Abstract
Oas1b was previously identified as the product of the Flvr allele that confers flavivirus-specific resistance to virus-induced disease in mice by an uncharacterized, RNase L-independent mechanism. To gain insights about the mechanism by which Oas1b specifically reduces the efficiency of flavivirus replication, cellular protein interaction partners were identified and their involvement in the Oas1b-mediated flavivirus resistance mechanism was analyzed. Initial difficulties in getting the two-hybrid assay to work with full-length Oas1b led to the discovery that this Oas protein uniquely has a C-terminal transmembrane domain that targets it to the endoplasmic reticulum (ER). Two peptides matching to oxysterol binding protein-related protein 1L (ORP1L) and ATP binding cassette protein 3, subfamily F (ABCF3), were identified as Oas1b interaction partners in yeast two-hybrid assays, and both in vitro-transcribed/translated peptides and full-length proteins in mammalian cell lysates coimmunoprecipitated with Oas1b. Knockdown of a partner involved in Oas1b-mediated antiflavivirus activity would be expected to increase flavivirus replication but not that of other types of viruses. However, RNA interference (RNAi) knockdown of ORP1L decreased the replication of the flavivirus West Nile virus (WNV) as well as that of other types of RNA viruses. This virus-nonspecific effect may be due to the recently reported dysregulation of late endosome movement by ORP1L knockdown. Knockdown of ABCF3 protein levels increased the replication of WNV but not that of other types of RNA viruses, and this effect on WNV replication was observed only in Oas1b-expressing cells. The results suggest that Oas1b is part of a complex located in the ER and that ABCF3 is a component of the Flvr-mediated resistance mechanism.
INTRODUCTION
The genus Flavivirus, in the family Flaviviridae, consists of ∼70 viruses and includes human pathogens such as dengue virus, yellow fever virus, tick-borne encephalitis virus, Japanese encephalitis virus, and West Nile virus (WNV) (20). WNV was first isolated in Uganda in 1937 and was previously reported to be endemic in Africa, Australia, and southern Asia (3); it has recently emerged in the Americas, with over 23,000 human infections reported in the United States as of late 2006 (2, 39). WNV is arthropod borne, with a natural transmission cycle typically between Culex mosquito species and birds, with occasional virus transmission by mosquitoes to horses and humans (3). Usually, WNV infections in humans are asymptomatic or cause mild flu-like symptoms. However, some infections cause more severe disease with symptoms such as meningitis, encephalitis, or paralysis, which can be fatal (3).
The 2′-5′ oligoadenylate synthetase (OAS) pathway functions as an innate host defense response against viral infections. OAS gene expression is upregulated by the signaling of interferons produced by cells in response to a viral infection (34). Viral double-stranded RNA (dsRNA) binds to and activates OAS, causing it to polymerize ATP into short 2′-5′-linked oligomers (2-5A) (14). These 2-5A oligomers bind to and activate latent endoribonuclease L (RNase L,) which is constitutively expressed in cells. Activated RNase L cleaves viral and cellular single-stranded RNAs.
Data from numerous studies indicate that both host factors and virus virulence factors determine the outcome of a virus infection. Genetically controlled resistance to flavivirus-induced central nervous system (CNS) disease in mice was first discovered in the 1920s and rediscovered several times in the 1930s because it was not appreciated that all of the viruses being tested belonged to the same virus genus and family (4). Breeding studies with mice displaying differential susceptibility to flavivirus-induced disease showed that the alleles of a single gene, Flv, controlled this phenotype and that resistance was inherited as a Mendelian dominant trait (44, 45). Oas1b was identified by a positional cloning strategy as the Flv gene (30). There are 8 adjacent orthologs of the OAS1 gene (Oas1a to Oas1h) on mouse chromosome 5 that were generated by gene duplication (15, 31). A full-length Oas1b protein is expressed by the Flvr allele, while the presence of a premature stop codon in the Flvs allele results in expression of a truncated Oas1b (Oas1bt) protein. When the Flvs (Oas1bs) allele in homozygous susceptible C57BL/6 mice was replaced with the resistance allele, Flvr (Oas1br), by a homologous recombination strategy, both the heterozygous and homozygous C57BL/6-Oas1bKI mice generated displayed the flavivirus resistance phenotype, confirming that only Oas1b is required for this phenotype (35).
Among the eight Oas1 genes in mice, only Oas1a and Oas1g have been reported to encode functional synthetases (7, 15). A recent study from our lab demonstrated that Oas1b is an inactive synthetase (7). Previous work from our lab showed that although the activation of RNase L had an antiviral effect on a flavivirus infection in both resistant (Flvr) and susceptible (Flvs) mouse embryo fibroblasts (MEFs), RNase L activity and viral titers were consistently higher in susceptible cells that do not express Oas1b (36). Expression of a dominant negative RNase L did not overcome the less efficient virus replication phenotype of resistant MEFs (36). Also, two of the six inactive Oas1 proteins, Oas1b and Oas1d, were reported to act as dominant negative inhibitors of Oas1a synthetase activity (7, 48). The Oas/RNase L pathway is involved in mRNA regulation during cell differentiation and apoptosis as well as during the innate immune response (8, 33), and it was postulated that the inactive mouse Oas1 proteins function to tightly regulate Oas1 production of 2-5A to prevent deleterious effects on the cell (7, 48). The data obtained to date indicate that Oas1b does not exert its flavivirus-specific antiviral activity through the Oas/RNase L pathway (36). The goal of the present study was to identify and characterize novel interaction partners of Oas1b as a means of gaining insights about the mechanism by which this protein specifically reduces the efficiency of flavivirus replication independent of the Oas/RNase L pathway.
MATERIALS AND METHODS
Cell lines.
C3H/He (He), C3H/RV (RV), C57BL/6 (B6), and C57BL/6-Oas1bKI (KI) simian virus 40 (SV40)-transformed mouse embryo fibroblast (MEF) lines, as well as BHK-21 WI2 cells, were maintained in minimal essential medium (MEM) supplemented with 5% heat-inactivated fetal bovine serum (FBS) and 10 μg/ml of gentamicin at 37°C in a 5% CO2 atmosphere.
Viruses.
A stock of lineage 1 WNV strain Eg101 was prepared by infecting a monolayer of BHK cells at a multiplicity of infection (MOI) of 0.1 and harvesting culture fluid at 32 h after infection. Clarified culture fluid (5 × 107 PFU/ml) was aliquoted and stored at −80°C. A Sindbis virus (SinV) strain SAAR339 stock was prepared as a clarified 10% (wt/vol) newborn mouse brain homogenate (7 × 109 PFU/ml). A stock of vesicular stomatitis virus (VSV) strain New Jersey was prepared by infecting BHK cells at an MOI of 1 and harvesting culture fluid at 9 h after infection (109 PFU/ml).
Plaque assay.
Infectivity titers were determined by infecting monolayers of BHK cells in duplicate wells of a six-well plate with serial 10-fold dilutions of virus samples. After adsorption for 1 h at room temperature, the virus inoculum was removed and the wells were overlaid with 1% SeaKem ME agarose (Bio-Whittaker Molecular Applications, Rockland, ME), mixed 1:1 with 2× MEM containing 5% FBS, and incubated for 72 h at 37°C in 5% CO2. Plaque assays for SinV and VSV were performed on BHK cells using the same protocol, but the plates were incubated for 48 h and 24 h, respectively. After removal of the agarose plugs, cells were stained with 0.05% crystal violet in 10% ethanol.
Transient expression of Oas1b in BHK cells.
Oas1b and Oas1bt cDNAs were amplified with primers that added BamHI and NotI restriction sites to the PCR fragment and then subcloned into the pEBG vector (a gift from Tom Hobman, University of Alberta, Edmonton, Alberta, Canada) that expresses a fusion protein with an N-terminal glutathione S-transferase (GST) tag (GST-Oas1b and GST-Oas1bt). BHK cell cultures at approximately 80% confluence were transiently transfected with cDNA constructs expressing GST-Oas1b, GST-Oas1bt, or GST using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. All constructs were verified by sequencing.
Yeast two-hybrid screen.
Yeast two-hybrid screens were performed according to the manufacturer's protocol (Clontech). Murine full-length Oas1b and truncated Oas1bt cDNAs were obtained as previously described (30). Oas1b cDNA was cloned into the pGBKT7 vector (Clontech), which was transformed into AH109 yeast cells. These transformed cells gave very poor mating efficiency. A construct expressing an Oas1b protein with the predicted C-terminal transmembrane domain (Oas1bΔTM) deleted (corresponding to amino acids 1 to 353 of the Oas1b protein) was subsequently generated by PCR amplification from the cDNA sequence using the forward primer 5′-CATATGATGGAGCAGGATCTG-3′ and the reverse primer 5′-GAATTCCTAATACTTCATTGG-3′ and then subcloned into pGBKT7 (Clontech) using the NdeI and EcoRI restriction enzyme sites. All constructs were verified by sequencing. Oas1bΔTM-transformed AH109 cells were mated with Y187 yeast cells pretransformed with a mouse brain cDNA library (prey) (Clontech). The yeast two-hybrid screen yielded a mating efficiency of ∼1.58%, and approximately 2.1 × 106 clones were screened. Diploid yeast cells were plated onto triple-dropout (TDO; -His/-Leu/-Trp) agarose selection plates and incubated at 30°C. Colonies selected from TDO plates were restreaked onto quadruple-dropout (QDO; -Ade/-His/-Leu/-Trp) agarose plates for a more stringent secondary selection. Colonies growing on QDO selection plates were further tested for Gal4-mediated MEL1 activation.
