it is expressed in hepatocytes, it could be used by HAV to infect liver cells. However, since it is not exactly the same receptor molecule, the binding efficiency is predicted to be low. Due to the extremely low rate of replication, the rat model is not recommended for studies focused on virus-cell interactions, to avoid misleading conclusions. A reliable small-animal model would require adaptation of HAV to grow in such a model or the use of transgenic animals with a humanized liver.
RNA viruses are highly error prone and can use their replication infidelity to adapt to complex environments within an infected host (38). However, viral populations may experience bottlenecks, such as natural host barriers, which limit their diversity (20). Given that the initiation of a HAV infection of the liver must overcome so many bottlenecks, the final outcome is predicted to be highly dependent on the initial dose reaching the organ. HAV has consequently evolved by building a highly stable capsid in and out of the host body that escapes at least one main mechanism of blood clearance. Any mutant failing in one or several of these features will show a decrease in fitness and will be negatively selected in nature. Consequently, these bottlenecks may contribute to the low antigenic diversity of HAV. In this sense, it was also suggested in an earlier study that the immunodominant site of the HAV capsid is involved in receptor interactions and that replacements around this site are prevented due to the lack of fitness of the resulting mutants (22). However, while the specific mutation in the immunodominant site inducing the loss of fitness in chimpanzees reported in the mentioned study was D3070H, some simian isolates able to produce hepatitis A in macaques and African green monkeys bear a D3070A mutation (26, 37). The strains used in the present study (parental and mutant) also bear the D3070A mutation. However, after sequencing of the corresponding genomic region of all viruses isolated from sera of all available rats inoculated with both the 43c parental and G1217D mutant strains at 2 weeks p.i., the reversion of this mutation was not detected (data not shown). The role of the mutation at residue 12 of VP3 detected in most viruses isolated at 2 weeks p.i. from rats inoculated with either the 43c parental strain or the G217D mutant is unknown, although this position is highly variable as concluded from in vitro quasispecies analyses (1). Altogether, these data indicate how critical the balance between antigenic variation, selective constraints, and virus fitness is.
Although it is tempting to speculate that the G1217D mutant may have an attenuated phenotype, the lack of clinical signs of disease in the rat model and the fact that there is not always a correlation between fitness and virulence (13) prevent us from drawing this conclusion.
Overall, this example reflects how a mutation may be far from neutral even when the virus does not present any disadvantage in terms of replication, due to the complex network of the biological cycle. Other examples probably exist regarding the loss of fitness due to less stable capsids in scenarios of different biological bottlenecks and prove once more that RNA viruses live on the edge of error catastrophe (3, 9).
ACKNOWLEDGMENTS
This work was supported in part by the Spanish Ministry of Science and Innovation and the Ministry of Economy (projects BIO2008-01312 and BIO2011-23461, respectively), by Generalitat de Catalunya project 2005SGR00966, and by the Biotechnology Reference Network (XRB).
Footnotes
Published ahead of print 16 May 2012
REFERENCES
- 1. Aragonès L, Bosch A, Pintó RM. 2008. Hepatitis A virus mutant spectra under the selective pressure of monoclonal antibodies: codon usage constraints limit capsid variability. J. Virol. 82:1688–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aragonès L, Guix S, Ribes E, Bosch A, Pintó RM. 2010. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis A virus capsid. PLoS Pathog. 6:e1000797 doi:10.1371/journal.ppat.1000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biebricher CK, Eigen M. 2005. The error threshold. Virus Res. 107:117–127 [DOI] [PubMed] [Google Scholar]
- 4. Bishop NE, Hugo DL, Borovec SV, Anderson DA. 1994. Rapid and efficient purification of hepatitis-A virus from cell-culture. J. Virol. Methods 47:203–216 [DOI] [PubMed] [Google Scholar]
- 5. Byrnes AP, Griffin DE. 2000. Large-plaque mutants of Sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J. Virol. 74:644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell JB, Buera JG, Tobias FM. 1970. Influence of blood clearance rates on interferon production and virulence of Mengo virus plaque mutants in mice. Can. J. Microbiol. 16:821–826 [DOI] [PubMed] [Google Scholar]
- 7. Costafreda MI, Bosch A, Pinto RM. 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 72:3846–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costa-Mattioli M, et al. 2002. Molecular evolution of hepatitis A virus: a new classification based on the complete VP1 protein. J. Virol. 76:9516–9525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Domingo E, Holland JJ. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151–178 [DOI] [PubMed] [Google Scholar]
