Abstract
Alphaviruses represent a highly important group of human and animal pathogens, which are transmitted by mosquito vectors between vertebrate hosts. The hallmark of alphavirus infection in vertebrates is the induction of a high-titer viremia, which is strongly dependent on the ability of the virus to interfere with host antiviral responses on both cellular and organismal levels. The identification of cellular factors, which are critical in orchestrating virus clearance without the development of cytopathic effect, may prove crucial in the design of new and highly effective antiviral treatments. To address this issue, we have developed a noncytopathic Venezuelan equine encephalitis virus (VEEV) mutant that can persistently replicate in cells defective in type I interferon (IFN) production or signaling but is cleared from IFN signaling-competent cells. Using this mutant, we analyzed (i) the spectrum of cellular genes activated by virus replication in the persistently infected cells and (ii) the spectrum of genes activated during noncytopathic virus clearance. By applying microarray-based technology and bioinformatic analysis, we identified a number of IFN-stimulated genes (ISGs) specifically activated during VEEV clearance. One of these gene products, the long isoform of PARP12 (PARP12L), demonstrated an inhibitory effect on the replication of VEEV, as well as other alphaviruses and several different types of other RNA viruses. Additionally, overexpression of two other members of the PARP gene superfamily was also shown to be capable of inhibiting VEEV replication.
INTRODUCTION
The Alphavirus genus is a group of arthropod-borne viruses in the Togaviridae family, many of which are important human and animal pathogens (54). In nature, these viruses are transmitted by mosquito vectors between vertebrate hosts (45). While mosquitoes remain mostly unaffected by alphavirus infection, vertebrates develop diseases of various severities. The hallmark of alphavirus replication in vertebrates is an induction of a high-titer viremia, which is required for further transmission of infection to feeding mosquitoes, thus perpetuating the cycle (24, 54). Alphaviruses are capable of reaching high titers due to highly efficient viral RNA replication and rapid production of viral nonstructural and structural proteins. However, the development of viremia is also strongly dependent on the virus' ability to interfere with the development of antiviral responses at the cellular level and in the organism as a whole (52, 53). Similar to the case for many other viruses, alphaviruses have developed a complex system of countermeasures, which promote their replication in the presence of cellular molecular pattern recognition receptors (PRRs) (17–19). These viruses downregulate induction of the antiviral response and/or become at least partially resistant to its activation. The ability of the alphaviruses to interact with host responses is a multicomponent process, with some of the constituents being specific to each member of the genus. However, some mechanisms appear to be common for several different members of the genus.
First, all characterized alphaviruses isolate their double-stranded RNA (dsRNA) replication intermediates from the cytoplasm into plasma and endosome membrane invaginations (spherules), in order to prevent these dsRNA molecules from being efficiently recognized by such known PRRs as RIG-I, MDA5, and protein kinase R (PKR) or Toll-like receptor 3 (TLR3) (14, 15, 23). These membrane spherules are connected to the cytosol by very narrow openings, which are associated with viral nsPs. The dsRNAs are sufficiently isolated to prevent their recognition by dsRNA-specific antibodies unless the spherule membrane is permeabilized by nonionic detergents (14). However, such dsRNA compartmentalization is likely to be incomplete, because cells release cytokines in response to replication of alphavirus mutants that are incapable of inhibiting cellular transcription (5, 13, 21).
Second, in cells of vertebrate origin, alphavirus replication induces transcriptional and translational shutoff (10, 20, 22). Previously, it was thought that inhibition of translation downregulates virus replication and represents a mechanism of host defense. However, an accumulating amount of data demonstrates that, at least in the case of Sindbis virus (SINV) and Semliki Forest virus (SFV), translational shutoff not only is mostly independent of PKR (22, 50) but also is highly beneficial for translation of viral structural proteins (11, 12, 44). These viruses have developed translational enhancers in the 5′ termini of their subgenomic RNAs (G-C-rich RNA sequences folded into stem-loops), which function only during virus-induced translational shutoff. The presence of these enhancers strongly increases expression of structural proteins by modified cellular translational machinery.
Inhibition of cellular transcription has been unambiguously demonstrated in cells of vertebrate origin infected by evolutionarily distinct alphaviruses, such as SINV, Venezuelan equine encephalitis virus (VEEV), and eastern equine encephalitis virus (EEEV) (2–4, 18). Each of these viruses uses different virus-specific proteins to achieve the same goal: to downregulate activation of antiviral genes. Cellular transcription inhibition is an efficient means of promoting virus replication but has some limitations. Both the SINV-specific nsP2 and the VEEV- and EEEV-specific capsid proteins, which demonstrate transcription-inhibitory activities, function in stoichiometric rather than catalytic, protease-dependent modes. This is likely to make them less efficient inhibitors during virus replication in vivo in less permissive cells, in which viral structural and nonstructural proteins are synthesized to lower levels. Previously described noncytopathic clearance of SINV from neuronal cells strongly supports the possibility that cells with incomplete transcription and translation inhibition might downregulate virus replication and ultimately clear replicating virus (6, 25).
Identification of such noncytopathic clearance mechanisms is critical for further understanding of alphavirus pathogenesis and development of new means of therapeutic intervention. However, the dissection and study of these processes in in vitro experiments are problematic. Most of the commonly used vertebrate cell lines are highly permissive for alphavirus replication and develop a profound cytopathic effect (CPE) within a few hours postinfection (p.i.). This phenomenon is a result of numerous, profound changes in cellular metabolism, manifested primarily by transcription and translation inhibition. This rapid CPE development could mask activation of the signaling pathways, which are better detectable in cells supporting lower levels of virus replication. Therefore, in our recent studies, we have developed variants of VEEV which exhibit a dramatically less cytopathic phenotype in all of the tested cell types but retain RNA replication at the wild-type (wt) level (5). These viruses produced the same amounts of virus-specific nonstructural and structural proteins, and in in vitro experiments, the infected cells released the same amounts of infectious virions as during wt virus infection. Therefore, it was concluded that the capsid-specific mutations introduced do not directly affect virus replication. The unique feature of these designed VEEV mutants was their noncytopathic, persistent phenotype in cells defective in type I interferon (IFN) signaling, such as IFN-α/βR−/− and STAT1−/− mouse embryonic fibroblasts (MEFs). However, cells competent in IFN production and signaling were able to clear these modified viruses. This clearance was a result of the virus' inability to effectively inhibit induction of the antiviral response, particularly the release of type I IFN into the medium and its autocrine signaling.
In this study, we used one of the noncytopathic VEEV variants to compare (i) the spectrum of cellular genes activated by virus replication in cells defective in IFN-α/β signaling and incapable of virus clearance to (ii) the spectrum of genes activated in cells with intact IFN-α/β signaling, which include the genes induced by both virus replication and the released type I IFN. The latter combined response was sufficient for virus clearance. Next, we identified a number of mouse IFN-stimulated genes (ISGs) which were differentially expressed during virus clearance and demonstrated that the product of one of them, a long isoform of PARP12, exhibits a strong negative effect on replication of VEEV, other alphaviruses, and viruses with both negative- and positive-strand RNA genomes.
MATERIALS AND METHODS
Cell cultures.
BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, MO). NIH 3T3 cells were obtained from the American Type Culture Collection (Manassas, VA). These cell lines were maintained at 37°C in alpha minimum essential medium (αMEM) supplemented with 10% fetal bovine serum (FBS) and vitamins. IFN-α/βR−/− mouse embryonic fibroblasts (MEFs) were kindly provided by Michael Diamond (Washington University, St. Louis, MO). They were propagated in Dulbecco's MEM supplemented with 10% FBS and nonessential amino acids.
Plasmid constructs.
Plasmids encoding VEEV genomes, pVEEV/GFP and pVEEV/GFP/C1 (Fig. 1) were described elsewhere (5). All other genes were synthesized by reverse transcription-PCR (RT-PCR) using specific primers and RNA isolated from NIH 3T3 cells. They were cloned into the plasmids, sequenced, and recloned into pVEEV/GFP and pVEEV/GFP/C1 to replace the green fluorescent protein (GFP)-coding sequence. VEEV replicon-carrying plasmids were described elsewhere (36). They were used to place heterologous genes of interest under the control of the subgenomic promoter carried by VEEV replicons. The schemes of the replicon and viral genomes are presented in the relevant figures. VEEV helper RNA-encoding plasmids were described elsewhere (51). All of the sequences and details of the cloning procedures can be provided upon request.