Yeast cotransformation.
A yeast cotransformation analysis was performed according to the manufacturer's protocol (Clontech). Oas1bΔTM-transformed AH109 yeast cells were mated with Y187 yeast cells transformed with an individual positive clone, and diploid cells were plated onto double-dropout (DDO; SD/-Leu/-Trp) selection plates to confirm cotransformation, as well as onto QDO plates to confirm a positive interaction.
In vitro transcription and translation.
Individual positive library clones and bait (Oas1bΔTM or Oas1bt) constructs were separately in vitro transcribed and translated in the presence of 20 μCi of 35S-labeled methionine (EasyTag Met L-[35S]; Perkin Elmer) according to the manufacturer's protocol (Promega).
In vitro coimmunoprecipitation.
Two 35S-methionine-labeled, in vitro-translated peptides were incubated together for 1 h at room temperature. The reaction was divided into two portions; one was incubated with either agarose alone or anti-IgG antibody as the control and the other with antihemagglutinin (anti-HA) antibody (Roche) for 1 h with rotation at 4°C, after which protein G agarose (Roche) was added and rotation was continued at 4°C overnight. The beads were then washed 3 times in lysis buffer (1% Triton X-100, 0.1% SDS, 150 mM NaCl, 50 mM Tris HCl [pH 7.4], and fresh protease inhibitor), 30 μl of 2× sample buffer (20% SDS, 25% glycerol, 0.5 M Tris HCl [pH 6.8], and 0.5% bromophenol blue) was added, and the samples were boiled at 95°C to denature protein complexes. Immunoprecipitated proteins were separated by SDS-PAGE on 10% gels. Gels were washed 3 times in fixing solution (10% acetic acid and 30% methanol) to remove free radiolabeled methionine, incubated for 30 min at room temperature in Autofluor (National Diagnostics), incubated for 5 min at room temperature in an anticracking buffer (7% acetic acid, 7% methanol, and 1% glycerol), and then dried onto 3MM chromatography paper (Whatman) using a model 543 gel dryer (Bio-Rad). Dried gels were exposed to HyBlot CL autoradiography film (Denville) at −80°C, and protein bands were detected by autoradiography.
Mammalian cell coimmunoprecipitation.
For mammalian cell expression, Oas1b cDNA was subcloned into the p3xFLAG-CMV-10 vector (Sigma) using the EcoRI and XbaI restriction sites (p3xFLAG-Oas1b). Full-length Orp1L cDNA was amplified from C3H/He cell RNA with the Orp1LF forward primer (5′-GCCATGAACACAGAAGCAGAACAGCAGCTTCTC-3′) and the Orp1LR reverse primer (5′-ATAAATATCAGGCAAATTGAAGTAGTTTCTGTC-3′) using a ThermoScript reverse transcriptase PCR (RT-PCR) kit with a Platinum Taq DNA high-fidelity polymerase (Invitrogen) and then cloned into the pEF6-V5/HIS-TOPO vector (Invitrogen) to create the pEF6-V5/HIS-TOPO-Orp1L (V5-Orp1L) construct for mammalian cell expression. All constructs were verified by sequencing.
He MEFs were transiently transfected with a p3xFLAG-Oas1b expression plasmid using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Transfected cells were selected with 250 μg/ml of G418 (Sigma) to create a heterogeneous population of cells stably expressing FLAG-Oas1b, referred to as He-Oas1b cells. Clonal cell lines were generated by plating drug-selected cells at a low density, harvesting colonies with cloning rings, and expanding the individual colony cell populations. Multiple clones were selected and analyzed for FLAG-Oas1b protein expression. Clone 12 (He-Oas1b-12) had the highest level of Oas1b expression. He-Oas1b-12 cells were transfected with a pEF6-V5/HIS-TOPO-Orp1L expression vector using Lipofectamine 2000 (Invitrogen). These cells were then selected using 5 μg/ml of blasticidin S hydrochloride (Sigma), as well as G418, to obtain a cell population that stably expressed both FLAG-Oas1b and V5-Orp1L (He-Oas1b/Orp1L). Approximately 2 × 107 cells were washed twice with 1× PBS and, in some experiments, were treated with 0.5 mM dithiobis[succinimidyl propionate] (DSP) for 30 min at room temperature to cross-link cellular proteins. The reaction was quenched with 50 mM Tris (pH 7.5) for 15 min on ice. The cells were rinsed in 1× phosphate-buffered saline (PBS), incubated on ice for 30 min after the addition of 1.5 ml of lysis buffer, and then passed 5 times through a 21-gauge needle. Cell lysates were clarified by centrifugation at 2,000 × g at 4°C for 5 min. The supernatant was divided into two and incubated with either control anti-mouse IgG antibody-conjugated protein G agarose beads (Sigma) or anti-FLAG antibody-conjugated protein G agarose beads (Sigma) at 4°C overnight. The beads were washed 3 times in lysis buffer to remove unbound proteins and centrifuged. Addition, of 2× sample buffer containing β-mercaptoethanol reversed the DSP cross-links. The beads were boiled at 95°C for 5 min, and the samples were sonicated prior to separation of the immunoprecipitated proteins by SDS-PAGE on 10% gels, followed by transfer to a nitrocellulose membrane for 1 h at 100 V. Membranes were blocked for 1 h at room temperature in 5% nonfat dry milk (NFDM), incubated with a primary antibody diluted in 5% NFDM overnight at 4°C, washed 3 times in 1× Tris-buffered saline (TBS)–Tween 20, and then incubated with a secondary antibody diluted in 5% NFDM for 1 h at room temperature. Membranes were washed twice in 1× TBS–Tween 20 and once in 1× TBS and then incubated in SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) for 5 min. Protein bands were detected by autoradiography.
Clonal cell lines expressing shRNA.
KI (Oas1b-expressing) (35) MEFs were transduced with a mixture of 4 lentivirus-packaged small hairpin RNAs (shRNAs) targeting either mouse Abcf3 (sc-140761-V), mouse Orp1L (sc-62716-V), or a nonspecific control shRNA (sc-108060-V) (Santa Cruz Biotechnology) according to the manufacturer's protocol. Heterogeneous cell populations stably expressing the targeted shRNAs were selected using 5 μg/ml of puromycin dihydrochloride (Santa Cruz Biotechnology). Clonal lines were generated by plating cells at a low density, harvesting colonies with cloning rings, and expanding the clonal cell populations. The knockdown efficiency in each clonal population was determined by Western blotting to detect protein and by real-time quantitative RT-PCR (qRT-PCR) to detect mRNA. KI shControl clone 7 (KI shCont-7) cells were used in experiments because they expressed levels of ATP binding cassette protein 3, subfamily F (ABCF3), and oxysterol binding protein (OSBP)-related protein 1L (ORP1L) that were similar to those in wild-type KI cells. The same protocol was used to generate B6 (Oas1bt-expressing) MEF lines stably expressing either a nonspecific shRNA control (shControl clone 3 [B6 shCont-3]) or shRNA targeting mouse Abcf3 mRNA.
To increase the efficiency of the knockdown of ABCF3, KI shCont-7 or KI shAbcf3-8 clonal cells were grown in 6-well plates to 50% confluence (∼1 × 106 cells) and then transduced a second time with lentivirus-packaged shRNAs targeting either mouse Abcf3 or with nonspecific control shRNAs, respectively, as described above. Twenty-four hours after transduction, cells were infected with either WNV (MOI of 1) or SinV (MOI of 1) and harvested at various times after infection.
Transient expression of ABCF3 in RV cells.
Abcf3 cDNA was amplified from C3H/He RNA with the forward primer 5′-CCACTAGTGCCATGGCGACTTGCGCTGATATCCTGCGAAGC-3′ and the reverse primer 5′-CCTCTAGACTGAGGAAGCCCTCCCGGCGGAACTGTTCTTGGAGG-3′ using a ThermoScript RT-PCR kit (Invitrogen) and a Platinum Taq DNA polymerase high-fidelity kit (Invitrogen) and then subcloned into the pEF6-V5/HIS-TOPO vector (Invitrogen) to create the pEF6-V5/HIS-TOPO-Abcf3 (V5-ABCF3) construct for mammalian cell expression. After verification by sequencing, the V5-ABCF3 construct was transiently transfected into RV MEF monolayers at 80% confluence using Lipofectamine LTX with Plus reagent (Invitrogen) according to the manufacturer's protocol.
Antibodies.
