Abstract
The maturation of the hepatitis C virus (HCV) core protein requires proteolytic processing by two host proteases: signal peptidase (SP) and the intramembrane-cleaving protease signal peptide peptidase (SPP). Previous work on HCV genotype 1a (GT1a) and GT2a has identified crucial residues required for efficient signal peptide processing by SPP, which in turn has an effect on the production of infectious virus particles. Here we demonstrate that the JFH1 GT2a core-E1 signal peptide can be adapted to the GT3a sequence without affecting the production of infectious HCV. Through mutagenesis studies, we identified crucial residues required for core-E1 signal peptide processing, including a GT3a sequence-specific histidine (His) at position 187. In addition, the stable knockdown of intracellular SPP levels in HuH-7 cells significantly affects HCV virus titers, further demonstrating the requirement for SPP for the maturation of core and the production of infectious HCV particles. Finally, our nuclear magnetic resonance (NMR) structural analysis of a synthetic HCV JFH1 GT2a core-E1 signal peptide provides an essential structural template for a further understanding of core processing as well as the first model for an SPP substrate within its membrane environment. Our findings give deeper insights into the mechanisms of intramembrane-cleaving proteases and the impact on viral infections.
INTRODUCTION
Hepatitis C virus (HCV) is a major global health problem, and current estimates suggest that 2 to 3% of the world population, corresponding to 130 to 170 million people, is infected (21). In the majority of cases, HCV establishes a chronic infection, which often progresses to liver fibrosis and cirrhosis and, in some cases, hepatocellular carcinoma (8). HCV is a highly variable virus that has been grouped into seven major genotypes (genotype 1 [GT1] to GT7), which differ by 31 to 33% at the nucleotide level (32, 48). In the United Kingdom, genotype 3 strains account for about 45% of all infections (11). Patients infected with HCV often present with an increased accumulation of lipids in the liver, referred to as steatosis. The incidence of steatosis is higher in genotype 3a infections than in infections by other genotypes and could arise from a virus-specific factor (12, 34, 42, 43).
HCV is a single-stranded positive-sense RNA virus that encodes a single polyprotein, which is processed both co- and posttranslationally by viral and endoplasmic reticulum (ER)-bound cellular proteases to produce mature viral proteins (31). These cleavage events result in the release of structural proteins (core protein and two envelope glycoproteins termed E1 and E2) and nonstructural proteins (p7 and NS2-NS5B). For the production of mature core protein, a signal peptide between core and E1 undergoes an initial cleavage by the cellular enzyme signal peptidase (SP), which is followed by a second proteolytic event mediated by the intramembrane protease signal peptide peptidase (SPP) (27, 39). The unequivocal identification of the C-terminal amino acid of mature core after SPP processing has proven problematic. Studies using insect cells have shown that the final residue in core is either Phe177 (35), Leu179, or Leu182 (16). More recent studies using mammalian cells indicated that Phe177 is the most probable C-terminal residue in the protein following SPP cleavage (36). The maturation of HCV core by SPP and the subsequent trafficking of the mature protein to lipid droplets (LDs) have been identified as crucial steps in virus production (3, 27, 45, 50).
SPP is an aspartyl protease that belongs to the family of intramembrane-cleaving proteases (I-CLiPs) (54). The protease is thought to be involved in the clearance of signal peptides and misfolded membrane-binding proteins from the ER membrane. SPP forms multimers, and recent evidence suggests that the active protease is a tetramer, which forms a bullet-like structure with concaves on the surface and a chamber in the center (29). The requirements for the proteolytic processing of the core-E1 signal peptide by SPP have been studied with a variety of HCV strains and in various systems (13, 18, 27, 37, 39, 50). Those reports identified residues both within the core-E1 signal peptide and elsewhere in the core coding region that influence cleavage. Moreover, within the core-E1 signal peptide, the effects of mutations at conserved residues on processing differ between strains. Such findings led us to conclude that several factors, particularly the structure of the core-E1 signal peptide combined with physicochemical characteristics at certain residues, are likely to contribute to optimal SPP cleavage.
To further investigate the factors that determine SPP cleavage, we have analyzed the structural features of the core-E1 signal peptide by circular dichroism (CD) and nuclear magnetic resonance (NMR) analyses in membrane-mimetic environments. In addition, we have studied the requirements for efficient SPP cleavage in the core-E1 signal peptide for a GT3 strain of HCV, which differs in sequence at key residues previously identified as being important for efficient cleavage in strains from other genotypes. Our findings provide further insight into the mechanism of SPP cleavage at the core-E1 signal peptide and the maturation of the HCV core protein.
MATERIALS AND METHODS
Plasmid construction.
pGEM-GT3a-wt was constructed by the PCR amplification of the sequence encoding the proteins core, E1, E2, and p7 and the N-terminal 90 amino acids (aa) of NS2 from isolate HCV3a-Gla (46). Following the insertion of BamHI sites at the termini, the resulting PCR fragment was cloned first into pGEM-T Easy (Promega), thereby generating pGEM-GT3a-wt, and then into the expression vector pSFV (Life Technologies); this strategy yielded plasmid pSFV-GT3a-wt. Mutations in the core-E1 signal peptide sequence were generated from pGEM-GT3a-wt by using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). Oligonucleotides for site-directed mutagenesis were designed according to the manufacturer's guidelines. The sequence of each core-E1 signal peptide mutant was verified prior to cloning into pSFV-GT3a-wt. Plasmid pJFH1 was a gift from Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan). The core-E1 signal peptide region of JFH1 (GT2a) was adapted to GT3a by the site-directed mutagenesis of a pJFH1 FspAI/BsiWI fragment in pTopo-CE1E2-JFH1 (4). The same approach was used to generate the respective JFH1/GT3a core-E1 signal peptide variants. After the verification of the sequence, the mutated FspAI/BsiWI fragments were inserted into the pJFH1 backbone.
Antibodies.
Antibodies used to detect HCV core (rabbit antisera R308 and 4210), E2 (monoclonal antibody ALP98, a gift from Arvind Patel, CVR, Glasgow University), NS5A (sheep antisera, a gift from Mark Harris, Leeds University), and human adipocyte differentiation-related protein (ADRP) were described previously (6, 14, 25, 49). Antibodies to detect human SPP (ab16080) and actin (clone AC-40) were purchased from Abcam and Sigma, respectively.
Maintenance of tissue culture cells.
HuH-7 and HEK293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS).
Generation of shSPP cell lines.
HuH-7 cells were transduced with lentiviral vectors expressing either a scrambled short hairpin RNA (shRNA) or SPP shRNA and were named sh-Scram-HuH-7 and sh-SPP-HuH-7, respectively. The human SPP target sequence was 5′-TCCTGCTGTCCATGTATTTCTTC-3′ (NCBI accession number NM_030789.2). The shRNA sequence was constructed as double-stranded oligonucleotides containing a BamHI site at the 5′ terminus and an EcoRI site at the 3′ terminus for cloning into the lentivirus plasmid vector pLKO-shPML (9), from which the shPML sequence was removed. Lentiviral vector stocks were prepared by the cotransfection of HEK293T cells with the relevant pLKO-shRNA plasmid, helper plasmid pCMVDR8.91 (kindly supplied by Didier Trono [http://tronolab.com/index.php]), and pVSV-G (BD Biosciences). Virus supernatants were harvested 48 h after transfection and passed through a 450-nm filter before the transduction of HuH-7 cells. Transduced cells were selected with puromycin at 5 μg/ml.
In vitro synthesis of RNA and electroporation of cells.
Plasmid pSFV-GT3a-wt and its respective mutant constructs were linearized with SapI. RNA was transcribed in vitro from linearized constructs by using SP6 RNA polymerase (Promega) according to the manufacturer's instructions. Constructs harboring JFH1 and the JFH1-GT3a variants were linearized with XbaI and treated with mung bean nuclease. RNA was transcribed in vitro from linearized constructs by using the T7 RiboMAX Express large-scale RNA production system (Promega). Transcripts were introduced into HuH-7 and HuH-7 shRNA cells by electroporation as described previously (14).
Infection of cells and determination of virus titers.
Supernatants from JFH1-electroporated cells were removed at the required time points and used to infect monolayers of naive HuH-7 cells. Infected cells were detected at 3 days postinoculation by indirect immunofluorescence using NS5A antisera. The 50% tissue culture infectious dose (TCID50) was determined by a limiting-dilution assay (24).
Preparation of cell extracts, SDS-PAGE, and Western blot analysis.
To prepare extracts, cell monolayers were washed with phosphate-buffered saline (PBS) and solubilized in sample buffer (160 mM Tris-HCl [pH 6.7], 2% SDS, 700 mM β-mercaptoethanol, 10% glycerol, bromophenol blue). Samples were heated at 95°C for 5 min prior to electrophoresis through a 10% or 12% SDS-polyacrylamide gel (acrylamide/bis-acrylamide ratio of 37.5:1). For Western blot analysis, proteins were transferred onto a nitrocellulose membrane. After blocking with PBS containing 5% milk powder (Marvel), membranes were incubated with the relevant primary and horseradish peroxidase (HRP)-linked secondary antibodies and diluted in PBS containing 5% milk powder and 0.05% Tween 20. After washing, bound antibody was detected by using ECL Plus (Amersham).