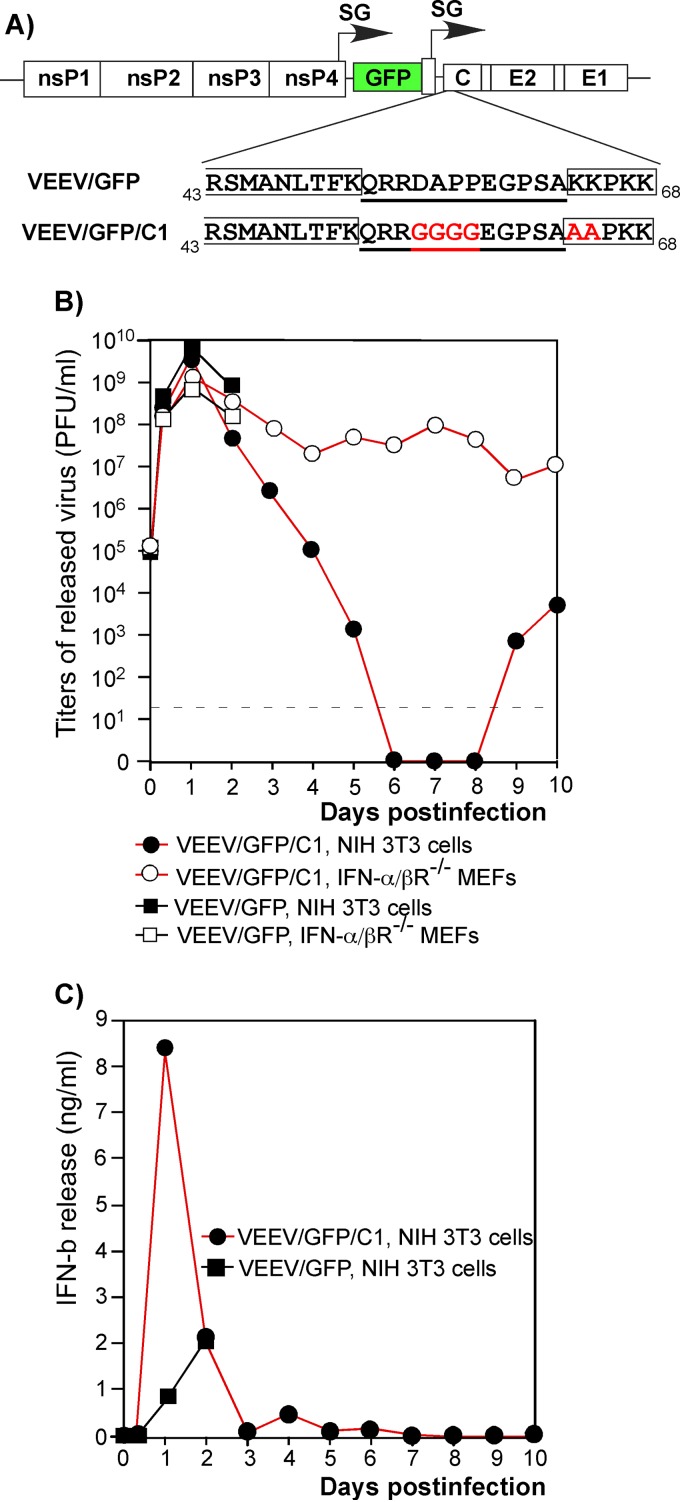
Fig 1.
The VEEV variant with a mutated capsid gene persistently replicates in IFN-α/βR−/− MEFs but is cleared from IFN-competent cells. (A) Schematic representation of the VEEV genomes encoding wt and mutated capsid proteins. The mutated amino acids are presented in red. The NLS and the carboxy terminus of the supra-NES are indicated by boxes (3). Underlined amino acids indicate connecting peptide. nsP1 to -4 indicate nonstructural genes. C, E2, and E1 indicate structural genes. SG indicates the subgenomic RNA promoter. (B) NIH 3T3 cells and IFN-α/βR−/− MEFs were infected with the indicated viruses at an MOI of 20 PFU/cell. Media were replaced at the indicated time points, and titers of the released viruses were measured by plaque assay on BHK-21 cells. (C) Concentrations of IFN-β were measured in the samples (see panel B) harvested from the infected NIH 3T3 cells.
RNA transcriptions.
Plasmids were purified by centrifugation in CsCl gradients. They were linearized using the MluI restriction site located downstream of the poly(A) sequence of the viral replicon and helper genomes. RNAs were synthesized using SP6 RNA polymerase in the presence of a cap analog using previously described conditions (37). The yield and integrity of transcripts were analyzed by gel electrophoresis under nondenaturing conditions, and transcription reaction products were used for electroporation without additional purification (32). Released viruses were harvested at 24 h postelectroporation, and titers were determined by plaque assay on BHK-21 cells (29). Replicon RNAs were coelectroporated into the cells together with helper RNAs, and released viral particles were harvested at 24 to 30 h postelectroporation. Titers were determined by infecting BHK-21 cells with different dilutions of harvested particles and staining them with VEEV nsP2-specific monoclonal antibody and secondary Alexa Fluor 555-labeled antibody. Numbers of infected cells were assessed by fluorescence microscopy.
Viral replication analysis.
Cells were seeded into 35-mm dishes. After a 4-h incubation at 37°C, monolayers were infected at the multiplicities of infection (MOIs) indicated in the figures or figure legends, washed with phosphate-buffered saline (PBS), and overlaid with 1 ml of complete medium. In some experiments, cells were initially infected with packaged replicons expressing heterologous genes of interest and at 1 h postinfection were superinfected with viruses. At the times indicated in the figures, medium was replaced by fresh medium, and virus titers were determined by a plaque assay on BHK-21 cells as previously described (29). VEEV TC-83 derivatives were found to be equally efficient in plaque formation in both the BHK-21 and NIH 3T3 cells used in this study (data not shown). Therefore, the applied MOIs are equally applicable to the experiments performed with both cell lines. All of the virus-related experiments were performed under biosafety level 2 (BSL2) conditions.
Microarray analysis.
NIH 3T3 cells and IFN-α/βR−/− MEFs were seeded at a concentration of 2 × 106 cells per 100-mm dish. After a 4-h incubation at 37°C, cells were infected with VEEV/GFP/C1 at an MOI of 20 PFU/cell or treated with 1,000 IU/ml of IFN-β. At the indicated time points, total RNA was isolated using TRIzol according to the manufacturer's instructions (Invitrogen). RNA was additionally purified with the RNeasy minikit (Qiagen). RNA quality was tested using an Agilent 2100 Bioanalyzer. cDNA synthesis, labeling, hybridization on mouse gene 1.0 ST array GeneChips (Affymetrix), and image processing were performed at the Heflin Center Genomics Core facility (University of Alabama, Birmingham). Two independent RNA samples were prepared for each indicated time point for VEEV/GFP/C1-infected cells and three RNA samples for IFN-β-treated cells. The robust multichip average (RMA) algorithm was used to normalize the raw intensity values using the GeneSpring software program, version GX 11.5 (Agilent). To find differentially expressed genes, a one-way analysis of variance (ANOVA) statistical test with Benjamini-Hochberg correction was applied to the normalized intensity data. Entries with more than a 2-fold change in expression and a P value of <0.05 were chosen for further analysis.
RT-qPCR.
RNA samples which were used for microarray analysis were also analyzed to compare concentrations of specific cellular RNAs by quantitative PCR (qPCR). cDNA was synthesized using 1 μg of total isolated RNA with a QuantiTect reverse transcription kit (Qiagen). qPCR primers were designed for the following genes: Bst2 (NM_198095), Trim30 (NM_009099), USP18 (NM_115783), Ifi27 (NM_029803), PARP12 (NM_172893), ISG20 (NM_020583), and MOV10 (NM_008619). qPCR was performed using SsoFast EvaGreen Supermix (Bio-Rad) in a CFX96 real-time PCR detection system (Bio-Rad) for 40 cycles with two steps per cycle, each step for 5 s (a denaturing step at 98°C and an annealing/extension step at 60°C). Results of the quantification were normalized to the amount of β-actin mRNA present in the same RNA samples. The RNA fold change was determined by comparing data for VEEV/GFP/C1-infected NIH 3T3 cells (24 h p.i.) to those for a mock-infected sample by the ΔΔCT method. Each qPCR was performed in triplicate, and means and standard deviations (SDs) were calculated.