Primary antibodies used for immunoblotting were mouse anti-c-Myc and mouse anti-HA (Roche); mouse anti-FLAG, rabbit anti-FLAG, and rabbit anti-ABCF3 (Sigma); goat anti-V5 (Bethyl Laboratories); rabbit anti-ORP1L (a gift from Vesa Olkkonen, Minerva Foundation Institute for Medical Research, Helsinki, Finland); and goat anti-NS3 (R&D Systems). The primary antibodies used for confocal microscopy were mouse anti-GST and rabbit anti-GST (Cell Signaling), rabbit anticalnexin (Sigma), mouse anti-CoxIV (Invitrogen), and mouse anti-dsRNA (English & Scientific Consulting, Szirak, Hungary). Secondary antibodies used for immunoblotting were anti-rabbit-horseradish peroxidase (anti-rabbit-HRP), anti-mouse-HRP (Cell Signaling), and anti-goat-HRP (SCBT). Secondary antibodies used for confocal microscopy were donkey anti-mouse Alexa Fluor 594, donkey anti-mouse Alexa Fluor 488, donkey anti-rabbit Alexa Fluor 488, and donkey anti-rabbit Alexa Fluor 594 (Invitrogen).
Confocal microscopy.
BHK cells (∼80% confluent) on 3-mm coverslips (Fisher Scientific) in 24-well plates, transfected as described above, were infected with WNV Eg101 at an MOI of 5. At 24 h after infection, cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, permeabilized with 0.1% Triton X-100 in PBS, rinsed three times with PBS, and incubated overnight at 4°C in blocking buffer (5% horse serum in PBS). Primary antibodies were diluted in blocking buffer and incubated with cells for 1 h. The cells were washed three times in PBS and then incubated with the secondary antibodies and 0.5 μg/ml of Hoechst 33258 dye (Invitrogen) for 1 h. Coverslips were mounted onto glass slides using ProLong Gold antifade reagent (Invitrogen), and images were taken with an LSM 510 confocal microscope (Zeiss, Oberkochen, Germany) using a 63× water immersion objective. Images were analyzed using Zeiss LSM Image Browser software.
qRT-PCR.
Total cellular RNA was isolated from B6 and KI cells with TriReagent (Molecular Research Center). Cellular and viral RNAs were quantified using a TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems) according to the manufacturer's protocol. Mouse intracellular Abcf3 mRNA levels were measured using predesigned primer and probe set Mm_00658695_m1 (Applied Biosystems). WNV Eg101 RNA was measured as previously described using WNV NS1 region-specific primers and probe (36). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an endogenous control for each sample and was detected using mouse GAPDH primers and probe (4352339E; Applied Biosystems). One-step RT-PCR was performed for each target gene and for the endogenous control in a singleplex format using 200 ng of RNA and the TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems). The cycling parameters were as follows: reverse transcription at 48°C for 30 min, AmpliTaq activation at 95°C for 10 min, denaturation at 95°C for 15 s, and annealing/extension at 60°C for 1 min (cycle repeated 40 times). Each experiment was done in triplicate and repeated at least three times. Triplicate cycle threshold (CT) values were analyzed with Microsoft Excel using the comparative CT (ΔΔCT) method of the SDS Applied Biosystems software, which also applied statistical analysis to the data (TINV test in Microsoft Excel). The values were normalized to those for GAPDH in the same sample and presented as the relative fold change compared to the 2-h infected-control calibrator sample in relative quantification (RQ) units. Error bars represent the standard error (SE) and indicate the calculated minimum (RQMin) and maximum (RQMax) of the mRNA expression levels based on an RQMin/RQMax of the 95% confidence level. The expression levels between two samples were considered statistically different (P value of <0.05) when the error bars did not overlap.
Cell viability [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT)] assay.
Cells at ∼100% confluence in 96-well plates were infected with WNV Eg101 at an MOI of 1. Cell viability was determined at 24 and 48 h after infection using a CellTiter 96 nonradioactive cell proliferation assay done according to the manufacturer's protocol (Promega).
RESULTS
Identification of putative Oas1b binding partners in a yeast two-hybrid screen.
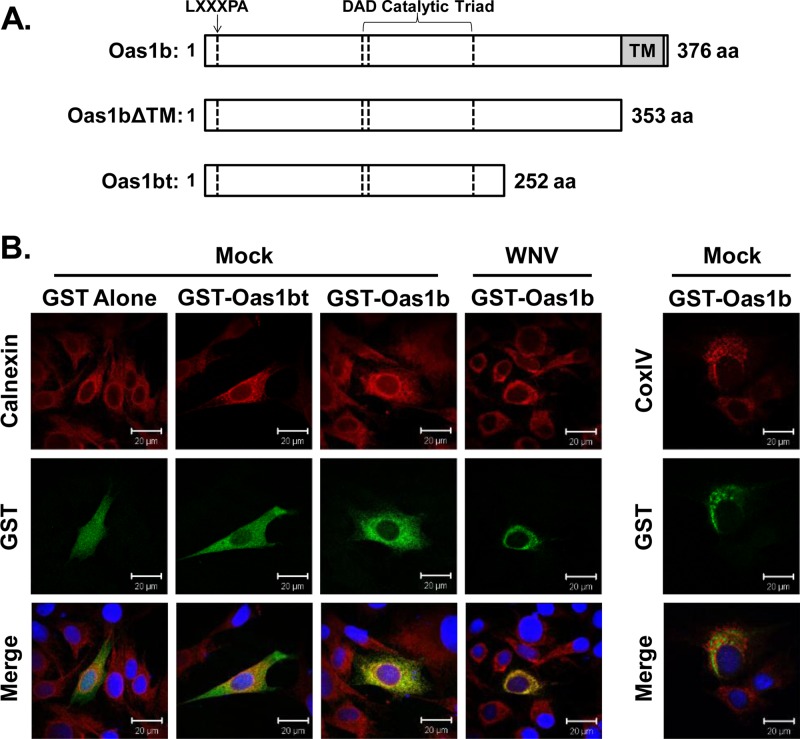
The conversion of B6 mice from a flavivirus-susceptible to a flavivirus-resistant phenotype by knocking in the Oas1br allele confirmed that resistance is mediated by Oas1b (35). However, the Oas1b protein was shown to be an inactive 2-5A synthetase and not to mediate its flavivirus-specific antiviral activity through an RNase L-dependent mechanism (7, 36). A yeast two-hybrid screen was used to identify candidate cell protein binding partners for Oas1b that might participate in the Oas1b-mediated antiflaviviral mechanism. In initial experiments, a pretransformed mouse brain cDNA library was screened against a full-length Oas1b bait as described in Materials and Methods, but no viable yeast colonies were detected on the selection plates. It was previously reported that fusion proteins with a transmembrane domain that tethers them to cytoplasmic membranes and prevents their translocation to the nucleus were not detectable as binding partners in yeast two-hybrid screens (10). Analysis of the Oas1b sequence using the PredictProtein software (www.predictprotein.org) predicted an 18-amino-acid transmembrane domain (aa 354 to 371) at the C terminus (Fig. 1A). To determine whether the predicted transmembrane domain was able to tether Oas1b to a cytoplasmic membrane, BHK cells were transfected with a vector expressing GST-tagged full-length Oas1b protein and the localization of the recombinant protein was analyzed by confocal microscopy. Oas1b colocalized with the endoplasmic reticulum (ER) marker calnexin, but not with the mitochondrial marker CoxIV (Fig. 1B). Oas1b remained localized to the ER in WNV-infected cells (Fig. 1B), even though the ER membrane was observed to condense in the perinuclear region of infected cells. In contrast, transient expression of GST-tagged truncated Oas1b (GST-Oas1bt) or GST alone produced protein that was dispersed throughout the cell with no evident ER colocalization (Fig. 1B). These results indicate that full-length Oas1b protein is anchored to the ER membrane by its C-terminal transmembrane domain. No other known OAS protein has a transmembrane domain.
Fig 1.
Full-length Oas1b contains a C-terminal transmembrane domain that localizes it to the ER membrane. (A) Schematic representation of the full-length Oas1b protein (product of the Flv resistance allele), Oas1bΔTM (a constructed deletion mutant that lacks the C-terminal transmembrane tail), and Oas1bt (product of the Flv susceptibility allele that has a premature stop codon). aa, amino acids. (B) Plasmids expressing GST, GST-Oas1bt, or GST-Oas1b were transfected into BHK cells, and 24 h later cells were either mock infected or infected with WNV Eg101 (MOI of 5). At 24 h after infection, cells were fixed, permeabilized, and analyzed by confocal microscopy. Anti-GST antibody (green) detected GST-tagged proteins, anticalnexin antibody (red) detected ER, anti-CoxIV antibody (red) detected mitochondria, and Hoechst stain (blue) detected nuclei.
A mutant Oas1b with the transmembrane tail deleted (Oas1bΔTM) (Fig. 1A) was next cloned into the bait vector pGBKT7 and analyzed in a yeast two-hybrid screen against the mouse brain cDNA library. Twenty-three unique candidate partner peptides were identified. The interaction between each of these peptides and Oas1bΔTM was then retested by yeast cotransformation as described in Materials and Methods. An interaction with Oas1bΔTM was confirmed for 8 of the original 23 putative peptide partners by yeast cotransformation on quadruple-dropout selection medium.