Indirect immunofluorescence.
Electroporated cells plated onto 13-mm coverslips were fixed in methanol at −20°C for 20 min. After washing with PBS, cells were incubated with primary antibody diluted in PBS-FCS (PBS containing 1% fetal calf serum) for 2 h at room temperature. Cells were washed extensively with PBS and then incubated with a secondary antibody conjugated to an Alexa fluorescent tag for 30 min at room temperature. After washing with PBS, cells were rinsed with distilled water before being mounted onto slides using Citifluor and examined with a Zeiss LSM510 Meta microscope. Images were recorded with a Plan-Apochromat 63× lens (numerical aperture [NA], 1.4). Adobe Photoshop (CS2) was used for further image processing.
Real-time quantification of RNA.
Total cellular and viral RNAs were extracted from HuH-7 cells by using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. Real-time reverse transcription quantitative PCRs (RT-qPCRs) were performed as a two-step process. First, cDNA was generated from total RNA by using a TaqMan kit (Applied Biosciences) and random hexamer oligonucleotides in accordance with the manufacturer's instructions. Reverse transcription was performed with a ThermoHybaid PX2 thermal cycler as follows: (i) primer annealing at 25°C for 10 min, (ii) strand elongation at 37°C for 1 h, and (iii) reverse transcriptase inactivation at 95°C for 5 min. For the second step, sample cDNAs were analyzed in triplicate by RT-qPCR using TaqMan Fast Universal PCR Master Mix, no ampErase UNG (ABI). Each sample was run as a singleplex reaction mixture containing the appropriate primers and a TaqMan probe or probe-primer mix. The HCV probe and primer sequences were located in the 5′ untranslated region (UTR) of the JFH1 genome (primers 5′-TCTGCGGAACCGGTGAGTAC-3′ [nucleotides {nt} 147 to 166] and 5′-GCACTCGCAAGCACCCTATC-3′ [nt 295 to 314] and probe 5′-GGCCTTGTGGTACTG-3′ [nt 277 to 291]). SPP probe-primer mix (assay ID Hs00604897_m1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe-primer mix (VIC/MGB probe) were purchased from ABI. Reactions were performed by using an Applied Biosciences 7500 Fast real-time PCR machine as follows: double-stranded DNA (dsDNA) strand separation at 95°C for 3 s (step 1) and primer annealing and strand elongation at 60°C for 30 s (step 2). Steps 1 and 2 were repeated 40 times. All RT-qPCRs were performed in triplicate, and values were normalized to endogenous GAPDH levels. Data were analyzed by using 7500 Fast system software (SDS v1.3.1; ABI).
Peptide synthesis and purification.
Peptide sp-E1 (KKGFPFSIFLLALLSCITVPVSAAQVK), which corresponds to aa 171 to 195 of the HCV strain JFH1 polyprotein (GenBank accession number AB047639), was synthesized in in-house facilities and purified by reverse-phase high-pressure liquid chromatography (RP-HPLC) on a Nucleosil C18 column (120 Å, 5 μm, and 10 by 250 mm) using a water-acetonitrile gradient containing 0.1% trifluoroacetic acid. The sp-E1 peptide was eluted as a single peak at 63% acetonitrile, and its purity (>98%) was determined by RP-HPLC, electrospray mass spectroscopy (observed mass, 2,891.79 ± 0.29 Da; calculated mass, 2,892.68 Da), and NMR spectroscopy. The sequence KK at the N terminus was added to increase peptide solubility.
Circular dichroism.
Far-UV CD spectra were recorded on a Chirascan spectrometer (Applied Photophysics, United Kingdom) calibrated with 1S-(+)-10-camphorsulfonic acid. Measurements were carried out at 298 K in a 0.1-cm-path-length quartz cuvette (Hellma), with peptide concentrations ranging from 14 to 150 μM. Spectra were measured in a 180-nm- to 260-nm-wavelength range with an increment of 0.2 nm, a bandpass of 0.5 nm, and an integration time of 1 s. Spectra were processed, baseline corrected, smoothed, and converted with Chirascan software. Spectral units were expressed as the mean molar ellipticity per residue. The secondary-structure content was estimated with various deconvolution methods by using the DICHROWEB Server (http://www.cryst.bbk.ac.uk/cdweb/) (55).
NMR spectroscopy.
The purified sp-E1 peptide was dissolved in 50% (vol/vol) 2,2,2-trifluoroethanol (TFE)-d2 (>99%) in H2O, and 2,2-dimethyl-2-silapentane-5-sulfonate was added to the NMR samples as an internal 1H chemical shift reference. Multidimensional experiments were performed at 25°C on Bruker Avance 500-MHz spectrometers using standard homonuclear pulse sequences such as nuclear Overhauser enhancement spectroscopy (NOESY) (with mixing times of between 100 and 250 ms) and clean total correlation spectroscopy (TOCSY) (with an isotropic mixing time of 75 ms), as detailed previously (see references 10 and 40 and references therein). Bruker Topspin software was used to process all data, and Sparky software was used for spectrum analyses (T. D. Goddard and D. G. Kneller, University of California, San Francisco [http://www.cgl.ucsf.edu/home/sparky/]). Intraresidue backbone resonances and aliphatic side chains were identified from homonuclear 1H TOCSY experiments and confirmed with 1H-13C heteronuclear single-quantum correlation (HSQC) experiments in a 13C natural abundance. Sequential assignments were determined by correlating intraresidue assignments with interresidue cross peaks observed by bidimensional 1H NOESY. NMR-derived 1H-α and 13C-α chemical shifts are reported relative to the random-coil chemical shifts in TFE (28, 51).
NMR-derived constraints and structure calculation.
The nuclear Overhauser effect (NOE) intensities used as the distance input for structure calculations were obtained from the NOESY spectrum recorded with a 150-ms mixing time and checked for spin diffusion on spectra recorded at lower mixing times (50 ms). NOEs were partitioned into three categories of intensities, which were converted into distances ranging from a common lower limit of 1.8 Å to upper limits of 2.8 Å, 3.9 Å, and 5.0 Å, respectively. Protons without stereospecific assignments were treated as pseudoatoms, and the correction factors were added to the upper distance constraints (57). The three-dimensional structures were generated from NOE distances and dihedral angle constraints (calculated with TALOS [47] from 1H and 13C chemical shifts) by means of a dynamic simulated annealing protocol, using the Xplor-NIH 2.21 program (44) and standard force field and default parameter sets. Sets of 50 structures were initially calculated to widely sample the conformational space, and the structures of low energy with no distance restraint (>0.5 Å) or dihedral-angle (>5°) restraint violations were retained. The selected structures were compared by pairwise root mean square deviation (RMSD) over the backbone atom coordinates (N, C-α, and C′). Local analogies were analyzed by calculating the local RMSD of a tripeptide window moved sequentially along the sequence. Statistical analyses, superimpositions of structures, and structural analyses were performed with MOLMOL, version 2.6 (19), and the quality of the selected structures was checked with the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) Validation Server.
Accession numbers.
The atomic coordinates for the NMR structure of the sp-E1 peptide (HCV polyprotein aa 171 to 195, strain JFH1) (GenBank accession number AB047639) and the NMR restraints in 50% TFE are available from the RCSB Protein Data Bank under accession number 2kqi and RCSB ID rcsb101447. The proton chemical shifts of all residues have been deposited in the BioMagResBank (BMRB) database under accession number 16601. The GenBank accession number for HCV3a-Gla is JQ664687.
RESULTS
Amino acid residues required for efficient SPP proteolysis in the HCV GT3a core-E1 signal peptide.