Western blotting.
Equal amounts of proteins were separated on a 4–12% gradient NuPAGE gel. After protein transfer, the membranes were treated with VEEV nsP2-specific monoclonal antibody (MAb) and β-actin-specific antibodies, followed by treatment with infrared dye-labeled secondary antibodies. For quantitative analysis, membranes were scanned on a Li-Cor imager.
Firefly luciferase activity.
Firefly luciferase activity was measured with the luciferase assay system (Promega) according to the manufacturer's instructions. Briefly, at the indicated time points, cells were washed with PBS, lysed with Reporter lysis buffer (Promega), and frozen overnight at −80°C. The luminescence was measured for 10 μl of 100-fold-diluted lysates using an FB12 luminometer (Berthold). All of the measurements were performed in triplicates, and means and SDs were calculated.
RESULTS
A VEEV variant with mutated capsid genes can be cleared from NIH 3T3 cells but not from IFN-α/βR−/− MEFs.
In our previous studies, we developed VEEV TC-83 variants containing a set of redundant, attenuating point mutations in the capsid gene (5). One of these variants, VEEV/GFP/C1, was used in this new study. It contained mutations in the capsid protein-specific nuclear localization signal (NLS) and in the peptide connecting the supraphysiological nuclear export signal (supra-NES) and the NLS (Fig. 1A) (3, 4). These mutations made the capsid protein incapable of interfering with nucleocytoplasmic trafficking and inhibiting cellular transcription (3, 5). The control virus, VEEV/GFP, was identical to VEEV/GFP/C1 in terms of genome organization but encoded the wt VEEV TC-83 capsid protein (Fig. 1A). Both recombinant viruses contained a GFP-coding sequence under the control of an additional subgenomic promoter.
In both NIH 3T3 cells, which have no defects in type I IFN production and signaling, and IFN-α/βR−/− MEFs, VEEV/GFP infection caused a cytopathic effect and total cell death within 24 to 48 h postinfection (Fig. 1B). In contrast, the VEEV/GFP/C1 mutant replicated without detectable CPE in both NIH 3T3 cells and IFN-α/βR−/− MEFs (Fig. 1B), as well as in STAT1−/− MEFs, Vero cells, and BHK-21 cells (reference 5 and data not shown). In NIH 3T3 cells, this mutant replicated to essentially the same titers as VEEV/GFP (Fig. 1B); however, the infected cells rapidly released high levels of IFN-β (Fig. 1C). Within 5 to 6 days postinfection, NIH 3T3 cells were able to downregulate virus replication to an undetectable level. Virus replication was then reactivated after day 9 (Fig. 1B). The latter stage of the infection was characterized by the presence of a low concentration of IFN-β in the medium and inefficient virus release by a small percentage of the cells (5). IFN-α/βR−/− MEFs were unable to clear the noncytopathic mutant and, in multiple, reproducible experiments, continued to produce infectious virus for at least 21 days, until termination of the experiments (Fig. 1B) (5).
Thus, the VEEV variant with a mutated capsid protein, VEEV/GFP/C1, induced an antiviral state in cells with intact type I IFN production and signaling, which led to the downregulation of viral replication to undetectable levels. However, the same virus was able to persistently replicate in cells defective in type I IFN signaling, suggesting that autocrine IFN signaling and activation of ISGs must play critical roles in noncytopathic VEEV/GFP/C1 clearance. Experiments similar to those presented above have been performed not only with NIH 3T3 cells but also with primary and immortalized MEFs. The latter cells also demonstrated VEEV/GFP/C1 clearance. However, it was almost impossible to synchronously infect these wt MEFs at reasonable MOIs (10 to 20 PFU/cell). This effect was observed with VEEV/GFP/C1 and particularly Sindbis virus (SINV) (reference 5 and data not shown). This strongly complicated interpretation of the data from the long-term-infection-based experiments and those based on microarray analysis. Therefore, the experiments presented in the following sections were performed using NIH 3T3 cells as a representative wt mouse cell line. They unambiguously demonstrated the ability of these cells to clear replication of noncytopathic alphaviruses.
Microarray analysis defined a set of genes differentially expressed during virus clearance but not in persistently infected cells.
The experiments described above suggested that in the absence of autocrine IFN signaling, the PRR-mediated antiviral response in IFN-α/βR−/− MEFs is insufficient for clearance of VEEV/GFP/C1 infection. This indicated that the infected NIH 3T3 cells activate an important component(s) of the cellular response that is induced by released type I IFN and that this cellular factor(s) plays a critical role in the downregulation of virus replication. Thus, the rationale of the following analysis, presented in Fig. S1 in the supplemental material, was to identify the cellular gene(s) that was (i) activated in NIH 3T3 cells by VEEV/GFP/C1 replication and secreted type I IFN, (ii) activated by IFN-β treatment itself, and (iii) not activated in the IFN-α/βR−/− MEFs by persistently replicating virus and to further assess its antiviral function.
To achieve this, we infected NIH 3T3 cells and IFN-α/βR−/− MEFs with VEEV/GFP/C1, isolated total cellular RNAs at different times postinfection, and analyzed changes in the mRNA profiles using a microarray-based approach. The same analysis was applied to NIH 3T3 cells that were treated with 1,000 IU/ml of IFN-β for 24 h (the first time point of RNA isolation). The IFN-β-treated cells were then incubated in IFN-free medium, and RNA was isolated at 24 h and 48 h after IFN-β withdrawal (time points 2 and 3, respectively, in Fig. 2). Mock-infected NIH 3T3 cells and IFN-α/βR−/− MEFs were used to isolate control RNAs. To ensure statistical significance, all of the microarray experiments were performed in duplicates or triplicates. Data from several time points were analyzed on both Affymetrix and Illumina gene chips, which generated very similar results (data not shown).
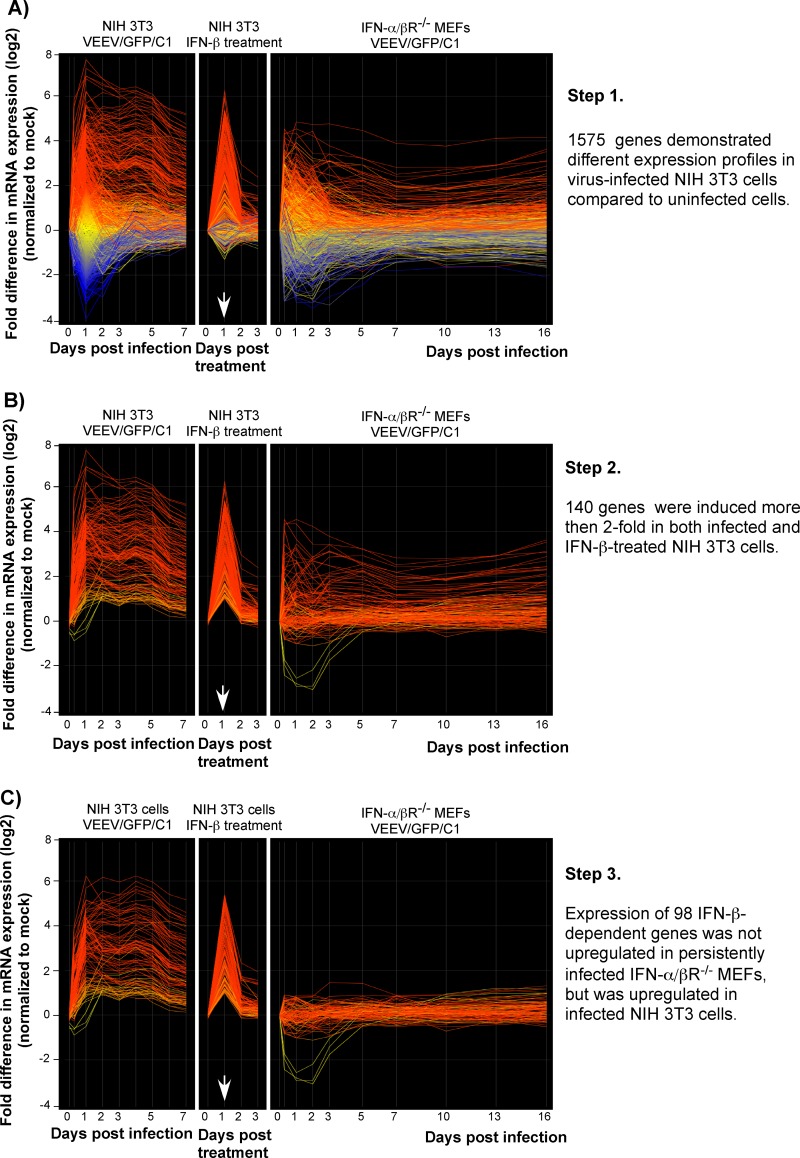
Fig 2.