Coimmunoprecipitation of putative binding partners and Oas1b.
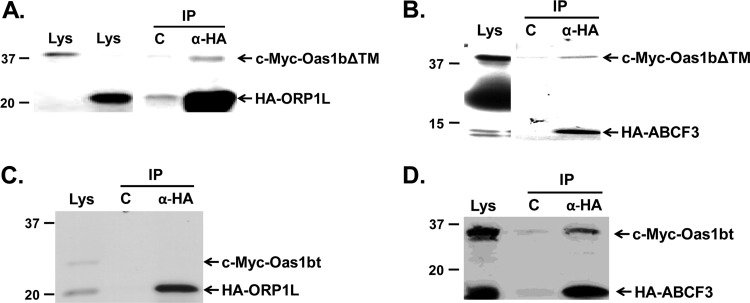
Since yeast two-hybrid analyses often produce false-positive results, in vitro coimmunoprecipitation assays done as described in Materials and Methods were next used to further confirm that the putative peptide partners interacted with Oas1bΔTM. Oas1bΔTM fused to an N-terminal c-Myc tag and one of the putative partners fused to an N-terminal HA tag were in vitro transcribed and translated in the presence of [35S]methionine, and the mixed products were reciprocally immunoprecipitated. Oas1bΔTM coimmunoprecipitated with only two of the eight peptides tested; one of these was predicted by a BLAST analysis to match with a region of oxysterol binding protein-related protein 1L (ORP1L) (Fig. 2A) and the other with a region of ATP binding cassette protein 3, subfamily F (ABCF3) (Fig. 2B). In vitro interaction between each of these peptides and N-terminal c-Myc-tagged Oas1bt was also tested. The ORP1L peptide did not coimmunoprecipitate with Oas1bt (Fig. 2C), suggesting that this peptide interacts with Oas1bΔTM in a region located between the premature stop codon of Oas1bt and the transmembrane domain of Oas1b (Fig. 1A). In contrast, the ABCF3 peptide coimmunoprecipitated with Oas1bt (Fig. 2D), suggesting that this peptide interacts with both Oas1b and Oas1bt in a region upstream of the premature stop codon of Oas1bt.
Fig 2.
In vitro coimmunoprecipitation of Oas1bΔTM and putative partner peptides ORP1L and ABCF3. Tagged Oas1bΔTM or Oas1bt and a putative tagged partner peptide were individually translated in vitro in the presence of [35S]methionine, mixed, and then immunoprecipitated. Immunoprecipitated (IP) protein complexes were separated by SDS-PAGE and detected by autoradiography. (A) 35S-labeled c-Myc-Oas1bΔTM and an HA-ORP1L peptide immunoprecipitated with anti-HA antibody. (B) 35S-labeled c-Myc-Oas1bΔTM and an HA-ABCF3 peptide immunoprecipitated with anti-HA antibody. (C) 35S-labeled c-Myc-Oas1bt and an HA-ORP1L peptide immunoprecipitated with anti-HA antibody. (D) 35S-labeled c-Myc-Oas1bt and an HA-ABCF3 peptide immunoprecipitated with anti-HA antibody. Each experiment was performed at least twice, and representative autoradiographs are shown. Lys, lysate; C, control (agarose beads alone [A and B] or anti-mouse IgG antibody [C and D]; α-HA, anti-HA antibody. The positions of protein size markers are indicated on the left side of each panel.
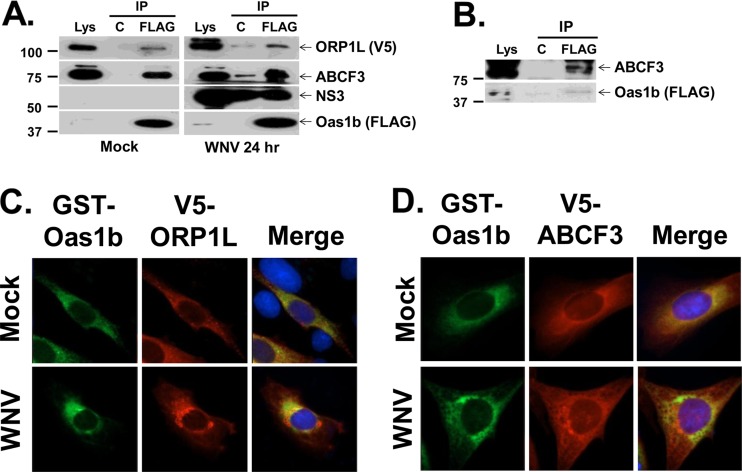
Because a peptide representing only a portion of each of the putative Oas1b partners had been tested in the nonmammalian cell interaction assays, the interaction between Oas1b and each of the two full-length partners was next tested in mammalian cells by coimmunoprecipitation. Because of the high degree of homology between the eight duplicated mouse Oas1 proteins, an antibody that can specifically recognize individual mouse Oas1 proteins is not available. Therefore, He MEFs stably expressing both 3× FLAG-tagged full-length Oas1b and V5-tagged ORP1L (He-Oas1b/Orp1L) were generated, and the interaction between Oas1b and either ORP1L or ABCF3 (endogenous) was assessed by coimmunoprecipitation using cell lysates as described in Materials and Methods. In some experiments, cells were treated, prior to lysis, with DSP, a thiol-reversible chemical protein-cross-linker, to stabilize transient or weak protein-protein interactions. Antibody to the Oas1b FLAG tag coimmunoprecipitated both ORP1L and ABCF3 from cross-linked He-Oas1b/Orp1L MEF lysates that were mock infected or infected for 24 h with WNV Eg101 at an MOI of 1 (Fig. 3A). In the absence of the cross-linker, ABCF3 consistently efficiently coprecipitated with Oas1b (Fig. 3B), while ORPL1 coprecipitation was less efficient and the extent was variable (data not shown). The anti-FLAG antibody also coimmunoprecipitated WNV NS3, suggesting the possibility that the Oas1b complex may interact with WNV replication complexes (Fig. 3A). However, NS3 was also immunoprecipitated by the control anti-mouse IgG antibody agarose.
Fig 3.
Coimmunoprecipitation of ORP1L and ABCF3 with Oas1b from mammalian cell lysates. (A) He-Oas1b/ORP1L MEFs that stably express FLAG-tagged Oas1b and V5-tagged ORP1L were either mock infected or infected with WNV Eg101 (MOI of 1), cross-linked with DSP, quenched, and lysed at 24 h after infection. Lysates were immunoprecipitated with either a control antibody or anti-FLAG antibody, and immunoprecipitated (IP) proteins were detected by Western blotting: ORP1L (anti-V5 antibody), ABCF3 (anti-ABCF3 antibody), NS3 (anti-NS3 antibody), and Oas1b protein (anti-FLAG antibody). Each experiment was repeated at least twice, and a representative blot is shown. Lys, lysate; C, control (anti-IgG antibody-conjugated agarose); FLAG, anti-FLAG antibody-conjugated agarose. The positions of protein size markers are indicated on the left. (B) Cell lysates from He-Oas1b cells that stably express FLAG-tagged Oas1b were immunoprecipitated with either a control antibody or anti-FLAG antibody, and immunoprecipitated proteins were detected by Western blotting. Plasmids expressing GST-Oas1b and V5-ORP1L (C) or V5-ABCF3 (D) were transiently cotransfected into BHK cells, and 24 h later cells were either mock infected or infected with WNV Eg101 (MOI of 5). At 24 h after infection, cells were fixed, permeabilized, and analyzed by immunofluorescence. Anti-GST antibody (green) detected GST-Oas1b, anti-V5 antibody (red) detected V5-tagged proteins, and Hoechst stain (blue) detected nuclei.
To further confirm the interaction of full-length mammalian Oas1b with ORP1L or ABCF3, GST-tagged Oas1b and either V5-tagged ORP1L or V5-tagged ABCF3 were cotransfected into BHK cells. Twenty-four hours after transfection, cells were either mock infected or infected with WNV Eg101 at an MOI of 5 and analyzed for colocalization. Consistent with the coimmunoprecipitation data, GST-Oas1b and V5-ORP1L partially colocalized (Fig. 3C), while GST-Oas1b and V5-ABCF3 strongly colocalized (Fig. 3D), in mock- and virus-infected cells.
Effect of reducing ORP1L protein levels on various single-stranded RNA virus infections.