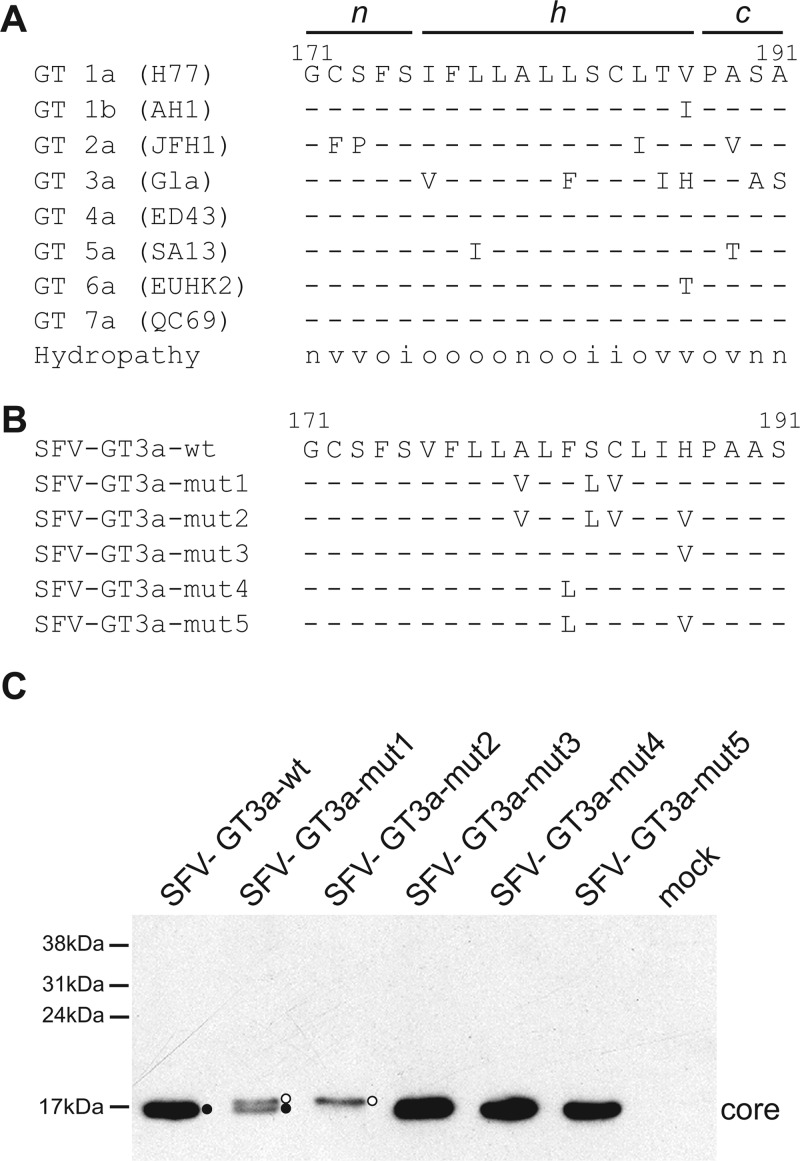
Previous studies, aimed at an understanding of the requirements for efficient SPP proteolysis that are needed to generate mature core protein, have focused primarily on the sequences in the core-E1 signal peptide region from HCV genotypes 1 and 2. In contrast, GT3 has received little attention aside from studies that indicated a link between polymorphisms in the core-E1 signal peptide of GT3 isolates, which may be linked to lipid accumulation (17). From sequence comparisons of the different segments that encompass the core-E1 signal peptide, the n region from the different HCV genotypes is highly conserved, with the exception of JFH1, which encodes Phe and Pro residues at positions 172 and 173 (Fig. 1A). The conversion of these amino acids to the consensus residues Cys and Ser increased titers of infectious progeny upon the propagation of the virus (7). In contrast to the n region, there is greater variability in the h and c regions (Fig. 1A). Interestingly, alignments of sequences from strains representing various HCV genotypes indicated that GT3a strains differed from the consensus at 3 to 4 positions in the h region (17, 18) (Fig. 1A). In addition, the final two amino acids in the c region of GT3a sequences (Ala and Ser) are inverted compared to the sequences of all other genotypes (Ser followed by Ala) (Fig. 1A). Given these differences, we wished to explore the impact that such variability may have on the processing of immature core protein by SPP.
Fig 1.
Effect of mutations in the GT3a core-E1 signal peptide on SPP proteolysis. (A) Amino acid sequences of the core-E1 signal peptide region for HCV genotypes 1 to 7. Representative strains for each genotype are shown in parentheses. The predicted n, h, and c regions within the signal peptide are indicated. The amino acid hydropathy at each position is indicated as neutral (n), hydrophobic (o), hydrophilic (i), or variable (v). (B) wt and mutant sequences for the core-E1 signal peptide from HCV3a-Gla examined with the SFV system. (C) HuH-7 cells were electroporated with in vitro-transcribed RNA from the indicated SFV constructs. Cell extracts were prepared at 16 h after electroporation and analyzed by Western blotting with an antibody against core. Open and closed circles indicate SP- and SPP-cleaved core species, respectively.
To examine the contribution of sequences in the HCV GT3a core-E1 signal peptide to the processing of immature core protein, we expressed wild-type (wt) and mutant variants of the core-E1-E2 region from an HCV GT3a isolate, called HCV3a-Gla, using the Semliki Forest virus (SFV) system. Compared to the h region encoded by strain JFH1, HCV3a-Gla differs at 4 amino acids (Ile→Val [position 176], Leu→Phe [position 182], Thr→Ile [position 186], and Val→His [position 187]). Previously, we established that the introduction of a hydrophobic residue at position 186 in the core-E1 signal peptide of strain JFH1 does not affect SPP proteolysis (50), and therefore, no mutations were made at this position. The Ile→Val polymorphism at position 176 occurs infrequently, and most GT3a sequences encode Ile at this site. In addition, both Ile and Val are β-branched hydrophobic residues that exhibit similar physicochemical features, notably an increased rigidity compared to those of other hydrophobic residues. Therefore, this amino acid was not mutated. Conversely, the other amino acid polymorphisms at positions 182 and 187 are unique to GT3a sequences (Phe and His, respectively) and represent the presence of aromatic and polar residues, respectively, at positions normally occupied by hydrophobic amino acids. Hence, their impact on the processing of core was examined by converting them to Leu and Val, thereby creating the constructs pSFV-GT3a-mut3, -mut4, and -mut5 (Fig. 1B).
Our previous studies also showed that amino acid residues at positions 180 (Ala), 183 (Ser), and 184 (Cys) were critical for the efficient proteolytic processing of the core-E1 signal peptide for GT1a strain Glasgow (13, 27). In strain JFH1 (GT2a), mutations at these 3 residues were insufficient to block SPP cleavage, and an additional substitution of a Thr residue at position 186 was required to disrupt proteolysis (50). The replacement of the 4 residues at positions 180, 183, 184, and 186 by the hydrophobic amino acids Val and Leu not only impaired SPP cleavage and the production of mature core but also prevented the targeting of the immature protein to LDs (50). To determine whether the SPP processing of the core-E1 signal peptide from GT3a sequences had similar requirements, we first created pSFV-GT3a-mut1, which has a triplet of mutations at positions 180, 183, and 184 (Fig. 1B). We also reasoned that the His residue at position 187 in the GT3a sequence may be functionally equivalent to the Thr residue at position 186 in other HCV sequences. Hence, we introduced a mutation at this position in pSFV-GT3a-mut1 to give pSFV-GT3a-mut2, which therefore contains a quadruple set of mutations in the h region of the core-E1 signal peptide (Fig. 1B). We elected not to examine other residues in the h region that could influence SPP proteolysis, in particular the Ile/Phe doublet at positions 176 and 177 (36), since mutations at these sites can lead to lower or barely detectable levels of core in the context of the expression of the HCV polyprotein (13, 18, 36). Moreover, previous studies have convincingly shown that the mutagenesis of these amino acids abolishes virion production (18, 36).
RNA from each of the SFV constructs was synthesized in vitro and then electroporated into HuH-7 cells. Western blot analysis revealed that the migration patterns for core protein expressed by SFV-GT3a-mut3, -mut4, and -mut5 were identical to that for core expressed from SFV-GT3a-wt (Fig. 1C). Hence, the mutation of Phe at position 182 and His at position 187, either individually or in combination, had no discernible effect on SPP proteolysis. In contrast, SFV-GT3a-mut1 gave two core species, presumed to represent SP- and SPP-cleaved forms of the protein. For SFV-GT3a-mut2, only a single species of core was detected, which comigrated with the SP-cleaved form of core produced by SFV-GT3a-mut1 (Fig. 1C), demonstrating that the introduction of four mutations prevents cleavage by SPP in the SFV system. From indirect immunofluorescence analyses, core expressed by SFV-GT3a-wt colocalized with ADRP, a marker of lipid droplets, whereas both SFV-GT3a-mut1 and SFV-GT3a-mut2 gave a more diffuse expression pattern of core localization, with no evidence for significant attachment to lipid droplets (data not shown). From these data, we concluded that, similar to our previous study of the JFH1 structural proteins expressed by the SFV vector (50), the mutation of the triplet Ala/Ser/Cys to Val/Leu/Val impaired but did not completely block SPP cleavage. However, the additional replacement of the His residue by Val at position 187 abrogated SPP proteolysis in the SFV system (Fig. 1C).