Microarray data analysis. Panels A, B, and C present the sequential analysis used to reduce the number of genes to analyze further in this study (see the text for details). In each, the first panel shows the gene expression profiles in the VEEV/GFP/C1-infected NIH 3T3 cells. The expression profiles of the same genes in IFN-β-treated NIH 3T3 cells and in the infected IFN-α/βR−/− MEFs are presented in the second and the third panels, respectively. Each line reflects expression levels of a single gene at different time points. The red lines represent the genes whose expression was activated the most, and the blue color indicates the genes whose expression was downregulated the most in the VEEV/GFP/C1-infected compared to mock-infected NIH 3T3 cells. Other colors indicate intermediate levels of activation and downregulation of gene expression. The arrow indicates the end of IFN-β treatment.
In the NIH 3T3 cells infected with VEEV/GFP/C1, gene expression profiles were assessed and analyzed starting from 8 h postinfection, until day 16. However, in order to simplify the data presentation, we present results only for samples harvested until day 7 (Fig. 2), when the virus was no longer detectable in the medium (Fig. 1B). The ANOVA statistical test revealed that the VEEV/GFP/C1 infection in the NIH 3T3 cells significantly changed expression of 1,575 genes. The first panel of Fig. 2A displays the expression profiles of these 1,575 genes. The second and third panels represent the expression profiles of the same genes in IFN-β-treated NIH 3T3 cells and in VEEV/GFP/C1-infected IFN-α/βR−/− MEFs, respectively. In all three cases, the highest activation of gene expression occurred by 24 h postinfection, and at this time, in the infected NIH 3T3 cells, 388 different mRNAs were present at 2- to 207-fold-higher levels than in the mock-infected cells.
Further analysis narrowed down the spectrum of genes of interest by identifying the subset of those activated more than 2-fold in both infected NIH 3T3 cells and cells treated with IFN-β (Fig. 2B; see Tables S1 and S2 in the supplemental material). This subset contained only 140 genes. Their expression reached the highest level by 24 h postinfection and then gradually decreased. The rates of decrease were higher in cells activated by IFN-β treatment only. In 24 h, after removal of IFN-β from the medium, the NIH 3T3 cells demonstrated a rapid return of gene expression back to normal, pretreatment levels (Fig. 2A and B). By this time, cells previously treated with type I IFN become susceptible to virus infection (5).
Next, we excluded from further analysis the genes that were activated by virus replication in IFN-α/βR−/− MEFs (Fig. 2C). We concluded that these genes were activated by virus infection only and expressed proteins that were incapable of stopping virus replication (at least at their observed levels of expression). After applying the above analytical criteria, 98 IFN-inducible genes that were activated by VEEV/GFP/C1 replication and produced IFN-α/β in the NIH 3T3 cells but not in the IFN-α/βR−/− MEFs were left (Fig. 2C).
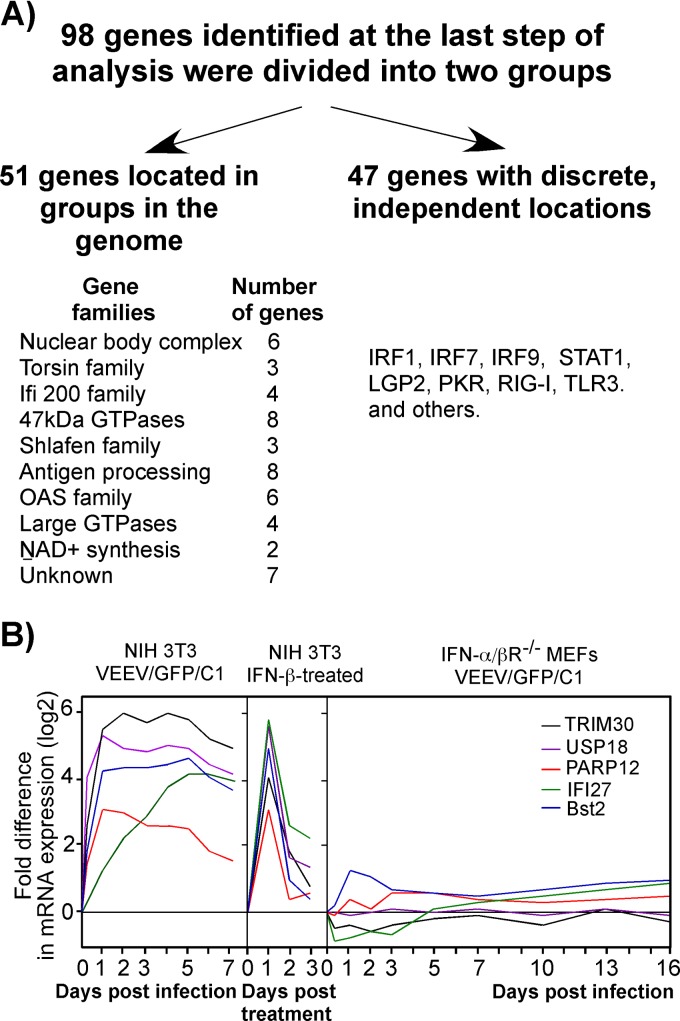
To further narrow down the analysis, we divided the remaining 98 genes into two groups (Fig. 3A). The first group included genes that are located close together in the mouse genome and expressed as families, such as the oligoadenylate synthetase (OAS) genes. We theorized that it was likely that these genes have related functions and if expressed separately should have no significant effects on viral replication. The second group contained genes that are separated in the genome and activated independently, such as the IRF7, STAT1, LGP2, PKR, RIG-I, and TLR3, genes (7, 30, 39, 46, 55). The latter group contained only 47 genes. We selected from them a small subset of genes demonstrating the most efficient induction and long-term activation in the VEEV/GFP/C1-infected NIH 3T3 cells and also excluded genes encoding proteins with previously characterized antiviral functions, such as PKR and RIG-I. We also excluded genes for proteins involved in type I IFN signaling pathways and those mediating the downregulation of the IFN response, such as Usp18. The latter gene was used in some of the experiments presented below but only as a control having no antiviral function.
Fig 3.
Four cellular genes were selected for further analysis. (A) Scheme used to define the final set of genes with putative antiviral functions. (B) Expression profiles of cellular genes that were selected for testing of their antiviral activities.
After applying these final exclusion filters, the final group of selected genes was represented by PARP12, Ifi27, Trim30, and Bst2. Their expression profiles in (i) VEEV/GFP/C1-infected NIH 3T3 cells, (ii) IFN-β-treated NIH 3T3 cells, and (iii) VEEV/C1/GFP-infected IFN-α/βR−/− MEFs are presented in Fig. 3B.
Murine PARP12 demonstrates antiviral activity.