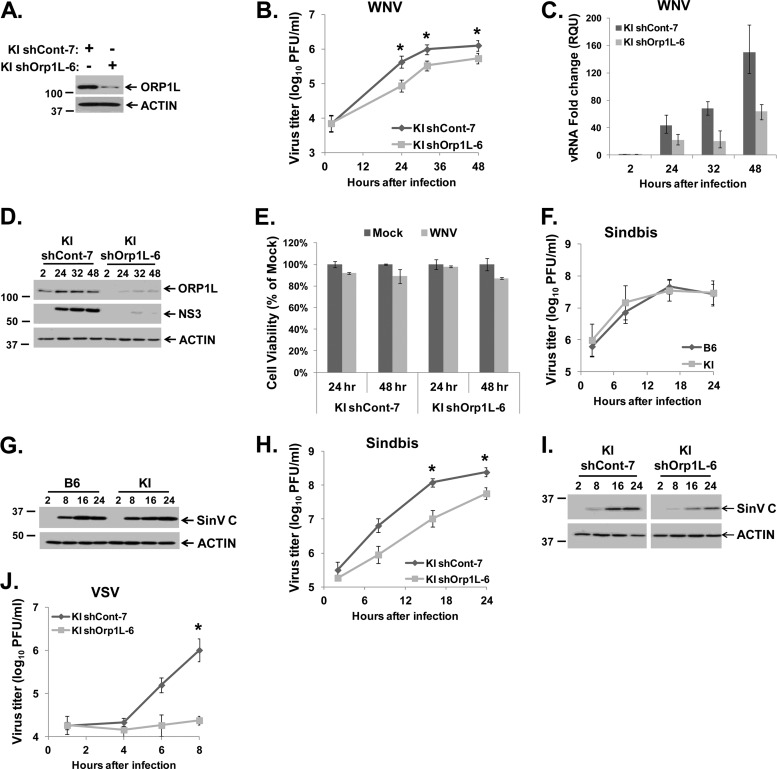
As one means of determining whether ORP1L is specifically involved in Oas1br-mediated flavivirus resistance, the effect of knocking down ORP1L protein levels with RNA interference (RNAi) on WNV replication was first analyzed. It was hypothesized that if ORP1L was required for the genetic resistance phenotype, the extracellular WNV yield as well as intracellular viral RNA and protein levels would increase when ORP1L levels were decreased. ORP1L protein expression was reduced by stable expression of an ORP1L-specific shRNA in resistant KI MEFs, and then clonal cell lines were selected as described in Materials and Methods. Among the 19 lines tested, the KI shOrp1L-6 clone had the lowest ORP1L protein levels compared to that in the nonspecific control shRNA clone, KI shCont-7 (Fig. 4A). KI shCont-7 and KI shOrp1L-6 cultures were infected with WNV Eg101 at an MOI of 1, and clarified culture fluid and either total cell RNA or cell lysates were harvested at various times after infection from duplicate wells. Virus yields were measured by plaque assay on BHK cells. Counter to what was expected, lower WNV yields were produced by KI shOrp1L-6 cells at 24 to 48 h after infection than from KI shCont-7 cells (Fig. 4B). Intracellular viral RNA levels were assessed by real-time qRT-PCR, and intracellular viral protein levels were detected by Western blotting. Both WNV RNA (Fig. 4C) and NS3 protein levels (Fig. 4D) were also reduced in KI shOrp1L-6 cells compared to KI shCont-7 at 24 through 48 h after infection. A loss in cell viability of approximately 15% was detected by an MTT assay at 48 h after WNV infection in both types of shRNA-expressing cell lines compared to mock-infected shRNA-expressing cells (Fig. 4E), suggesting that the decreased viral yields observed were not due to off-target cytotoxic effects of the shRNA. It was noted that the efficiency of ORP1L knockdown by shRNA decreased with time after WNV infection, suggesting that the RNAi pathway may be antagonized in infected cells (Fig. 4D).
Fig 4.
Effect of ORP1L knockdown on WNV, SinV, and VSV infections in Oas1b-expressing MEFs. (A) Western blot analysis of ORP1L protein levels in KI shCont-7 and KI shOrp1L-6 MEFs with anti-ORP1L R247 antibody. (B to E) KI shCont-7 and KI shOrp1L-6 MEFs were infected with WNV Eg101 at an MOI of 1. (B) Extracellular virus yields from infected KI shCont-7 and KI shOrp1L-6 MEFs were measured by plaque assay on BHK cells. (C) Intracellular viral RNA levels were assessed by real-time qRT-PCR. Each experiment was done in triplicate and repeated at least twice. Representative data from one experiment are shown. The viral RNA levels were normalized to GAPDH mRNA in the same sample and are shown as fold change over the amount of viral RNA at 2 h after infection. Error bars represent SEs. (D) Intracellular viral protein (NS3) levels were detected by Western blotting. (E) MTT assay of the viability of KI shCont-7 and KI shOrp1L-6 MEFs mock infected or infected with WNV for 24 or 48 h. Each sample was normalized to the mock from the same time. Error bars represent standard deviations. (F and G) Wild-type B6 (Oas1bt-expressing) and KI (Oas1b-expressing) MEFs were infected with SinV at an MOI of 20. (F) Extracellular SinV yields from infected B6 and KI MEFs were determined by plaque assay on BHK cells. (G) Intracellular SinV capsid (C) protein levels in B6 and KI-infected MEF lysates were detected by Western blotting. (H and I) KI shCont-7 and KI shOrp1L-6 cells were infected with SinV at an MOI of 20. (H) Extracellular SinV yields from infected KI shCont-7 and KI shOrp1L-6 MEFs were measured by plaque assay on BHK cells. (I) Intracellular SinV capsid protein levels in infected KI shCont-7 and KI shOrp1L-6 MEF lysates were detected by Western blotting. (J) KI shCont-7 and KI shOrp1L-6 cells were infected with VSV at an MOI of 5, and extracellular VSV yields were measured by plaque assay on BHK cells. (B, F, H, and J) Each data point is the average of duplicate titrations from at least two experiments. Error bars represent SEs. (A, D, G, and I) Each experiment was performed at least twice, and a representative blot is shown. *, statistical significance compared to the control from the same time after infection was determined at a 95% confidence interval (P < 0.05).
The antiviral activity of Oas1b was previously reported to be specific to flaviviruses (4). To confirm this, susceptible wild-type B6 and resistant KI MEFs were infected with the alpha togavirus SinV, a member of another positive-strand RNA virus family, at an MOI of 20, and clarified culture fluid and infected cell lysates were harvested at various times after infection. As expected, virus yields and intracellular viral protein levels from the two types of cells were similar (Fig. 4F and G). There was also no difference in virus yields from the two cell types after infection with an MOI of 1 (data not shown). If ORP1L is involved in the Oas1b-mediated resistance mechanism, its reduction would not be expected to affect an infection with a virus from another family. KI shCont-7 and KI shOrp1L-6 MEFs were infected with SinV at an MOI of 20. SinV yields from KI shOrp1L-6 MEFs were reduced by ∼0.5 to 1 log compared to those from KI shCont-7 MEFs (Fig. 4H), and SinV capsid protein levels were lower in KI shOrp1L-6 MEFs than in KI shCont-7 MEFs (Fig. 4I). The yields of the negative-strand RNA virus vesicular stomatitis virus (VSV) (MOI of 5) were also reduced in KI shOrp1L-6 MEFs compared to KI shCont-7 MEFs (Fig. 4J). Similar results were also obtained for each of these viruses after infections with an MOI of 1 (data not shown). Knockdown of ORP1L was recently reported to enhance the movement of late endosomes (43). This effect would be expected to negatively impact the efficiency of infection and replication of viruses that enter cells by endocytosis. Decreased replication of three viruses that enter cells by endocytosis was observed in this study. Interestingly, VSV, which has the fastest fusion time of the three viruses (25), was affected to the greatest extent.
Effect of reducing ABCF3 protein levels on single-stranded RNA virus infections.
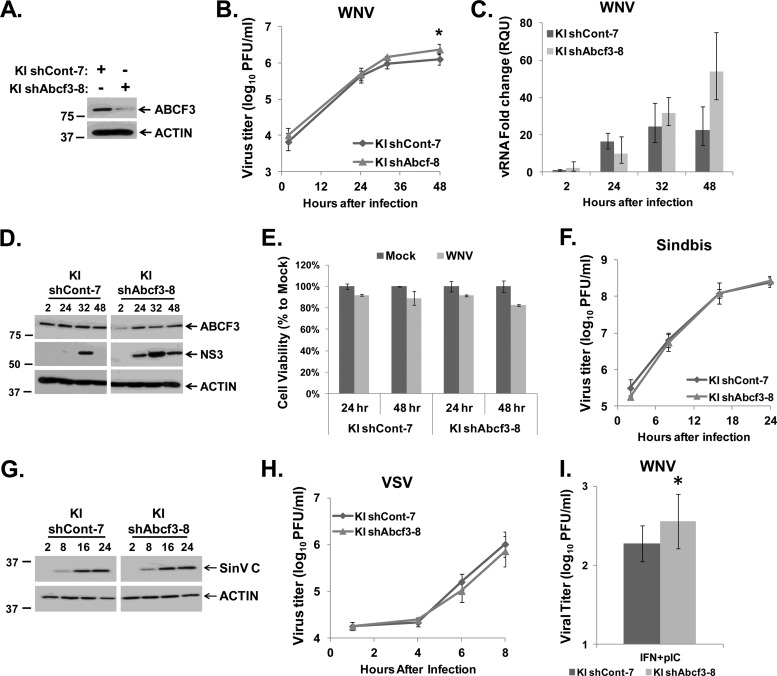
The effect of reducing ABCF3 protein levels on WNV infection was also analyzed using RNAi. shRNAs targeting ABCF3 or nonspecific control shRNAs were transduced into resistant KI MEFs, and then stably expressing clonal cell lines were selected as described in Materials and Methods. Of the 21 lines tested, the KI shAbcf3-8 clonal cell line showed the most efficient knockdown of ABCF3 protein compared to the control cell line KI shCont-7 (Fig. 5A). Replicate cultures of KI shCont-7 and KI shAbcf3-8 MEFs were infected with WNV Eg101 at an MOI of 1, and clarified culture fluid and either total cell RNA or cell lysates were harvested at various times after infection from duplicate wells. A slight but statistically significant increase in WNV yields from the KI shAbcf3-8 cells was consistently observed at 32 and 48 h after infection compared to those from KI shCont-7 cells (Fig. 5B). An increase in the levels of intracellular WNV viral RNA and NS3 protein was also observed in KI shAbcf3-8 MEFs compared to those in KI shCont-7 cells (Fig. 5C and D), suggesting that ABCF3 is antiviral. An MTT assay on the KI shCont-7 and KI shAbcf3-8 cells at 24 and 48 h after infection with WNV Eg101 at an MOI of 1 detected about a 15% decrease in cell viability at 48 h after infection in both cell lines compared to mock-infected cells (Fig. 5E). These results indicate that the increase in WNV replication was due to knockdown of ABCF3 and that the shRNAs were not cytotoxic.