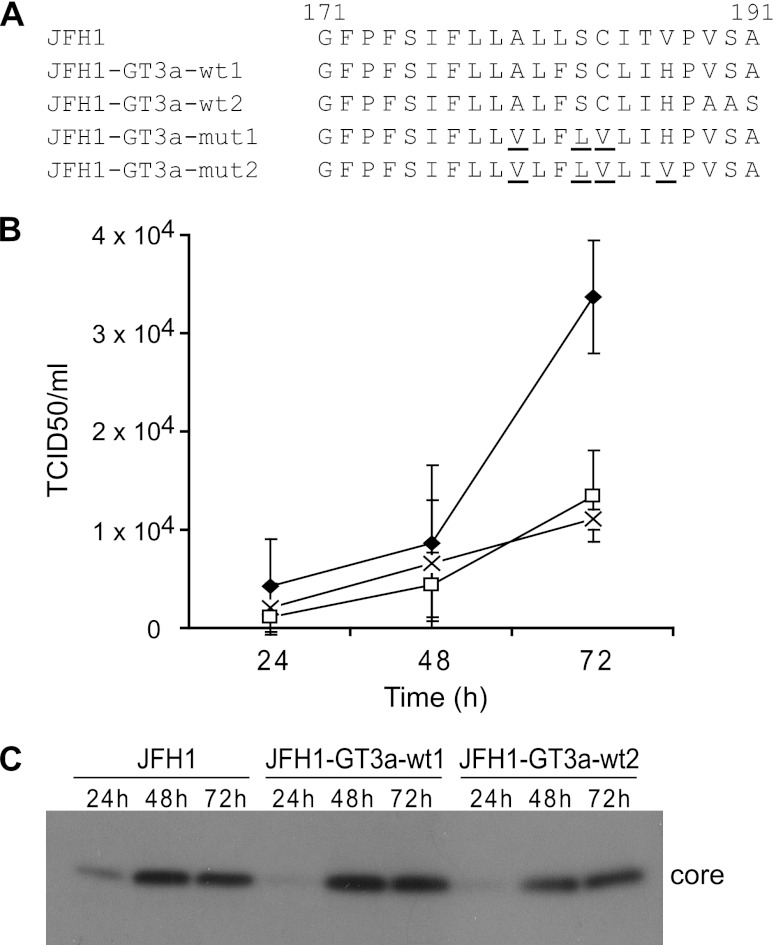
The JFH1 core-E1 signal peptide can be adapted to encode GT3a sequences without loss of virus production.
Prior to examining the sequence requirements for SPP processing at the GT3a core-E1 signal peptide in an infectious system, the JFH1 signal peptide was adapted to encode the GT3a sequence. Attempts to determine sequence requirements by transferring the entire GT3a core-E1-E2-p7-NS2 coding region into JFH1, thereby creating GT3a-JFH1 chimeras, did not produce any infectious virus, in agreement with data from previous studies (41; data not shown). Hence, the core-E1 signal peptide in JFH1 was modified to encode the sequence for the h region between positions 182 and 187 from strain HCV3a-Gla, creating pJFH-GT3a-wt1 (Fig. 2A). Since the c region of the GT3a core-E1 signal peptide differs at amino acid positions 189, 190, and 191 compared to strain JFH1 (Fig. 1A), these residues were converted to also encode GT3a sequences (pJFH-GT3a-wt2) (Fig. 2A). In vitro-transcribed RNA from these modified JFH1 constructs and from wt JFH1 was electroporated into HuH-7 cells, and the amount of virus released was determined at 24, 48, and 72 h by TCID50 determinations. Our results indicated that virus production by variants containing either the GT3a h region or the combined h and c regions was comparable to that by JFH1 (Fig. 2B). We also found comparable levels of core protein expressed by wt JFH1 and the GT3a variants at each time point (Fig. 2C).
Fig 2.
Adaptation of the JFH1 core-E1 signal peptide to encode wt and mutant GT3a sequences. (A) Sequences encoded in the core-E1 signal peptide region for wt JFH1 and wt and mutant GT3a constructs. (B) HuH-7 cells were electroporated with in vitro-transcribed RNA, and cell supernatants were harvested at 24-h intervals. Virus titers were determined by measurements of TCID50 values. Data shown represent the average values from three independent experiments for the following constructs: JFH1-GT3a-wt1 (□), JFH1-GT3a-wt2 (◆), and JFH1 (×). Error bars indicate standard deviations. (C) HuH-7 cells were electroporated with in vitro-transcribed RNA from the indicated constructs. Cell extracts were prepared at the times indicated after electroporation and analyzed by Western blotting with an anti-core antibody.
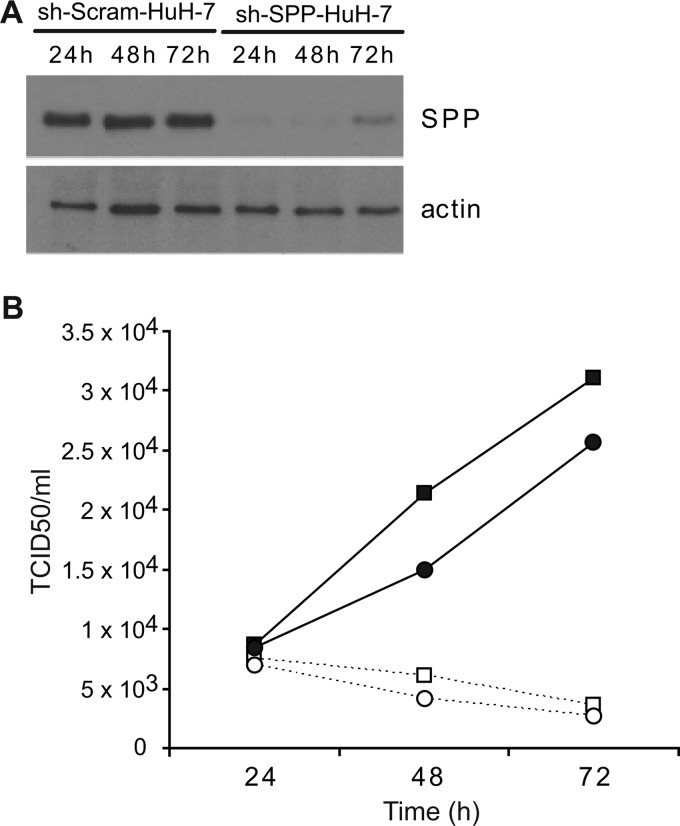
To analyze the contribution of SPP proteolysis to virus production by JFH-GT3a-wt1 and JFH1, the amount of virus released from sh-SPP-HuH-7 cells, an HuH-7 cell line that expressed shRNA targeting SPP mRNA, was analyzed. Results were compared with results for virus production from sh-Scram-HuH-7 cells, a control cell line that expressed a scrambled shRNA sequence. In sh-SPP-HuH-7 cells, SPP mRNA levels were reduced by 5- to 10-fold compared to levels in sh-Scram-Huh-7 cells (data not shown), and the amount of SPP protein detected by Western blot analysis was lowered to similar levels (Fig. 3A). The electroporation of JFH1-GT3a-wt1 and JFH1 RNAs into these cell lines showed that the level of virus production rose between 24 and 72 h in sh-Scram-HuH-7 cells, whereas the sh-SPP-HuH-7 cells yielded much less virus such that by 72 h, the reduction in virus yield was about 9-fold for both constructs (Fig. 3B).
Fig 3.
Virus production in HuH-7-derived shRNA cell lines. (A) HuH-7 cells expressing either a scrambled (sh-Scram-HuH-7) or SPP (sh-SPP-HuH-7) short hairpin RNA were analyzed by Western blotting with antibodies to SPP and actin. (B) Cells were electroporated, and cell supernatants were harvested at 24-h intervals. Virus titers were determined by measurements of TCID50 values; values represent the averages of data from three independent experiments. Data are shown as follows: ■, JFH1-GT3a-wt1 in sh-Scram-HuH-7 cells; ●, JFH1 in sh-Scram-HuH-7 cells; □, JFH1-GT3a-wt1 in sh-SPP-HuH-7 cells; ○, JFH1 in sh-SPP-HuH-7 cells.
From the above-described data, we concluded that the amino acid differences between the h and c regions of GT3a and GT2a sequences did not exert any impact on the contribution of SPP to virion production.
Mutations in the core-E1 signal peptide that impair virus production also block efficient SPP proteolysis of HCV core.