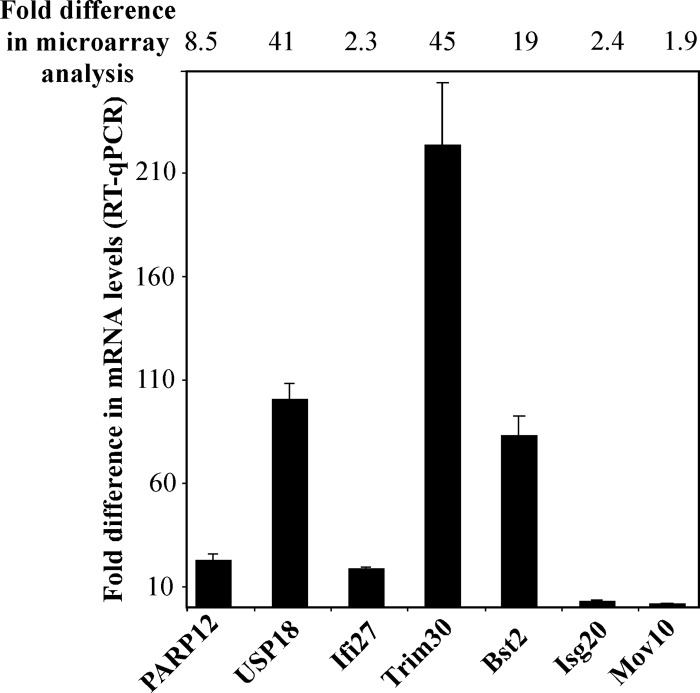
The first step in further analysis was to confirm gene activation by RT-qPCR. In these experiments, we used the RNA samples isolated from the NIH 3T3 cells at 24 h postinfection with VEEV/GFP/C1 and previously used for the microarray analysis presented in Fig. 2. RT-qPCR was performed using gene-specific primers (see Materials and Methods for details). Data were normalized to the concentrations of β-actin mRNA. The results, presented in Fig. 4, confirmed activation of the genes which had previously been defined using gene-chip analysis (Fig. 3B). However, for each of the 4 genes tested, the induction levels determined by RT-qPCR were higher than those found in the microarray experiments. Usp18 and ISG20 mRNA templates, used as additional controls, were also found at higher concentrations than those expected from the microarray data. Thus, the expression of PARP12, Ifi27, Trim30, and Bst2 is activated in NIH 3T3 cells during VEEV/GFP/C1 replication.
Fig 4.
qPCR analysis confirms the microarray data. The selected genes demonstrate higher levels of activation in RT-qPCR tests than those detected in the microarray experiments. The experiments were performed in triplicates (see Materials and Methods for details).
Alphaviruses represent a convenient system for simultaneous expression of the genes of interest and testing their effects on virus replication. The genes can be introduced under the control of an additional subgenomic promoter into the viral genome, and the effects of their products on replication can then be evaluated by comparing the titers of released viruses (30, 55). While it is true that cloned heterologous genes are not expressed at very early times postinfection and their effects on early events of virus replication cannot be tested, this system mimics the events occurring during ISG induction and allows the experiments to be performed in different cell types. To prevent argument over the validity of the experimental system, a number of different approaches were undertaken to confirm the antiviral effect (see the following sections for details). Thus, we synthesized murine Ifi27, Trim30, Bst2, and PARP12 genes by RT-PCR and cloned them into the VEEV/GFP/C1 genome under the control of an additional subgenomic promoter, replacing the GFP-coding sequence (Fig. 5A). Sequencing of PARP12-specific cDNA revealed that its mRNA is present in the cells in two splice forms (Fig. 5B), and thus, two corresponding proteins are likely to accumulate. We termed them long and short isoforms (PARP12L and PARP12S, respectively) (Fig. 5A and B). The completely spliced PARP12L RNA encoded a 711-amino-acid (aa) protein, and PARP12S-specific RNA encoded a 485-aa polypeptide that contained the same five Zn fingers as PARP12L but lacked the PARP domain. In order to detect the antiviral functions of these two forms, both the PARP12L and PARP12S genes were separately cloned into the viral genome. Because the insertion size might affect RNA replication and/or packaging, we used cDNA fragments of the same length. Thus, in the case of PARP12S, the cloned gene contained a large, 3′-terminal nucleotide sequence which was not translated due to presence of a short intron-mediated frameshift. The recombinant viral genomes were synthesized in vitro and transfected into BHK-21 cells. All of the designed double-subgenomic viruses were viable, and most of them produced homogenous plaques with a size approximating that of VEEV/GFP/C1. The only two viruses that produced noticeably smaller plaques were those expressing PARP12L and Bst2 proteins (data not shown). The smaller-plaque-forming phenotype was unlikely to be attributable to the insertion sizes, as the PARP12S-specific sequence was of the same length as that of PARP12L but did not affect plaque size and virus titers. The Bst2 gene was only 519 nucleotides (nt) long and thus was even smaller than the control GFP-coding sequence.
Fig 5.
PARP12L and Bst2 expression has a negative effect on VEEV mutant replication. (A) Schematic representation of recombinant viral genomes. HG indicates a position of cloned heterologous genes. (B) Schematic representation of long and short isoforms of PARP12 protein. (C) Replication of recombinant VEEV variants expressing the indicated heterologous genes in NIH 3T3 cells. Cells were infected at an MOI of 10 PFU/cell, media were replaced at the indicated times, and virus titers were determined by plaque assay on BHK-21 cells.
From all of the tested constructs, only PARP12L- and Bst2-encoding viruses reproducibly demonstrated lower rates of replication in NIH 3T3 cells (Fig. 5C). In this and numerous other tests, viruses expressing PARP12S, Trim30, and Ifi27 exhibited growth rates almost identical to those of VEEV/GFP/C1 (Fig. 5C and data not shown). The experiments with double-subgenomic viruses could not rule out the possibility that in order to demonstrate an antiviral effect, Trim30, Ifi27, and PARP12S need to be overexpressed before infection or at very early times postinfection. However, in other experiments, based on transient expression of these proteins from plasmid DNA, they also failed to demonstrate replication-inhibitory functions (data not shown). Thus, taken together, the data suggested that expression of Trim30, Ifi27, and PARP12S to higher levels alone has no detectable antiviral function.
During the course of the above-described experiments, we noticed that while the MOI used for controls had no effect on final virus titers, the inhibitory effect of PARP12L and Bst2 was dependent on the MOI and the cell type used, suggesting that the antiviral effect may depend on the time of expression during VEEV replication. To clearly demonstrate this and express the proteins of interest at earlier times postinfection, we infected NIH 3T3 and BHK-21 cells at different MOIs with VEEV/PARP12L/C1 and VEEV/Bst2/C1 viruses and assessed the rates of virus release. In these experiments, virus carrying the entire PARP12L gene in reverse orientation under control of the duplicated subgenomic promoter, VEEV/PARPrev/C1, was used as a control. The later time points of virus replication were excluded from the analysis in these and most of the other experiments, because PARP12L expression could potentially reach nonphysiological levels. We were unable to confirm this conjecture, as the commercially available PARP12-specific Abs that we tested were unable to recognize either PARP12L or PARP12S on Western blots. Thus, we could not analyze the levels of endogenous and exogenous PARP expression. Therefore, we focused our analysis of virus replication on the early times postinfection, i.e., 12 h p.i.
The results presented in Fig. 6 clearly demonstrate more efficient and earlier antiviral activities of both PARP12L and Bst2 proteins at higher MOIs, which appeared to promote earlier expression of PARP12L and Bst2 from VEEV genomes. The antiviral effect was also more prominent in BHK-21 cells, which, based on our previous experience, support more efficient viral RNA replication and higher levels of heterologous protein expression from the alphavirus genome-based constructs than do NIH 3T3 cells (data not shown). This suggested that inhibition of virus replication depends on the concentration and/or rates of accumulation of the studied proteins in the cells.
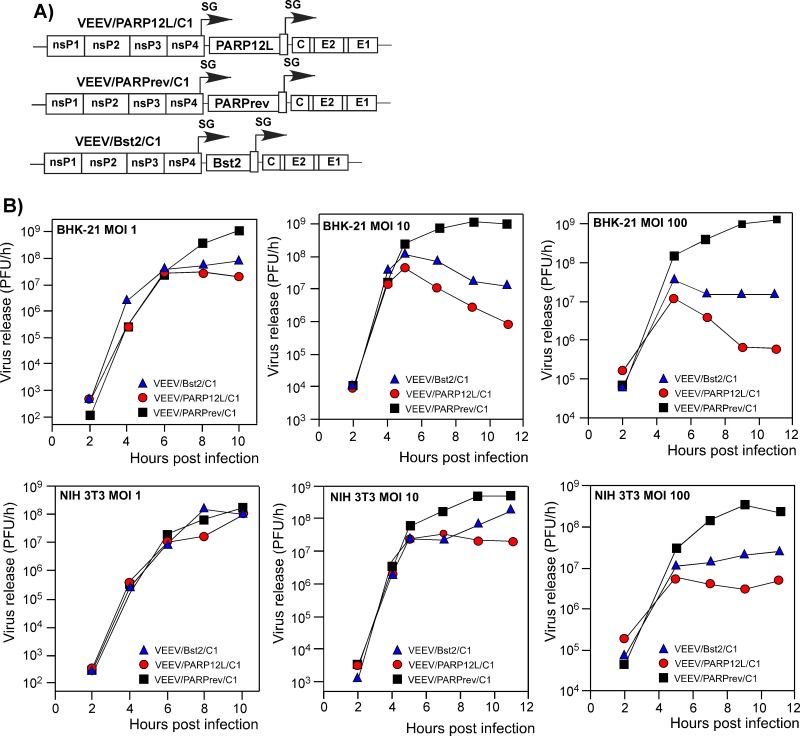
Fig 6.