Fig 5.
Effect of ABCF3 knockdown on WNV, SinV, and VSV infections in Oas1b-expressing MEFs. (A) Western blot analysis of ABCF3 protein levels in KI shCont-7 and KI shAbcf3-8 MEFs. (B to E) KI shCont-7 and KI shAbcf3-8 MEFs were infected with WNV Eg101 at an MOI of 1. (B) Extracellular virus yields from infected KI shCont-7 and KI shAbcf3-8 MEFs were measured by plaque assay on BHK cells. (C) Intracellular viral RNA levels were assayed by real-time qRT-PCR. Each experiment was done in triplicate and repeated at least twice, and representative data from one experiment are shown. The viral RNA levels were normalized to GAPDH mRNA in the same sample and are shown as fold change over the amount of viral RNA at 2 h after infection. Error bars represent SEs. (D) Intracellular viral protein (NS3) levels were detected by Western blotting. (E) MTT assay of the viability of KI shCont-7 and KI shAbcf3-8 MEFs mock-infected or infected with WNV for 24 or 48 h. Each sample was normalized to the mock from the same time. Error bars represent standard deviations. (F and G) KI shCont-7 and KI shAbcf3-8 MEFs were infected with SinV at an MOI of 20. (F) Extracellular SinV yields from infected KI shCont-7 and KI shAbcf3-8 MEFs were measured by plaque assay on BHK cells. (G) Intracellular SinV capsid protein levels in KI shCont-7 and KI shAbcf3-8 MEF lysates were detected by Western blotting. (H) KI shCont-7 and KI shAbcf3-8 MEFs were infected with VSV at an MOI of 5, and extracellular virus yields were measured by plaque assay on BHK cells. (I) KI shCont-7 and KI shAbcf3-8 MEFs were pretreated with 1,000 IU/ml of IFN-β for 4 h and 10 μg/ml of poly(I·C) for 1 h prior to infection with WNV Eg101 at an MOI of 1. Forty-eight hours after infection, culture fluid was harvested and extracellular virus yields were measured by plaque assay on BHK cells. (B, F, H, and I) Each data point is the average of duplicate titrations from at least two experiments. Error bars represent SEs. (A, D, and G) Each experiment was performed at least twice, and a representative blot is shown. *, statistical significance compared to the control from the same time after infection was determined at a 95% confidence interval (P < 0.05).
If ABCF3 is involved in the flavivirus-specific, Oas1b-mediated resistance mechanism, then reducing ABCF3 would not affect the efficiency of an infection with a virus from another family. To test this hypothesis, the effect of ABCF3 protein knockdown on SinV and VSV infections was analyzed. KI shCont-7 and KI shAbcf3-8 MEF cultures were infected with SinV (MOI of 20). Neither virus yields (Fig. 5F) nor intracellular SinV capsid protein levels (Fig. 5G) changed when ABCF3 protein levels were reduced. Similarly, no effect on virus yields was observed when KI shAbcf3-8 MEFs were infected with VSV (MOI of 5) (Fig. 5H). Similar results were obtained when infections were done with SinV or VSV at an MOI of 1 (data not shown). These data indicate that the antiviral activity of ABCF3 is specific to flaviviruses.
The effect of interferon (IFN) and/or poly(I·C) treatment on WNV replication in susceptible and resistant MEFs was previously reported (36). Treatment of the two types of cells with either IFN-β (1,000 IU/ml for 4 h prior to infection) or poly(I·C) (10 μg/ml for 1 h prior to infection) alone reduced virus yield ∼10-fold. However, when susceptible He MEFs were treated with both IFN-β and poly(I·C), a 2-log reduction in virus yield was observed, whereas virus yields were reduced by more than 3 logs in resistant RV MEFs. These results suggested that Oas1b contributed to the increased effect seen in resistant cells. It was expected that if ABCF3 is a component of the Oas1b resistance mechanism, then reducing ABCF3 protein levels in cells pretreated with IFN-β and poly(I·C) would result in an increase in viral yields. KI shCont-7 and KI shAbcf3-8 cells were treated with 1,000 IU/ml of IFN-β for 4 h prior to infection and with 10 μg/ml of poly(I·C) for 1 h prior to infection with WNV Eg101 at an MOI of 1. The results showed that reducing ABCF3 protein levels led to a small but significant increase in viral titers at 48 h after infection (Fig. 5I).
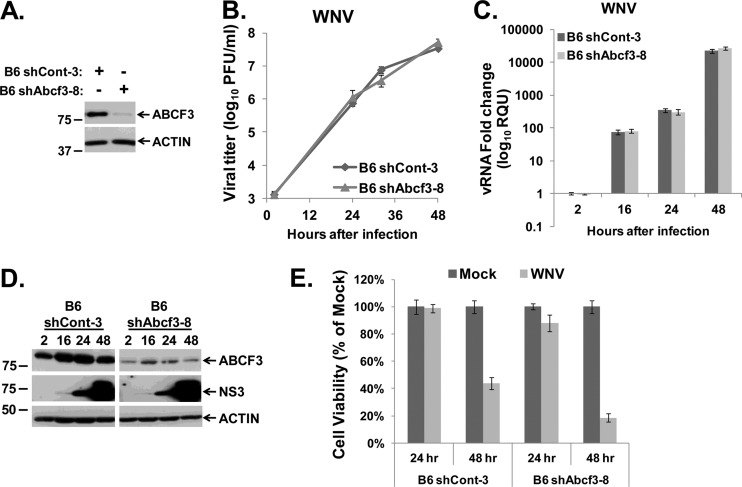
To test whether the antiflavivirus activity of ABCF3 requires the presence of Oas1b, ABCF3 protein levels were reduced in susceptible B6 MEFs that express Oas1bt. B6 MEFs were transduced with shRNAs targeting ABCF3 or with a nonspecific control shRNA, and then clonal cell lines were selected as described in Materials and Methods. Of the 8 lines tested, the B6 shAbcf3-8 clonal cell line had the most efficient reduction of ABCF3 protein compared to the control B6 shCont-3 cells (Fig. 6A). B6 shCont-3 and B6 shAbcf3-8 MEFs were infected with WNV Eg101 at an MOI of 1, and samples were collected as described above. As reported previously (36), WNV replicated more efficiently in susceptible B6 MEFs than in resistant KI MEFs (Fig. 5B and 6B). Neither the virus yield nor the intracellular viral RNA or protein levels were affected by ABCF3 knockdown in B6 MEFs (Fig. 6B to D). The higher level of virus replication in B6 cells resulted in more cytopathic effect (CPE). An MTT assay showed a 50% or greater decrease in cell viability in both B6 shCont-3 and B6 shAbcf3-8 MEFs infected with WNV Eg101 at 48 h after infection compared to mock-infected cells, suggesting that the results observed were virus specific and not due to off-target cytotoxic effects of the shAbcf3 shRNA (Fig. 6E). These results indicate that the flavivirus-specific, antiviral activity of ABCF3 requires the presence of Oas1b and provide additional evidence that ABCF3 is a component of the Oas1b-mediated, flavivirus-specific resistance mechanism.
Fig 6.