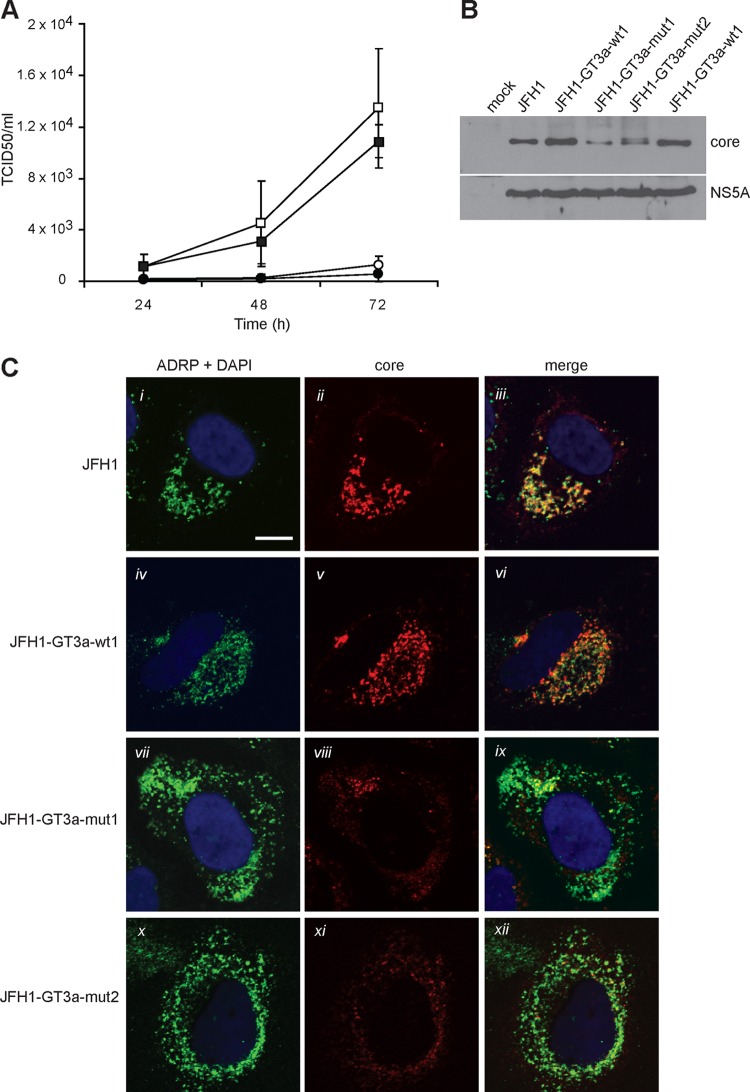
Based on the data obtained with the SFV system, the mutations in the GT3a core-E1 signal peptide that impaired SPP proteolysis were introduced into JFH1-GT3a-wt1. Thus, two mutants carrying three mutations (Ala180, Ser183, and Cys184 to Val180, Leu183, and Val184) and four mutations (Ala180, Ser183, Cys184, and His187 to Val180, Leu183, Val184, and Val187) in pSFV-GT3a-mut1 and pSFV-GT3a-mut2, respectively, were inserted into JFH1-GT3a-wt1 to give JFH1-GT3a-mut1 and JFH1-GT3a-mut2, respectively, (Fig. 2A). Based on results from three independent experiments, JFH1-GT3a-mut1 yielded barely detectable amounts of virus at 24 h after electroporation, while JFH1-GT3a-mut2 produced no detectable virus (Fig. 4A). In comparison, JFH1 and JFH1-GT3a-wt1 yielded TCID50 values of 2.6 × 103 and 3.2 × 103, respectively, at the same time point (Fig. 4A). At 48 and 72 h after electroporation, JFH1-GT3a-mut1 and JFH1-GT3a-mut2 did produce detectable amounts of virus, but these amounts remained more than 10-fold lower than those produced by JFH1-GT3a-wt1. Moreover, virus yields from JFH1-GT3a-mut2 were about three times lower than those for JFH1-GT3a-mut1 at 72 h (Fig. 4A). This pattern for the wt and mutant constructs was also found with sh-Scram-HuH-7 cells (data not shown). These data demonstrate that mutations in the h region of the core-E1 signal peptide of a GT3a sequence severely impair virus production. Indeed, TCID50 values for both mutants in sh-Scram-HuH-7 cells were considerably lower than those for JFH1-GT3a-wt1 in sh-SPP-HuH-7 cells (data not shown). Thus, the mutations have a greater impact on virus production than lowering the levels of SPP in cells.
Fig 4.
Effects of mutations in the core-E1 signal peptide from GT3a on virus production and proteolysis of core. (A) HuH-7 cells were electroporated with in vitro-transcribed RNA, and cell supernatants were harvested at 24-h intervals. Virus titers were determined by measurements of TCID50 values. Data shown represent the average values from three independent experiments and are for the following constructs: JFH1-GT3a-wt1 (□), JFH1 (■), JFH1-GT3a-mut1 (○), and JFH1-GT3a-mut2 (●). Error bars indicate standard deviations. (B and C) HuH-7 cells were electroporated with in vitro-transcribed RNAs as indicated. (B) Cell extracts were harvested at 72 h and analyzed by Western blotting with antibodies against core and NS5A. (C) Cells were prepared for immunofluorescence at 72 h after electroporation and analyzed by confocal microscopy with antibodies against core and ADRP. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and the scale bar represents 10 μm.
To determine whether the mutations in JFH1-GT3a-mut1 and JFH1-GT3a-mut2 affected the SPP proteolysis of core, cell extracts were examined by Western blot analysis. For JFH1-GT3a-wt1 and JFH1, only one core species was detected, which represented an SPP-cleaved protein (Fig. 4B). In the case of JFH1-GT3a-mut1, we also detected only a single species that comigrated with core made by JFH1-GT3a-wt1 and JFH1. However, the level of protein was about 4-fold lower than that with the wt constructs (Fig. 4B). We deduce that the lower abundance of core made by JFH1-GT3a-mut1 is due to degradation, since the amount of NS5A detected for the mutant construct was equivalent to those for both JFH1-GT3a-wt1 and JFH1. Interestingly, two core species were detected for ‘JFH1-GT3a-mut2’, presumed to represent SPP- and SP-cleaved forms of the protein for high- and low-molecular-weight (MW) species, respectively (Fig. 4b). Again, the abundance of core was somewhat lower than that detected for JFH1 and JFH1-GT3a-wt1. The detection of core at earlier time points for JFH1-GT3a-mut1 and JFH1-GT3a-mut2 was not reproducible over several experiments, consistent with the low abundance of the protein at 72 h after RNA electroporation. We note that the patterns for core produced by mut1 and mut2 differ between the SFV and JFH1 expression systems (compare Fig. 1C and 4B). These differences are likely to arise from the different times at which the samples were harvested (16 h for the SFV system compared to 72 h for the HCV system). The expression of the two mutants for longer periods in infected HuH-7 cells is likely to account for these differences, since the immature core generated by mut1 and mut2 would be exposed to potential cleavage by SPP for a longer time period; a similar phenomenon was found in our previous studies on the core-E1 signal peptide from JFH1 (50). Overall, our results show that SPP proteolysis is dependent on the sequences in the h region of the core-E1 signal peptide.
To further examine the characteristics of core made by JFH1-GT3a-mut1 and JFH1-GT3a-mut2, the intracellular localization of the protein was analyzed at 72 h after electroporation and compared to the distributions for JFH1 and JFH1-GT3a-wt1. The core protein made by JFH1 and JFH1-GT3a-wt1 colocalized with ADRP, indicating an association with lipid droplets (Fig. 4C). In contrast, JFH1-GT3a-mut1 and JFH1-GT3a-mut2 produced far less detectable core, which only partially localized with ADRP. The remainder of the detected protein had a diffuse pattern of staining.
The above-described results reaffirm our previous conclusions that mutations in the h region of the core-E1 signal peptide play a critical role in determining the efficiency of SPP proteolysis and that there are similar requirements in the signal peptide sequences from other genotypes. These results also confirm that the efficient SPP processing of core is required for virus production.
Conformational analyses of a synthetic core-E1 signal peptide by circular dichroism.