The antiviral effects of PARP12L and Bst2, expressed from the viral genome, is dependent on the applied MOI and cell type. (A) The schematic representation of recombinant VEEV genomes encoding mutated capsid protein, Bst2 and the PARP12L genes in direct or reverse orientations. (B) NIH 3T3 and BHK-21 cells were infected with recombinant viruses encoding PARP12L, Bst2, or PARP12L in reverse orientation at the indicated MOIs. Media were harvested at the indicated times postinfection, and virus titers were determined by plaque assay on BHK-21 cells.
Bst2 protein is known to affect release of some enveloped viruses from the cell surface (40, 41). Therefore, in these experiments, its expression was mostly used as a positive control to additionally demonstrate that the applied virus-based expression system is able to detect the inhibitory activities of the studied gene products. Moreover, the experiments described above demonstrated that PARP12L possesses a more efficient inhibitory function(s) than Bst2. Therefore, the experiments described in the following sections were focused exclusively on further analysis of the PARP12L-specific antiviral effect.
To rule out the possibility that the virus replication-inhibitory functions of PARP12L are specific to replication of VEEV with mutated capsid protein, we cloned the corresponding gene and PARP12L in reverse orientation into VEEV TC-83 containing the wt capsid protein, VEEV/PARP12L and VEEV/PARPrev, respectively (Fig. 7A). As we expected, expression of PARP12L affected VEEV replication in both BHK-21 and NIH 3T3 cells (Fig. 7B). This was a strong indication that the inhibitory activity of PARP12L is not limited to VEEV variants encoding mutated capsid protein.
Fig 7.
Expression of PARP12L affects replication of VEEV encoding wt capsid protein. (A) Schematic representation of recombinant VEEV genomes encoding wt capsid protein and the PARP12L genes in direct or reverse orientations. (B) Analysis of replication of the recombinant viruses in BHK-21 and NIH 3T3 cells. Media were harvested at different times postinfection, and virus titers were determined by plaque assay on BHK-21 cells.
Expression of PARP12L in trans downregulates virus replication.
To further assess the antiviral activity of PARP12L, we attempted to produce recombinant lentiviruses carrying the PARP12L and PARP12S genes. However, the PARP12L-encoding constructs, but not the constructs encoding PARP12S or other gene products, were packaged to very low titers (data not shown), suggesting that the antiviral effect of PARP12L might be broader than expected and affect replication of other viruses.
We also transiently expressed the PARP12L, PARP12S, Bst2, Trim30, and GFP genes in NIH 3T3 cells from RNA polymerase II promoters and tested their effects on virus replication. Plasmid DNAs carrying the genes of interest under the control of the cytomegalovirus (CMV) promoter were electroporated into NIH 3T3 cells. At 24 h posttransfection, cells were infected with the VEEV/GFP/C1 mutant, and replication rates of this virus were assessed. The results were encouraging. PARP12L expression led to a 4- to 6-fold decrease in virus titers (data not shown). However, plasmid transfection of NIH 3T3 cells is not very efficient, and the CMV promoter normally generates a very high level of heterologous gene expression, which could be higher than the standard physiological level.
In order to exclude potential effects of high PARP12L expression on cell metabolism indirectly affecting virus replication, we applied another experimental system, which allowed the study of the antiviral effect of PARP12L at early times after the initiation of its expression. PARP12L and PARPrev sequences were cloned into VEEV replicons to generate the constructs VEErep/PARP12L and VEErep/PARPrev, respectively (Fig. 8A). The in vitro-synthesized RNAs were cotransfected into BHK-21 cells together with helper RNAs (see Materials and Methods for details), and replicon-containing viral particles were harvested at 24 h posttransfection. Titers of packaged VEErep/PARP12L replicons were at least 10-fold lower than those of VEEV replicons carrying other heterologous genes. However, they were sufficiently high to infect all of the cells in the monolayers in the following experiments.
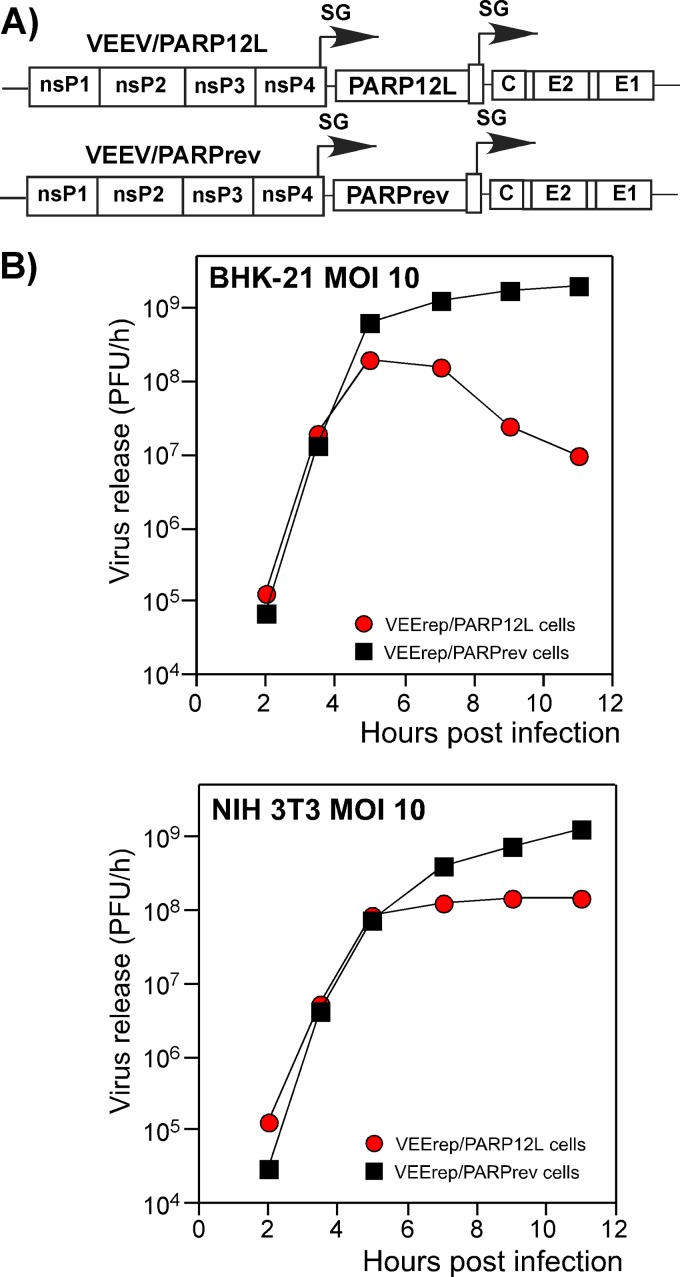
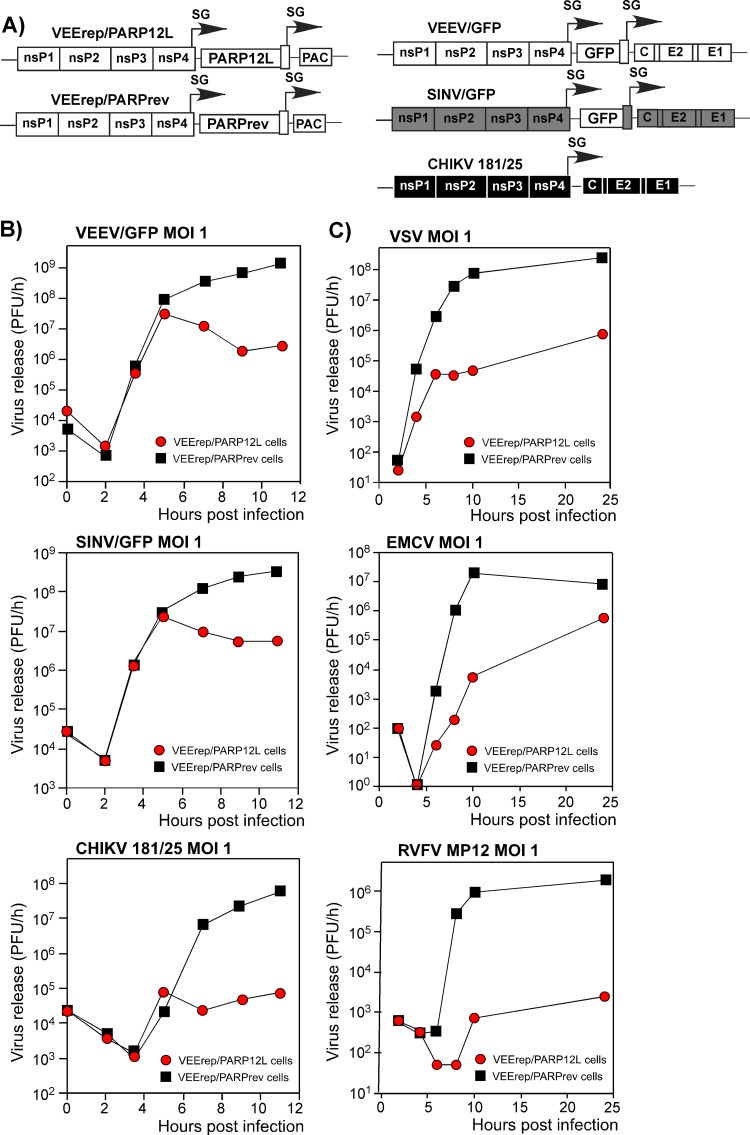
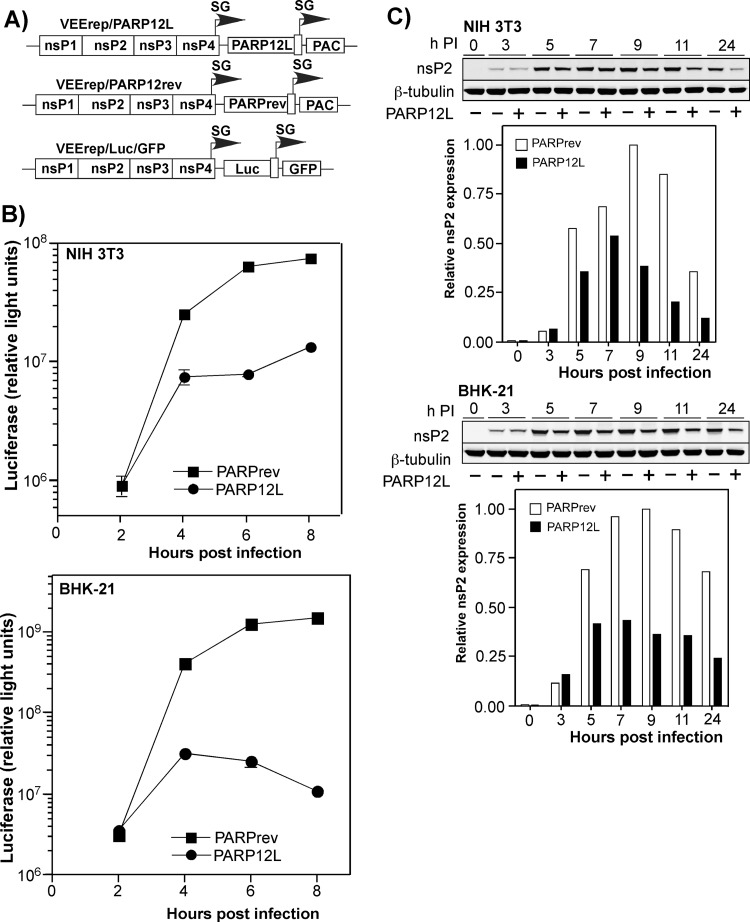
Fig 8.
Expression of PARP12L affects replication of different alphaviruses and other RNA viruses. (A) Schematic representation of the VEEV replicon expressing the PARP12L gene, the control VEEV replicon, and the genomes of VEEV, SINV, and CHIKV used for superinfection. PAC indicates a puromycin acetyltransferase gene. (B and C) BHK-21 cells were infected with packaged VEEV replicons encoding PARP12L in either the direct or reverse orientation at an MOI of 20 infectious units/cell. One hour later, they were infected with the indicated viruses at an MOI of 1 PFU/cell, and media were replaced at the indicated times. Titers were determined by plaque assay on BHK-21 cells for all viruses.