- 10. Dotzauer A, et al. 2000. Hepatitis A virus-specific immunoglobulin A mediates infection of hepatocytes with hepatitis A virus via the asialoglycoprotein receptor. J. Virol. 74:10950–10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gabrieli R, et al. 2004. Hepatitis in Albanian children: molecular analysis of hepatitis A virus isolates. J. Med. Virol. 72:533–537 [DOI] [PubMed] [Google Scholar]
- 12. Gagneux P, Varki A. 1999. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 9:747–755 [DOI] [PubMed] [Google Scholar]
- 13. Herrera M, Grande-Perez A, Perales C, Domingo E. 2008. Persistence of foot-and-mouth disease virus in cell culture revisited: implications for contingency in evolution. J. Gen. Virol. 89:232–244 [DOI] [PubMed] [Google Scholar]
- 14. Hollinger FB, Emerson SU. 2007. Hepatitis A virus, p 911–947 In Knipe DM, et al. (ed), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 15. Hornei B, et al. 2001. Experimental hepatitis A virus infection in guinea pigs. J. Med. Virol. 64:402–409 [DOI] [PubMed] [Google Scholar]
- 16. International Committee on Taxonomy of Viruses, Van Regenmortel MHV, International Union of Microbiological Societies Virology Division 2000. Family: Picornaviridae, p 657–678 In Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses Academic Press, San Diego, CA [Google Scholar]
- 17. Kaplan G, et al. 1996. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 15:4282–4296 [PMC free article] [PubMed] [Google Scholar]
- 18. Khansari N, Fudenberg HH. 1983. Immune elimination of autologous senescent erythrocytes by Kupffer cells in vivo. Cell. Immunol. 80:426–430 [DOI] [PubMed] [Google Scholar]
- 19. Kotchey NM, et al. 2011. A potential role of distinctively delayed blood clearance of recombinant adeno-associated virus serotype 9 in robust cardiac transduction. Mol. Ther. 19:1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuss SK, Etheredge CA, Pfeiffer JK. 2008. Multiple host barriers restrict poliovirus trafficking in mice. PLoS Pathog. 4:e1000082 doi:10.1371/journal.ppat.1000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lanford RE, et al. 2011. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc. Natl. Acad. Sci. U. S. A. 108:11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemon SM, et al. 1990. In vivo replication and reversion to wild type of a neutralization-resistant antigenic variant of hepatitis A virus. J. Infect. Dis. 161:7–13 [DOI] [PubMed] [Google Scholar]
- 23. Martin A, Lemon SM. 2006. Hepatitis A virus: from discovery to vaccines. Hepatology 43:S164–S172 [DOI] [PubMed] [Google Scholar]
- 24. McIntire JJ, et al. 2001. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat. Immunol. 2:1109–1116 [DOI] [PubMed] [Google Scholar]
- 25. Nainan OV, Brinton MA, Margolis HS. 1992. Identification of amino-acids located in the antibody-binding sites of human hepatitis A virus. Virology 191:984–987 [DOI] [PubMed] [Google Scholar]
- 26. Nainan OV, Margolis HS, Robertson BH, Balayan M, Brinton MA. 1991. Sequence analysis of a new hepatitis A virus naturally infecting cynomolgus macaques (Macaca fascicularis). J. Gen. Virol. 72:1685–1689 [DOI] [PubMed] [Google Scholar]
- 27. Perez-Sautu U, et al. 2011. Hepatitis A virus vaccine escape variants and potential new serotype emergence. Emerg. Infect. Dis. 17:734–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfeiffer JK, Kirkegaard K. 2006. Bottleneck-mediated quasispecies restriction during spread of an RNA virus from inoculation site to brain. Proc. Natl. Acad. Sci. U. S. A. 103:5520–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ping LH, Lemon SM. 1992. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J. Virol. 66:2208–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pinto RM, et al. 1998. Enhancement of the immunogenicity of a synthetic peptide bearing a VP3 epitope of hepatitis A virus. FEBS Lett. 438:106–110 [DOI] [PubMed] [Google Scholar]
- 31. Pinto RM, et al. 2002. Hepatitis A virus polyprotein processing by Escherichia coli proteases. J. Gen. Virol. 83:359–368 [DOI] [PubMed] [Google Scholar]
- 32. Pinto RM, Costafreda MI, Bosch A. 2009. Risk assessment in shellfish-borne outbreaks of hepatitis A. Appl. Environ. Microbiol. 75:7350–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanchez G, et al. 2004. Capsid region involved in hepatitis A virus binding to glycophorin A of the erythrocyte membrane. J. Virol. 78:9807–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanchez G, Bosch A, Gomez-Mariano G, Domingo E, Pinto RM. 2003. Evidence for quasispecies distributions in the human hepatitis A virus genome. Virology 315:34–42 [DOI] [PubMed] [Google Scholar]
- 35. Sanchez G, Bosch A, Pinto RM. 2003. Genome variability and capsid structural constraints of hepatitis A virus. J. Virol. 77:452–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanchez G, Pinto RM, Vanaclocha H, Bosch A. 2002. Molecular characterization of hepatitis A virus isolates from a transcontinental shellfish-borne outbreak. J. Clin. Microbiol. 40:4148–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsarev SA, Emerson SU, Balayan MS, Ticehurst J, Purcell RH. 1991. Simian hepatitis A virus (HAV) strain AGM-27: comparison of genome structure and growth in cell culture with other HAV strains. J. Gen. Virol. 72:1677–1683 [DOI] [PubMed] [Google Scholar]
- 38. Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wargo AR, Kurath G. 2011. In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding. J. Virol. 85:3959–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]