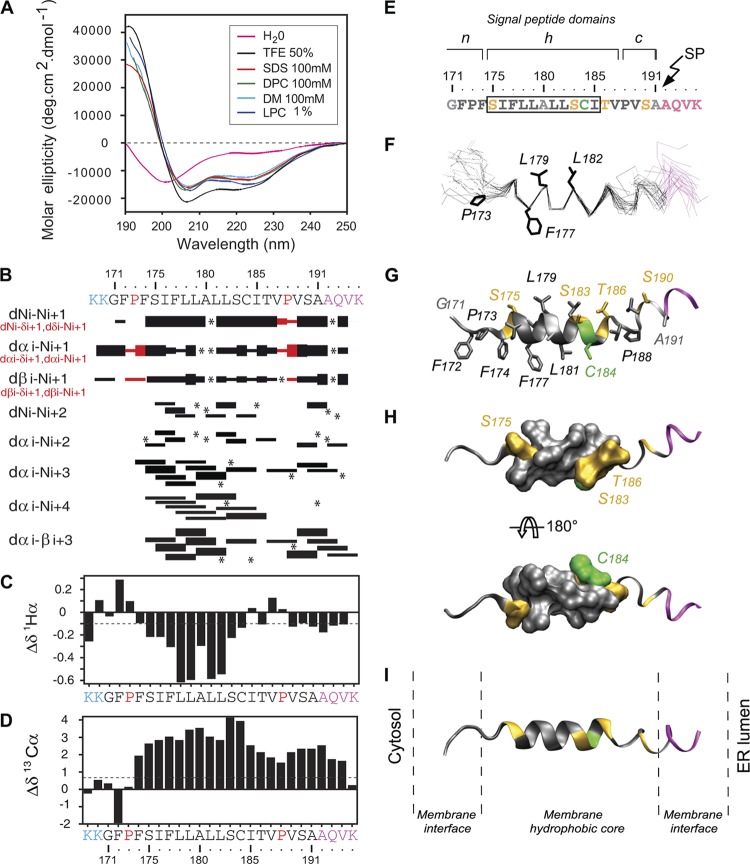
Given the similarities in the sequence requirements for SPP proteolysis for genotypes 1, 2, and 3 and the conserved physicochemical characteristics of the core-E1 signal sequence (Fig. 1A), we wished to gain deeper insights into the possible mechanism of cleavage by determining the structural characteristics and lipotropic properties of the core-E1 signal peptide region. Hence, we synthesized and purified a peptide of 27 residues, named sp-E1, which represented the uncleaved form of the core-E1 signal peptide. sp-E1 included the signal sequence at the C terminus of immature core (aa 171 to 191 from HCV strain JFH1) and the first four residues of E1 (Fig. 5B). The solubility of sp-E1 in water was limited, and it displayed a complex spectrum with a large negative band at around 200 nm and a negative shoulder at around 222 nm (Fig. 5A). The various CD deconvolution methods used indicated a mixture of random coil structures (50 to 70%) with some poorly defined and/or residual secondary structures (around 10% α-helix content and 7 to 18% extended structures). The secondary structure of sp-E1 was also examined in the presence of either lysophospholipid (lysophosphatidyl choline [LPC]) or a variety of detergents (SDS, N-dodecyl-β-d-maltoside [DM], and dodecyl phosphocholine [DPC]) or cosolvents (TFE-water mixture) that mimic a membranous environment (Fig. 5A). These conditions were selected to reflect the various conditions in an authentic membrane, to gain a more comprehensive picture of the peptide's conformational preferences. In the presence of the different detergents or TFE-water mixtures, the peptide exhibited typical CD spectra for α-helical folding, with a maximum at 190 nm and two minima at 208 and 222 nm (Fig. 5A). Analysis by various CD deconvolution methods indicated that sp-E1 was predominantly α-helical (44 to 52%), irrespective of the detergent used. The potential conformational preferences of the sp-E1 peptide were also probed in the presence of TFE, which can stabilize the folding of peptidic sequences, especially those exhibiting an intrinsic propensity to adopt an α-helical structure (5, 30). A titration of peptide folding with increasing proportions of TFE gave spectra that were characteristic of α-helical folding, and a maximal amplitude was reached at 40% TFE, which corresponds to an α-helical content of 55% (data not shown). An isodichroic point was observed at 204 nm (data not shown), indicating that the peptide undergoes a simple transition from a random coil to an α-helix, and according to the two-state model, equilibrium exists between the two conformers. This equilibrium, together with the α-helix content, which was comparable to that observed with detergents, is consistent with a stabilization of a helical region of about 13 amino acids. In summary, CD spectral analyses indicated the high propensity of the sp-E1 peptide to interact with lipids and to adopt an α-helical structure upon lipid binding.
Fig 5.
Structural analyses of the sp-E1 synthetic peptide. (A) Far-UV circular dichroism (CD) analysis of sp-E1 in various environments (the sp-E1 amino acid sequence is shown at the top of panel B). CD spectra were recorded in water (H2O), complemented with either 50% 2,2,2-trifluoroethanol (TFE), 1% l-α-lysophosphatidyl choline (LPC), or the following detergents: 100 mM sodium dodecyl sulfate (SDS), 100 mM N-dodecyl-β-d-maltoside (DM), or 100 mM dodecyl phosphocholine (DPC). (B to D) NMR analysis of the sp-E1 peptide in 50% TFE. The two lysine residues in blue at the N terminus were added to increase the solubility of the peptide. The four C-terminal residues in magenta correspond to the first residues of the E1 glycoprotein. (B) Summary of sequential (i, i + 1) and medium-range (i, i + 2 to i, i + 4) NOEs. Sequential NOEs allowing the assignment of proline residues are indicated in red. Asterisks indicate that the presence of an NOE cross peak was not confirmed because of overlapping resonances. Intensities of NOEs are indicated by the heights of the bars. (C and D) NMR-derived 1H-α (C) and 13C-α (D) chemical shift differences were calculated by the subtraction of the experimental values from the reported random-coil-conformation values in TFE (28). The dashed lines indicate the standard threshold value of ΔH-α (−0.1 ppm) (C) or ΔC-α (0.7 ppm) (D) for an α-helix. (E) Amino acid sequence of the core-E1 signal peptide from strain JFH1 (amino acid residues 171 to 191), including the first four residues of the E1 glycoprotein (shown in magenta at the C terminus). The box indicates α-helix residues 175 to 185, revealed by NMR. Residues are color coded according to their physicochemical properties: hydrophobic residues are black, neutral residues (Ala and Gly) are gray, polar residues (Ser and Thr) are yellow, and Cys is green. The characteristic structural topology of signal peptide domains is indicated at the top, including the N-terminal domain (n), the hydrophobic core region (h), and the C-terminal domain (c) (53) (see the text for details). (F) Superimposition of the backbone heavy atoms (N, C′, and C-α) of the 27 final structures (PDB accession number 2kqi), calculated by using the standard simulated annealing protocol in the Xplor-NIH 2.2.1 program (44). The 27 structures were superimposed for the best overlap of residues 175 to 185. Average positions of side-chain residues reported as potential C-terminal residues of the mature core protein (see the text) are displayed (stick representation). (G) Ribbon and stick models of a representative experimental structure of the sp-E1 peptide. Residues are colored based on the chemical properties of their side chains, as indicated above. (H) Two views of the surface of α-helix residues 175 to 185 showing the relatively polar face versus the highly hydrophobic side rotated by 180° (top and bottom views, respectively). (I) Tentative position of the sp-E1 peptide within a phospholipid bilayer of POPC (1-palmitoyl-2-oleoyl-3-sn-glycero-3-phosphocholine) on the same scale. Figures were generated from structure coordinates using VMD (http://www.ks.uiuc.edu/Research/vmd/) (15) and rendered with POV-Ray (http://www.povray.org/).
NMR analysis of the sp-E1 peptide.
To obtain detailed information at the atomic level for secondary structures formed by the sp-E1 peptide, its conformation was investigated by NMR. Deuterated micellar SDS and DPC are popular membrane-mimetic media for structural analyses of membrane-bound peptides by liquid NMR (38). Unfortunately, samples of sp-E1 peptide prepared in SDS and DPC displayed broad, poorly resolved NMR spectra (data not shown). Since approximately the same α-helical contents were observed for the peptide in TFE and in detergents, 50% TFE was considered to be an appropriate medium for such analyses. The sp-E1 peptide dissolved in 50% TFE-d2 yielded quite well-resolved NMR spectra (data not shown), and the sequential attribution of all spin systems was complete. An overview of the sequential and medium-range NOE connectivities is shown in Fig. 5B. The NOE connectivity patterns demonstrated that the central part of the peptide, including residues 175 to 185, displayed mostly characteristics of helix folding, including strong dNN(i, i + 1) and medium dN(i, i + 1) sequential connectivities as well as weak dN(i, i + 2), medium or strong dN(i, i + 3) and dα(i, i + 3), and weak dN(i, i + 4) medium-range connectivities. Apart from this central helix, typical connectivities of the helical fold are also present at the C terminus of the peptide (aa 188 to 194), including the SP cleavage site. The NOE-based indications of the helical conformation were supported by the deviation of the 1H and 13C chemical shifts from random-coil values (56), as shown in Fig. 5C and D. The long series of negative variation of 1H chemical shifts (Δδ1H⟨ ≤ −0.1 ppm) as well as the positive variation of 13C chemical shifts (Δδ13C⟨ ≥ 0.7 ppm) are indeed typical of an α-helical conformation. Based on the NOE-derived interproton distance constraints obtained with 50% TFE, a set of 50 structures was calculated with X-PLOR, and a final set of 27 low-energy structures that fully satisfied the experimental NMR data was retained. The numbers and types of NOE constraints used for the structure calculations as well as the statistics for this final set of 27 structures are given in Table S1 in the supplemental material. The superimposition of the 27 structures (Fig. 5F) shows that a central helical segment from aa 175 to 185 is well defined, with an RMSD of 0.24 Å (see Table S1 in the supplemental material). From the representative NMR structure shown in Fig. 5G, this α-helix includes two fully conserved Ser polar residues at positions 175 and 183, which are asymmetrically distributed on one side of the helix (Fig. 5G and H). The structure of the region downstream from the α-helix is poorly determined due to the lack of distance restraints (Fig. 5B), which is indicative of some structural flexibility. Interestingly, this region tends to adopt some helical folding in most calculated structures, as can be seen in Fig. 5G to I. However, the C-terminal α-helix from aa 189 to 194, which includes the SP cleavage site, is not well defined. The instability of this helix as well as that of the upstream segment between aa 186 and 188 are consistent with the necessity of an extended conformation of this region to allow cleavage by SP (52). In addition, there is a lack of any obvious structure for the n region of sp-E1 (Fig. 5F to H). These extended-conformation features at the n and c regions of sp-E1 are consistent with the need for the signal peptide, which is relatively short, to span the ER membrane (Fig. 5I).