Effect of ABCF3 knockdown on WNV infection in Oas1bt-expressing MEFs. (A) Western blot analysis of ABCF3 protein levels in B6 shCont-3 and B6 shAbcf3-8 MEFs. (B to E) B6 shCont-3 and B6 shAbcf3-8 MEFs were infected with WNV Eg101 at an MOI of 1. (B) Extracellular virus yields from infected B6 shCont-3 and B6 shAbcf3-8 MEFs were determined by plaque assay on BHK cells. Each data point is the average of duplicate titrations from at least two experiments. Error bars represent SEs. (C) Intracellular viral RNA levels were assayed by real-time qRT-PCR. Each experiment was done in triplicate and repeated at least twice, and representative data from one experiment are shown. The viral RNA levels were normalized to GAPDH mRNA in the same sample and are shown as fold change over the amount of viral RNA at 2 h after infection. Error bars represent SEs. (D) Intracellular viral protein (NS3) levels were detected by Western blotting. Each experiment was performed at least twice, and a representative blot is shown. (E) MTT assay of the viability of B6 shCont-7 and B6 shAbcf3-8 MEFs mock infected or infected with WNV for 24 or 48 h. Each sample was normalized to the mock sample from the same time. Error bars represent standard deviations.
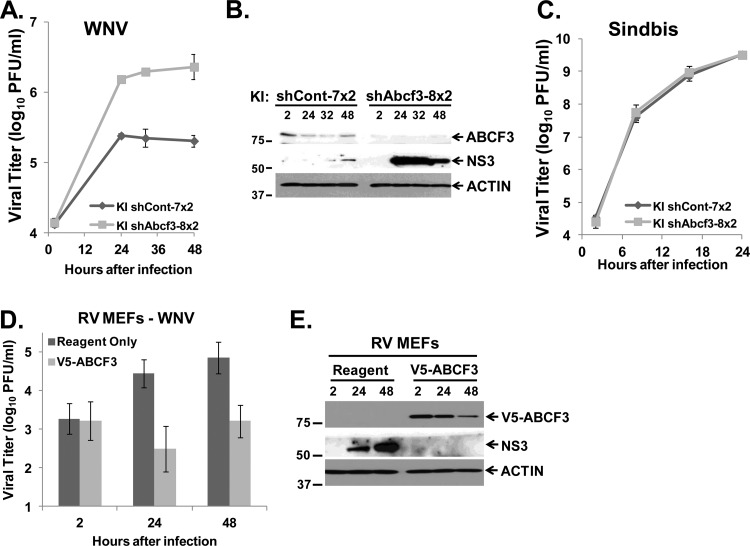
The knockdown of ABCF3 protein levels was only partial and appeared to slightly recover during the course of a WNV infection. To obtain a greater level of ABCF3 knockdown, KI shCont-7 and KI shAbcf3-8 clonal cells were transduced a second time with control and Abcf3 lentiviral shRNAs, respectively, and then infected with WNV Eg101 at an MOI of 1. Compared to the original clonal cells (Fig. 5B and D), a >3-fold increase in extracellular virus yield as well as increased intracellular viral protein levels were observed in the KI cells retransduced with shABCF3 (Fig. 7A and B). However, no difference was observed in the viral yields from these retransduced cells after infection with SinV at an MOI of 1 (Fig. 7C).
Fig 7.
Further analysis of the involvement of ABCF3 in Oas1b-expressing MEFs. KI shCont-7 and KI shAbcf3-8 MEFs were transduced a second time with control and Abcf3-specific lentiviral shRNAs, respectively. Twenty-four hours after transduction, cells were infected with WNV (A and B) or SinV (C) at an MOI of 1. Culture fluid and cell lysates were harvested at the indicated times after infection. (A) Extracellular virus yields from WNV-infected KI shCont-7x2 and KI shAbcf3-8x2 MEFs were measured by plaque assay on BHK cells. Error bars represent standard deviations. (B) Intracellular viral protein (NS3) levels from WNV-infected KI shCont-7x2 and KI shAbcf3-8x2 MEFs were detected by Western blotting. Each experiment was performed at least twice, and a representative blot is shown. (C) Extracellular virus yields from SinV-infected KI shCont-7x2 and KI shAbcf3-8x2 MEFs were determined by plaque assay on BHK cells. Each data point is the average of duplicate titrations from at least two experiments. Error bars represent SEs. (D and E) A plasmid expressing V5-ABCF3 was transiently transfected into resistant RV MEFs using Lipofectamine LTX with PLUS reagent according to the manufacturer's protocol. Transfection reagent alone was used as a control. Twenty-four hours after transfection, cells were infected with WNV Eg101 at an MOI of 1, and culture fluid and cell lysates were harvested at the indicated times. (D) Extracellular virus yields from WNV-infected RV MEFs treated with reagent only or expressing V5-ABCF3 were measured by plaque assay on BHK cells. Each data point is the average of duplicate titrations from at least two experiments. Error bars represent SEs. (E) Intracellular viral protein (NS3) levels from WNV-infected RV MEFs treated with reagent only or expressing V5-ABCF3 were detected by Western blotting. The experiment was performed at least twice, and a representative blot is shown. All proteins were detected with SuperSignal West Pico chemiluminescent substrate except for NS3 in panel E, which was detected with Femto substrate.
Effect of ABCF3 overexpression on WNV-infected Oas1b-expressing cells.
Since the previous experiments showed that ABCF3 has antiflaviviral activity in resistant cells, overexpressing ABCF3 would be expected to decrease virus yields if uncomplexed Oas1b were available in the resistant cells to interact with ABCF3. RV cells were transiently transfected with V5-ABCF3 cDNA. RV cells treated with the transfection reagent alone were used as a control. Twenty-four hours after transfection, cells were infected with WNV Eg101 at an MOI of 1. A decrease in virus yield as well as in the levels of intracellular viral protein was observed (Fig. 7D and E).
DISCUSSION
Oas1b was previously identified as the product of the Flvr allele that confers resistance to flavivirus-induced disease (30, 35). Previous studies showed that Oas1b was not an active 2′-5′ oligodenylate synthetase and that the Oas1b-mediated, flavivirus-specific resistance phenotype was independent of the RNase L pathway (7, 36). ABCF3 and ORP1L were identified in the present study as binding partners of Oas1b. The ABC superfamily of ATP-binding proteins is one of the largest protein families known. ABC proteins are highly conserved and are found in archaea, prokaryotes, and eukaryotes (9, 21, 22, 38, 40). Most ABC proteins are transporters that export various substrates, including drugs and lipids. Some prokaryotic ABC proteins can also function as importers (9, 22). Seven subfamilies of ABC proteins (A to G) have been identified in humans. The ABCE and ABCF subfamilies lack transmembrane domains and are thought not to function as transporters (16). ABCF3 has not yet been functionally analyzed in mammalian cells. Some of the other eukaryotic ABCE and ABCF proteins have been reported to regulate translation initiation and/or elongation, ribosome assembly, and RNase L activity (5, 16, 24, 29, 40, 42). Although ABCF3 is constitutively expressed in most mammalian cells and to similar levels in resistant and susceptible MEFs, its flavivirus-specific antiviral function was only observed when full-length Oas1b was also present, even though ABCF3 was also shown to interact with the product of the susceptible allele Oas1bt. Oas1b is the only member of the OAS family analyzed to date in any species that has a transmembrane domain, and this domain was shown to localize the full-length Oas1b to the ER membrane. Oas1bt, which lacks the transmembrane region, has a diffuse cytoplasmic distribution. The ER localization of the Oas1b complex therefore appears to be essential for its antiflavivirus activity. Flaviviruses translate their proteins, replicate their genomic and antigenomic RNAs, and assemble nascent virions on proliferated ER membranes (17). The identified interaction between Oas1b and ABCF3 would localize ABCF3 in close proximity to flavivirus translation and replication sites. Knockdown of ABCF3 in resistant MEFs increased the replication of the flavivirus WNV but not that of RNA viruses from other families. These observations strongly suggest that ABCF3 is a component of the Oas1b-mediated, flavivirus-specific resistance mechanism, since its antiflaviviral activity was observed only in Oas1b-expressing cells. Exactly how ABCF3 functions within the Oas1b complex to decrease flavivirus replication is under investigation.
ORP1L was also shown to interact with Oas1b. ORP1L belongs to the oxysterol-binding protein (OSBP)-related protein family, the members of which are involved in cholesterol maintenance, sterol sensing, and lipid transport (18, 19, 28, 46, 47). ORP1L localizes primarily to the surface of late endosomes (LEs), colocalizes with LE proteins such as Rab7, Rab9, Rab7-interacting lysosomal protein (RILP), and lysosome-associated membrane protein 1 (LAMP1) (11–13), and was recently reported to regulate LE movement along microtubules by sensing cholesterol levels in LE membranes (12, 13, 32, 43). The binding site for ORP1L on Oas1b is located in the region between the C terminus of Oas1bt and the beginning of the Oas1b C-terminal transmembrane domain, which would position an interacting ORP1L close to the ER membrane. Oas1bt does not have this region and so does not interact with ORP1L. The region of ORP1L that interacts with Oas1b is the OSBP domain, which senses cholesterol. It is possible that the interaction between ORP1L and the Oas1b-ABCF3 complex provides a lipid sensing function that may be involved in directing these antiflaviviral protein complexes to regions of virus replication in the proliferated ER of flavivirus-infected cells. The need for DSP cross-linking to enhance the efficiency of ORP1L (but not of ABCF3) pulldown by Oas1b from mammalian cell lysates suggested that the interaction between ORP1L and Oas1b may be transient. It was previously reported that knockdown of ORP1L expression results in increased LE motility, while ORP1L overexpression leads to clustering of LEs in the perinuclear region of the cell (32, 43). In the present study, knockdown of ORP1L protein levels negatively affected WNV, SinV, and VSV infections. Since each of these viruses uses the host endosomal pathway for entry (6, 23, 41), the dysregulation of LE motility by knockdown of ORP1L could explain the observed negative effect. Because of the effects on endosomes, experimental strategies other than protein knockdown or overexpression will be required to further investigate the role of ORP1L in the flavivirus resistance phenotype.