BHK-21 cells were infected with packaged replicons at an MOI of 20 infectious units/cell (inf.u./cell). One hour later, before PARP12L expression reached a high level, they were infected with VEEV/GFP, SINV/GFP (double-subgenomic Sindbis virus expressing GFP), and attenuated Chikungunya virus strain 181/25 (CHIKV 181/25) at an MOI of 1 PFU/cell. In the preliminary experiments, 1 h between replicon and virus infections was found to be an insufficient time for the development of the superinfection exclusion phenomenon, which occurs in vertebrate cells during double infection with alphaviruses. In multiple, reproducible experiments, PARP12L expression had a strong negative effect on the rates of VEEV/GFP release (Fig. 8B presents the results of one of the reproducible experiments). Similarly, it inhibited replication of SINV/GFP and CHIKV 181/25. At 6 h postinfection, the CHIKV titers were more than three orders of magnitude lower than those found in the samples harvested from VEErep/PARPrev replicon-containing cells, superinfected with the indicated CHIKV. A similar negative effect of PARP12L expression on virus replication was also detected in NIH 3T3 cells (data not shown).
Importantly, the antiviral effect of PARP12L expression was found not to be limited to alphavirus infections. In another set of experiments, the VEErep/PARP12L-containing cells were superinfected with (i) vesicular stomatitis virus (VSV), a negative-strand RNA genome virus, (ii) encephalomyocarditis virus (EMCV), a positive-strand RNA genome virus, and (iii) Rift Valley fever virus (RVFV) MP12, an ambisense RNA genome virus. All of the indicated viruses replicated slower in the presence of VEErep/PARP12L replicon than in cells containing the control VEErep/PARPrev (Fig. 8C). This was particularly evident at early times postinfection, where a difference of a few orders of magnitude was observed. Thus, PARP12L protein expression appears to have a broad antiviral effect.
In additional experiments, we investigated whether a lower level of expression than that achieved in the experiments with VEErep/PARP12L would be sufficient to inhibit virus replication. To achieve this, the PARP12L sequence was cloned in direct and reverse orientations under the control of the subgenomic promoter into the VEEV replicon VEEVrepL, containing a point mutation in the nsP2-coding sequence (Q739L). This mutation was previously shown to have a strong negative effect on replicon RNA replication and decreased expression of the replicon-carried, heterologous genes 10- to 20-fold (36). Upon transfection into the cells, replication of VEEVrepL/PARP12L did not noticeably alter either cell growth or morphology, but replication of the tested viruses was strongly affected (see Fig. S2 in the supplemental material), indicating that even lower levels of PARP12L expression are sufficient for developing the antiviral effect.
PARP12L expression affects intracellular replication rather than alphavirus release.
The PARP12L inhibitory function could potentially result from its ability to interfere with either intracellular virus replication or release. In our earlier experiments (Fig. 8), VEEV/GFP and SINV/GFP demonstrated very poor GFP expression in cells expressing PARP12L but not in the VEErep/PARPrev-containing cells. To additionally analyze this, we infected BHK-21 and NIH 3T3 cells with packaged VEErep/PARP12L or VEErep/PARPrev replicons, and at 1 h postinfection we infected them with packaged VEE replicons carrying luciferase and GFP genes under control of the subgenomic promoters (VEErep/Luc/GFP) (Fig. 9A). At all times after superinfection, luciferase activity in PARP12L-expressing cells was lower than that detected in the VEErep/PARPrev replicon-containing cells. This was an indication that PARP12L expression strongly affected expression of the replicon-encoded proteins, most likely due to its effect on viral RNA replication/transcription (Fig. 9B). We also indirectly measured the effect of PARP12L expression on RNA replication by assessing the accumulation of the nsP2 protein in the cells infected with packaged VEErep/PARP12L and VEErep/PARPrev. Starting from 5 h postinfection, the replicon-encoded nsP2 was always found in VEErep/PARP12L-infected cells at a lower concentration, indicating a negative effect of PARP12L expression on replicon replication itself (Fig. 9C). These experiments successfully demonstrated PARP12L's effect on alphavirus replication. However, they were unable to ascertain whether the difference was mediated by direct effects of PARP12L on the function of virus-specific replication complexes, an indirect negative effect on replication through inhibition of translation of virus-specific RNA, or both. These possibilities are now under detailed investigation.