DISCUSSION
In this report, we have identified the amino acids that contribute to SPP cleavage in GT3a strains of HCV, thereby extending studies on the requirements for proteolysis. Moreover, we have established the structure of the core-E1 signal peptide from HCV strain JFH1, which constitutes an essential framework for a further understanding of the molecular mechanisms for cleavage by both SP and SPP. Importantly, this represents the first substrate for SPP whose structure has been determined and therefore provides new insights into cleavage by the SPP enzyme.
The core-E1 signal peptide is mainly a hydrophobic segment, which directs the nascent HCV polyprotein to the ER membrane. This signal sequence presumably plays a key part in arresting translation by host cell ribosomes and targeting the polyprotein to the translocon at the ER membrane. In addition, it serves as a recognition site for SP and SPP to enable the maturation of the HCV core protein. It is considered that the cleavage of signal peptides by SPP is linked to degradative processes in the cell to prevent the accumulation of signal sequences within translocons, which may impair the efficiency of translation of proteins bound for secretion. The maturation of core by SPP must avoid the exposure of the protein to such processes; otherwise, insufficient core will accumulate to facilitate capsid assembly. These characteristics indicate that critical coding information is embedded within the short core-E1 signal sequence in the HCV polyprotein.
Additional evolutionary pressure may be exerted on sequence variability and SPP cleavage within the core-E1 signal peptide in order to avoid recognition by the immune system. It was reported recently that signal peptides are intrinsically immunogenic and have a high epitope density (20); thus, they possess functions beyond protein targeting to the translocon (26). Moreover, epitopes generated from major histocompatibility complex (MHC) class I signal sequences are produced by cleavage events that include SPP proteolysis before loading onto the nonclassical MHC class I molecule HLA-E (2, 22). This process generates a complex that is subsequently transported to the cell surface for presentation to natural killer cells. In silico analyses of HCV sequences (http://www.cbs.dtu.dk/services/NetCTL/) predict the presence of cytotoxic T lymphocyte (CTL) epitopes across the core-E1 signal peptide region, some of which would be substrates for SPP cleavage and therefore not presented to the immune system (Table 1). Thus, SPP cleavage could be part of a strategy adopted by the virus to limit immune recognition.
Table 1.
Predicted CTL epitopes across the core-E1 signal peptide region
| CTL epitopea (positions) | Substrate for SPP cleavage atb: |
||
|---|---|---|---|
| 177 | 179 | 182 | |
| LPGFPFSIF (169–177) | N | N | N |
| GFPFSIFLL (171–179) | Y | N | N |
| FSIFLLALL (174–183) | Y | Y | Y |
| FLLALLSCI (177–185) | N | Y | Y |
| LLSCITVPV (182–190) | N | N | Y |
See http://www.cbs.dtu.dk/services/NetCTL/. Y, yes; N, no.
Positions of the C-terminal amino acids of mature core.
Our study demonstrates that the core-E1 signal sequence exhibits lipid-binding properties and that this binding is necessary for its folding. The overall secondary structure corresponds to that expected for a typical signal sequence. Hence, the n region between amino acids 171 and 174 is unfolded, and the segment between residues 175 and 186 forms an α-helix and is the hydrophobic h region; the remaining stretch of the signal peptide from amino acids 187 to 191 is more polar than the h region and forms the flexible c region. The c region includes the recognition site for SP, which requires small neutral residues at positions −1 and −3 relative to the cleavage site (Ala and Val, respectively, in strain JFH1 and Ser and Ala, respectively, in GT3a strains). We conclude that these structural characteristics of the core-E1 signal sequence are preserved in all HCV genotypes, since the variable amino acids do not change the overall physicochemical properties of the peptide (Fig. 1A). Compared to classical signal peptides, the core-E1 signal peptide is shorter and does not include positively charged residues in the n domain, which generally interact with the membrane interface to allow the positioning of the signal peptide within the membrane. The extended conformation propensities of both the n and c regions flanking the more rigid helix core (located between amino acids 176 and 182) may overcome any limitations on membrane spanning imposed by the short length of the core-E1 signal sequence in order to facilitate sliding within the membrane and, thus, the accessibility of the cleavage site at positions 191 and 192 to SP on the luminal side of the ER membrane. After processing by SP, the lack of any charged amino acids in the n region could also promote the sliding of the signal sequence in the membrane to aid the recognition of and interaction with SPP.
Apart from the lack of any positively charged residues in the n region, the h region also contains polar residues that are atypical for signal sequences (Ser, Cys, and Thr). The Ser/Cys doublet at positions 183 and 184 is conserved across all HCV genotypes, as is Ser175 and Ala180, suggesting that these residues have a critical function(s). This is supported by the fact that the replacement of these polar residues (Ala180, Ser183, Cys184, and Thr186) by the hydrophobic amino acids Val, Leu, and Ile severely impairs SPP cleavage (construct SFV-GT3a-mut1) (Fig. 1A), while the additional mutation of His187 is required to fully abrogate SPP cleavage (construct SFV-GT3a-mut2) (Fig. 1). These features suggest that the recognition of the signal peptide by SPP might extend across the length of the sequence and could occur in sequential stages. From a structural point of view, the introduction of bulky hydrophobic side chains at these positions is expected to rigidify and extend the h-domain helix by suppressing the flexibility introduced into this region by the small polar residues. Thus, we postulate that the flexibility of the h-domain helix is an important feature for SPP cleavage, perhaps to allow the appropriate positioning and conformational adaption of the signal peptide to the SPP cleavage site. In addition, the location of polar residues Ser175, Ser183, and Thr186 (His187 in GT3a) on the same helix face could play a role in recognition by SPP and the targeting of the signal peptide to the enzyme's active site. The position of the SPP cleavage site within the core-E1 signal sequence has not been unequivocally determined but was reported to lie in the h region at position 177, 179, or 182 (16, 35). In line with other proteolytic events, cleavage is presumed to require local helix unfolding to an extended conformation at the cleavage site as well as a contribution from amino acids on either side of this site. In this context, the unfolding of the rigid helix turn at position 177 due to the bulky hydrophobic side chains of neighboring residues appears to be energetically less favorable than that at position 179 because of the presence of the small alanine side chain at position 180, which creates a “hole” in the helix side and thus induces some flexibility.
The requirements of the core-E1 signal peptide for SPP processing and the production of infectious virus have been the subject of a large number of studies (1, 7, 13, 17, 18, 23, 27, 33, 36, 37, 39, 50). In this report, we have extended analyses of the requirements for SPP proteolysis to include a GT3a core-E1 signal peptide, since the sequences for this genotype have greater variability than those for other genotypes (Fig. 1A) (18). The results with GT3a support our previous conclusion that the disruption of virion production and SPP proteolysis requires up to 4 amino acid mutations that target the polar residues and Ala180 in the h region downstream from the putative SPP cleavage site (50). This illustrates the cumulative importance of these residues for cleavage and virion production, but the requirement for multiple mutations in this segment to mediate these effects also highlights the redundancy that is apparently inherent to the SPP recognition and cleavage process. In contrast, the Ala substitution of single residues between amino acids 174 and 177, which lie upstream from the SPP cleavage site and border the n and h regions, abolishes virion assembly (18). However, these alterations either decreased the stability of the protein or did not change the apparent molecular weight of core, and so the lack of SPP cleavage could not be determined. Moreover, the substitution of amino acids Ile and Phe at positions 176 and 177 with Ala and Leu, respectively, eliminated virus production, but again, core was barely detected when these mutants were expressed in the context of the full-length polyprotein (36); the lack of SPP cleavage for this mutant could be detected, however, by the expression of a truncated polyprotein that terminated at the SP cleavage site at position 191 (36). Although we (50) and others (39) have detected immature core (i.e., either not cleaved or aberrantly cleaved by SPP), the recurring characteristic for many mutations introduced into the core-E1 signal peptide of full-length infectious genomes is the reduced abundance of the core protein. It is probable that the reduced stability of core is a consequence of mutations in the signal peptide, which perturb SPP cleavage. Such a phenotype has hampered the interpretation of data from studies of SPP proteolysis, since it is possible that lower virion production levels are a consequence of the instability of immature core as well as impaired SPP cleavage. The characterization of the factors that link or regulate SPP cleavage and degradative pathways may help to give a more complete understanding of the interplay between the maturation of core and the regulation of its abundance.