Even though the inactive synthetase Oas1b was shown to act as a dominant negative for the 2-5A synthetase activity of Oas1a (7) and so would be “proviral,” the noncanonical, flavivirus-specific antiviral activity mediated by Oas1b is very effective in overriding this proviral activity. Resistant mice survive infections delivered by the intracerebral route that are 100% lethal in congenic susceptible mice (4). Although a number of loci, each controlling resistance or susceptibility to a particular type of virus, have been identified by breeding studies in mice, the gene(s) for only a few of these loci has been identified and the resistance mechanism(s) characterized (26, 27). Previous studies showed that viral RNA synthesis was less efficient starting at early times after infection in resistant cells (1, 37). The identification of cell partners for Oas1b in the present study opens new avenues for gaining an understanding of this novel natural flavivirus-specific antiviral mechanism. The data suggest that a complex consisting of Oas1b, ABCF3, and ORP1L and possibly additional proteins that is associated with the ER mediates the flavivirus resistance phenotype. Further study of this resistance mechanism is also expected to provide new insights about the virus-mediated host cell modification step during the flavivirus replication cycle that it targets.
ACKNOWLEDGMENTS
This work was supported by Public Health Service research grant AI045135 to M.A.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. S.C.C., H.D., and H.L. were supported by Molecular Basis of Disease Fellowships from Georgia State University.
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Brinton MA. 1983. Analysis of extracellular West Nile virus particles produced by cell cultures from genetically resistant and susceptible mice indicates enhanced amplification of defective interfering particles by resistant cultures. J. Virol. 46:860–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinton MA. 2008. Molecular biology of West Nile virus, p 97–136 In Diamond MS. (ed), West Nile encephalitis virus infection: viral pathogenesis and the host immune response. Springer, New York, NY [Google Scholar]
- 3. Brinton MA. 2002. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371–402 [DOI] [PubMed] [Google Scholar]
- 4. Brinton MA, Perelygin AA. 2003. Genetic resistance to flaviviruses. Adv. Virus Res. 60:43–85 [DOI] [PubMed] [Google Scholar]
- 5. Chen ZQ, et al. 2006. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J. Biol. Chem. 281:7452–7457 [DOI] [PubMed] [Google Scholar]
- 6. Chu JJ, Ng ML. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78:10543–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elbahesh H, Jha BK, Silverman RH, Scherbik SV, Brinton MA. 2011. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology 409:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghosh A, Sarkar SN, Rowe TM, Sen GC. 2001. A specific isozyme of 2′-5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl-2 family. J. Biol. Chem. 276:25447–25455 [DOI] [PubMed] [Google Scholar]
- 9. Hollenstein K, Dawson RJ, Locher KP. 2007. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17:412–418 [DOI] [PubMed] [Google Scholar]
- 10. Iyer K, et al. 2005. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci. STKE 2005:pl3. [DOI] [PubMed] [Google Scholar]
- 11. Johansson M, et al. 2003. The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol. Biol. Cell 14:903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johansson M, Lehto M, Tanhuanpaa K, Cover TL, Olkkonen VM. 2005. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol. Biol. Cell 16:5480–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johansson M, et al. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 176:459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Justesen J, Hartmann R, Kjeldgaard NO. 2000. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell. Mol. Life Sci. 57:1593–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kakuta S, Shibata S, Iwakura Y. 2002. Genomic structure of the mouse 2′,5′-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. 22:981–993 [DOI] [PubMed] [Google Scholar]
- 16. Kerr ID. 2004. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem. Biophys. Res. Commun. 315:166–173 [DOI] [PubMed] [Google Scholar]
- 17. Knipe DM, et al. (ed). 2007. Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 18. Lehto M, et al. 2001. The OSBP-related protein family in humans. J. Lipid Res. 42:1203–1213 [PubMed] [Google Scholar]
- 19. Lehto M, Olkkonen VM. 2003. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta 1631:1–11 [DOI] [PubMed] [Google Scholar]
- 20. Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21. Linton KJ, Higgins CF. 2007. Structure and function of ABC transporters: the ATP switch provides flexible control. Pflugers Arch. 453:555–567 [DOI] [PubMed] [Google Scholar]
- 22. Locher KP. 2009. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu YE, Cassese T, Kielian M. 1999. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J. Virol. 73:4272–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinand C, et al. 1999. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J. Virol. 73:290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833 [DOI] [PubMed] [Google Scholar]
- 26. Nathanson N, et al. 1997. Viral pathogenesis and immunity, p 303–328 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 27. Nathanson N, et al. 2007. Viral pathogenesis and immunity, 2nd ed, p 174–184 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 28. Olkkonen VM, et al. 2006. The OSBP-related proteins (ORPs): global sterol sensors for co-ordination of cellular lipid metabolism, membrane trafficking and signalling processes? Biochem. Soc. Trans. 34:389–391 [DOI] [PubMed] [Google Scholar]
- 29. Paytubi S, Morrice NA, Boudeau J, Proud CG. 2008. The N-terminal region of ABC50 interacts with eukaryotic initiation factor eIF2 and is a target for regulatory phosphorylation by CK2. Biochem. J. 409:223–231 [DOI] [PubMed] [Google Scholar]
- 30. Perelygin AA, et al. 2002. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. U. S. A. 99:9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perelygin AA, Zharkikh AA, Scherbik SV, Brinton MA. 2006. The mammalian 2′-5′ oligoadenylate synthetase gene family: evidence for concerted evolution of paralogous Oas1 genes in Rodentia and Artiodactyla. J. Mol. Evol. 63:562–576 [DOI] [PubMed] [Google Scholar]
- 32. Rocha N, et al. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150Glued and late endosome positioning. J. Cell Biol. 185:1209–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salzberg S, et al. 1997. Ectopic expression of 2-5A synthetase in myeloid cells induces growth arrest and facilitates the appearance of a myeloid differentiation marker. Cancer Res. 57:2732–2740 [PubMed] [Google Scholar]
- 34. Sarkar SN, Sen GC. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245–259 [DOI] [PubMed] [Google Scholar]
- 35. Scherbik SV, Kluetzman K, Perelygin AA, Brinton MA. 2007. Knock-in of the Oas1b(r) allele into a flavivirus-induced disease susceptible mouse generates the resistant phenotype. Virology 368:232–237 [DOI] [PubMed] [Google Scholar]
- 36. Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scherbik SV, Stockman BM, Brinton MA. 2007. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. J. Virol. 81:12005–12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schriml LM, Dean M. 2000. Identification of 18 mouse ABC genes and characterization of the ABC superfamily in Mus musculus. Genomics 64:24–31 [DOI] [PubMed] [Google Scholar]
- 39. Sejvar JJ. 2007. The long-term outcomes of human West Nile virus infection. Clin. Infect. Dis. 44:1617–1624 [DOI] [PubMed] [Google Scholar]
- 40. Sturm A, Cunningham P, Dean M. 2009. The ABC transporter gene family of Daphnia pulex. BMC Genomics 10:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Superti F, et al. 1987. Entry pathway of vesicular stomatitis virus into different host cells. J. Gen. Virol. 68(Pt 2):387–399 [DOI] [PubMed] [Google Scholar]
- 42. Tyzack JK, Wang X, Belsham GJ, Proud CG. 2000. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 275:34131–34139 [DOI] [PubMed] [Google Scholar]
- 43. Vihervaara T, et al. 2011. Sterol binding by OSBP-related protein 1L regulates late endosome motility and function. Cell. Mol. Life Sci. 68:537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Webster LT, Clow AD. 1936. Experimental encephalitis (St. Louis type) in mice with high inborn resistance: a chronic subclinical infection. J. Exp. Med. 63:827–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Webster LT, Johnson MS. 1941. Comparative virulence of St. Louis encephalitis virus cultured with brain tissue from innately susceptible and innately resistant mice. J. Exp. Med. 74:489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu Y, Liu Y, Ridgway ND, McMaster CR. 2001. Novel members of the human oxysterol-binding protein family bind phospholipids and regulate vesicle transport. J. Biol. Chem. 276:18407–18414 [DOI] [PubMed] [Google Scholar]
- 47. Yan D, et al. 2007. Oxysterol binding protein induces upregulation of SREBP-1c and enhances hepatic lipogenesis. Arterioscler. Thromb. Vasc. Biol. 27:1108–1114 [DOI] [PubMed] [Google Scholar]
- 48. Yan W, et al. 2005. Mice deficient in oocyte-specific oligoadenylate synthetase-like protein OAS1D display reduced fertility. Mol. Cell. Biol. 25:4615–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]