In conclusion, our mutational analysis of GT3a and our structural characterization of the JFH1 core-E1 signal peptides provide the basis for future studies of the mechanism for SPP proteolysis and its role in HCV infection. Moreover, few substrates for the enzyme have been identified, and apart from the HCV core-E1 signal peptide, the characterization of known substrates has received little attention. Thus, further studies of HCV as a model for SPP cleavage are likely to deepen our insights into intramembrane proteolysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Diesis for sp-E1 peptide chemical synthesis and purification, Takaji Wakita for providing JFH1, and Mark Harris and Stephen Griffin for the NS5A antiserum.
This work was supported by grants from the MRC Development Pathway Funding Scheme (grant number G0801769), the French National Agency for Research on AIDS and Viral Hepatitis (ANRS), the FINOVI foundation, and the TGE RMN THC Fr3050 for NMR studies. CD experiments were performed on the platform Production et Analyze de Protéines from the UMS BioSciences Gerland-Lyon Sud.
Footnotes
Published ahead of print 16 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ait-Goughoulte M, et al. 2006. Core protein cleavage by signal peptide peptidase is required for hepatitis C virus-like particle assembly. J. Gen. Virol. 87:855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bland FA, Lemberg MK, McMichael AJ, Martoglio B, Braud VM. 2003. Requirement of the proteasome for the trimming of signal peptide-derived epitopes presented by the nonclassical major histocompatibility complex class I molecule HLA-E. J. Biol. Chem. 278:33747–33752 [DOI] [PubMed] [Google Scholar]
- 3. Boulant S, et al. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236–22247 [DOI] [PubMed] [Google Scholar]
- 4. Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204–2213 [DOI] [PubMed] [Google Scholar]
- 5. Buck M. 1998. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31:297–355 [DOI] [PubMed] [Google Scholar]
- 6. Clayton RF, et al. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delgrange D, et al. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495–2503 [DOI] [PubMed] [Google Scholar]
- 8. Di Bisceglie AM. 1997. Hepatitis C and hepatocellular carcinoma. Hepatology 26:34S–38S [DOI] [PubMed] [Google Scholar]
- 9. Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Favier A, et al. 2002. Solution structure and dynamics of crh, the Bacillus subtilis catabolite repression HPr. J. Mol. Biol. 317:131–144 [DOI] [PubMed] [Google Scholar]
- 11. Health Protection Agency 2011. Hepatitis C in the UK: 2011 report. Health Protection Agency, London, United Kingdom [Google Scholar]
- 12. Hofer H, et al. 2002. Hepatocellular fat accumulation and low serum cholesterol in patients infected with HCV-3a. Am. J. Gastroenterol. 97:2880–2885 [DOI] [PubMed] [Google Scholar]
- 13. Hope RG, McElwee MJ, McLauchlan J. 2006. Efficient cleavage by signal peptide peptidase requires residues within the signal peptide between the core and E1 proteins of hepatitis C virus strain J1. J. Gen. Virol. 87:623–627 [DOI] [PubMed] [Google Scholar]
- 14. Hope RG, McLauchlan J. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913–1925 [DOI] [PubMed] [Google Scholar]
- 15. Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 16. Hussy P, Langen H, Mous J, Jacobsen H. 1996. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 224:93–104 [DOI] [PubMed] [Google Scholar]
- 17. Jhaveri R, McHutchison J, Patel K, Qiang G, Diehl AM. 2008. Specific polymorphisms in hepatitis C virus genotype 3 core protein associated with intracellular lipid accumulation. J. Infect. Dis. 197:283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kopp M, Murray CL, Jones CT, Rice CM. 2010. Genetic analysis of the carboxy-terminal region of the hepatitis C virus core protein. J. Virol. 84:1666–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koradi R, Billeter M, Wuthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 20. Kovjazin R, et al. 2011. Signal peptides and trans-membrane regions are broadly immunogenic and have high CD8+ T cell epitope densities: implications for vaccine development. Mol. Immunol. 48:1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 17:107–115 [DOI] [PubMed] [Google Scholar]
- 22. Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. 2001. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 167:6441–6446 [DOI] [PubMed] [Google Scholar]
- 23. Lemberg MK, Martoglio B. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735–744 [DOI] [PubMed] [Google Scholar]
- 24. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 25. Macdonald A, et al. 2003. The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signaling. J. Biol. Chem. 278:17775–17784 [DOI] [PubMed] [Google Scholar]
- 26. Martoglio B. 2003. Intramembrane proteolysis and post-targeting functions of signal peptides. Biochem. Soc. Trans. 31:1243–1247 [DOI] [PubMed] [Google Scholar]
- 27. McLauchlan J, Lemberg MK, Hope G, Martoglio B. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merutka G, Dyson HJ, Wright PE. 1995. ‘Random coil’ 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J. Biomol. NMR 5:14–24 [DOI] [PubMed] [Google Scholar]
- 29. Miyashita H, et al. 2011. Three-dimensional structure of the signal peptide peptidase. J. Biol. Chem. 286:26188–26197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Montserret R, et al. 1999. Structural analysis of the heparin-binding site of the NC1 domain of collagen XIV by CD and NMR. Biochemistry 38:6479–6488 [DOI] [PubMed] [Google Scholar]
- 31. Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 32. Murphy D, Chamberland J, Dandavino R, Sablon E. 2007. A new genotype of hepatitis C virus originating from central Africa. Hepatology 2007:623A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murray CL, Jones CT, Tassello J, Rice CM. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Negro F, Sanyal AJ. 2009. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int. 29(Suppl 2):26–37 [DOI] [PubMed] [Google Scholar]
- 35. Ogino T, Fukuda H, Imajoh-Ohmi S, Kohara M, Nomoto A. 2004. Membrane binding properties and terminal residues of the mature hepatitis C virus capsid protein in insect cells. J. Virol. 78:11766–11777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okamoto K, et al. 2008. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J. Virol. 82:8349–8361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okamoto K, Moriishi K, Miyamura T, Matsuura Y. 2004. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J. Virol. 78:6370–6380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Opella SJ. 1997. NMR and membrane proteins. Nat. Struct. Biol. 4(Suppl):845–848 [PubMed] [Google Scholar]
- 39. Pene V, Hernandez C, Vauloup-Fellous C, Garaud-Aunis J, Rosenberg AR. 2009. Sequential processing of hepatitis C virus core protein by host cell signal peptidase and signal peptide peptidase: a reassessment. J. Viral Hepat. 16:705–715 [DOI] [PubMed] [Google Scholar]
- 40. Penin F, et al. 1997. Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR. J. Mol. Biol. 270:496–510 [DOI] [PubMed] [Google Scholar]
- 41. Pietschmann T, et al. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rubbia-Brandt L, et al. 2001. Liver steatosis in chronic hepatitis C: a morphological sign suggesting infection with HCV genotype 3. Histopathology 39:119–124 [DOI] [PubMed] [Google Scholar]
- 43. Rubbia-Brandt L, et al. 2000. Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J. Hepatol. 33:106–115 [DOI] [PubMed] [Google Scholar]
- 44. Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160:65–73 [DOI] [PubMed] [Google Scholar]
- 45. Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169 [DOI] [PubMed] [Google Scholar]
- 46. Shaw ML, McLauchlan J, Mills PR, Patel AH, McCruden EA. 2003. Characterisation of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70:361–372 [DOI] [PubMed] [Google Scholar]
- 47. Shen Y, Delaglio F, Cornilescu G, Bax A. 2009. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simmonds P, et al. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973 [DOI] [PubMed] [Google Scholar]
- 49. Targett-Adams P, et al. 2003. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 278:15998–16007 [DOI] [PubMed] [Google Scholar]
- 50. Targett-Adams P, Hope G, Boulant S, McLauchlan J. 2008. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J. Biol. Chem. 283:16850–16859 [DOI] [PubMed] [Google Scholar]
- 51. Thanabal V, Omecinsky DO, Reily MD, Cody WL. 1994. The 13C chemical shifts of amino acids in aqueous solution containing organic solvents: application to the secondary structure characterization of peptides in aqueous trifluoroethanol solution. J. Biomol. NMR 4:47–59 [DOI] [PubMed] [Google Scholar]
- 52. von Heijne G. 1998. Protein transport: life and death of a signal peptide. Nature 396:111–113 [DOI] [PubMed] [Google Scholar]
- 53. von Heijne G. 1990. The signal peptide. J. Membr. Biol. 115:195–201 [DOI] [PubMed] [Google Scholar]
- 54. Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215–2218 [DOI] [PubMed] [Google Scholar]
- 55. Whitmore L, Wallace BA. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32:W668–W673 doi:10.1093/nar/gkh371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wishart DS, Sykes BD, Richards FM. 1992. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31:1647–1651 [DOI] [PubMed] [Google Scholar]
- 57. Wüthrich K. 1986. NMR of proteins and nucleic acids. John Wiley & Sons, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.