Fig 9.
Expression of PARP12L strongly affects expression of genes carried by the recombinant VEEV genome. (A) Schematic representation of VEEV replicons encoding PARP12L in either the direct or reverse orientation or firefly luciferase (Luc). (B) NIH 3T3 and BHK-21 cells were infected with packaged VEEV replicons expressing PARPrev and PARP12L at an MOI of 20 (inf.u./cell). After incubation for 1 h at 37°C, they were superinfected with packaged VEErep/Luc/GFP replicon at an MOI of 1 (inf.u./cell). Luciferase activity was measured at the indicated time points. (C) BHK-21 and NIH 3T3 cells were infected with packaged VEErep/PARP12L or VEErep/PARPrev replicons at an MOI of 20. Intracellular accumulation of VEEV nsP2 was measured at the indicated time points by Western blotting as described in Materials and Methods. The data were normalized to the level of β-tubulin.
Other PARPs demonstrate a negative effect on VEEV replication.
PARP12L is a member of a reasonably large family of proteins containing the PARP domain (42) in the subfamily of CCCH-type Zn finger-containing PARPs (CCCH PARPs). Other members of this subfamily include PARP7 and PARP13. The short splice form of PARP13 was previously termed ZAP (42). In the above-described microarray experiments, the PARP7 and PARP13 genes became activated in NIH 3T3 cells upon their treatment with IFN-β or during infection with VEEV/GFP/C1 (Fig. 10B). Further analysis of their antiviral functions was not given the highest priority because of detectable activation of the same genes in the IFN-α/βR−/− MEFs upon their infection with the same VEEV mutant. However, this does not necessarily mean that one or more other PARP proteins have no anti-alphavirus functions.
Fig 10.
Expression of some other PARP proteins also downregulates VEEV replication. (A) Schematic representations of the VEEV genome encoding different PARP proteins and of different PARPs used in this study. Zn fingers and domains are indicated. (B) Activation of different PARP genes in NIH 3T3 cells infected with VEEV/GFP/C1 or treated with IFN-β for 24 h. Activation of these genes was measured in the microarray experiments presented in Fig. 2 and 3. (C) Replication of recombinant PARP-expressing viruses and VEEV/PARPrev in BHK-21 and NIH 3T3 cells after infection at an MOI of 10 PFU/cell. Media were replaced at the indicated time points, and virus titers were determined by plaque assay on BHK-21 cells.
Therefore, to test the possible antiviral activity of other PARPs, we replaced the GFP gene in the VEEV/GFP/C1 genome with PARP7- and PARP13-coding sequences (Fig. 10A). In addition to CCCH PARPs, we also cloned a PARP10-coding sequence into the viral genome under the control of an additional subgenomic promoter (Fig. 10A and B). PARP10 also contains a putative, albeit different, RNA-binding sequence (42) and showed an increase in transcriptional activity following treatment of NIH 3T3 cells with IFN-β or infection with VEEV/GFP/C1 (Fig. 10B). We then tested the effects of the expressed proteins on the rates of virus replication (Fig. 10C). VEEV/PARPrev/C1, containing the PARP12 gene in the reverse orientation, was used as a control. Expression of PARP7 inhibited VEEV replication with an efficiency similar to that described above for PARP12L (Fig. 6B), while PARP13 expression had no effect on VEEV replication. PARP10 demonstrated intermediate inhibitory activity. Thus, the ability to interfere with alphavirus replication appears not to be limited to PARP12L, and other members of the PARP superfamily may also be involved in development of the antiviral response.
DISCUSSION
The identification of new cellular genes whose products demonstrate anti-alphavirus activities is critical for further understanding of alphavirus pathogenesis and virus-host interactions. Traditionally, replication of alphaviruses is considered to be highly cytopathic for cells of vertebrate origin (45). This cytopathic phenotype is determined by a combination of different factors, such as high-level expression of viral structural proteins (10), rapidly induced transcriptional shutoff (10), and the ability to inhibit translation and transcription in infected cells (20). Some of the cell lines used in laboratory practice also develop apoptotic changes (31). However, current data suggest that a cytopathic effect is not necessarily an inevitable result of infection, and at least in the murine brain, almost complete clearance of some alphaviruses can occur without widespread neuronal death and pathological changes (6). Moreover, in spite of the virus' ability to efficiently inhibit transcription and translation in vitro, infected animals are capable of responding with type I and type II IFN production, which for some alphavirus infections can reach very high levels (13, 28, 48, 49). This suggests that during in vivo replication or in less permissive cells in vitro, alphavirus replication might not completely inhibit transcription and translation or profoundly affect activation of the antiviral response. As a result, the release of IFN and other cytokines and chemokines can be detected.
Our previous studies (5, 18, 19), as well as those of other research groups (1, 8, 9), demonstrated that point mutations in the alphavirus genes encoding proteins with transcription-inhibitory functions make mutants dramatically less pathogenic and less cytopathic. Old World alphaviruses containing point mutations in nsP2, New World alphaviruses with mutations in the capsid-specific amino-terminal peptide, and chimeric viruses carrying the Old World alphavirus structural protein genes and the New World alphavirus-derived nsPs can persistently replicate in cells defective in IFN-α/β production or signaling without developing a profound CPE (5, 13, 27). However, in type I IFN-competent cells, replication of such mutated or chimeric viruses is downregulated to undetectable levels within a few days postinfection (5). Moreover, addition of IFN-α/β-specific Abs to the medium of infected type I IFN signaling-competent cells also makes replication persistent and similar to that observed in IFN-α/βR−/− or STAT1−/− MEFs (5, 13). This is a strong indication that the cellular antiviral response developed in the absence of autocrine IFN signaling is insufficient for virus clearance, even though these cells contain endogenous PRRs. In the experiments presented in this study, IFN-α/βR−/− MEFs demonstrated activation of numerous cellular genes, including those encoding proteins with known antiviral activities, in response to mutant virus replication (Fig. 2). However, this response was insufficient for virus clearance (Fig. 1). In NIH 3T3 cells, which have no defects in type I IFN production and signaling, VEEV variants with mutated capsid protein induced type I IFN at readily detectable levels by 4 to 6 h postinfection, and it reached a maximum level by 24 h postinfection (Fig. 1) (5). The autocrine IFN signaling leads to a gradual decrease in VEEV/GFP/C1 replication, and within 5 to 6 days postinfection, replication becomes undetectable and cells remain viable (Fig. 1). This is a strong indication that the IFN treatment induces an additional, and most likely a multicomponent, response that plays a critical role in the inhibition of virus replication. Therefore, in this study we made an attempt to define the previously underappreciated type I IFN-inducible genes that are involved in VEEV clearance from IFN-competent cells.
The microarray-based profiling and subsequent bioinformatic analysis pointed out the set of genes expressed in the IFN-competent but not in the IFN-α/βR−/− cells during VEEV/GFP/C1 mutant replication. These genes were also activated in uninfected IFN-β-treated cells, which demonstrated complete protection against subsequent viral infection (Fig. 2 and 3). Many of the 47 selected genes were previously shown to express proteins with virus-inhibitory functions, and this was a good indication that the bioinformatic analysis was correct. After further analysis of the identified group of genes, Bst2 and PARP12 attracted more attention. Bst2, a tetherin, was previously found to have an anti-HIV-1 function. Bst2 interferes with release of HIV-1, as well as other enveloped viruses, from the cell surface (40, 41). Thus, its anti-VEEV activity detected in the experimental tests was expected and served mostly as a control aimed at showing that the double-subgenomic viruses and VEEV replicons represent adequate experimental systems for demonstrating antiviral functions of the genes of interest. In contrast, the antiviral function of PARP12 has not been previously described. From the PARP superfamily, only PARP13 (and its short splice form) was previously characterized as having an intrinsic immunological function and was found in organisms of broad taxonomic range (26, 34). The inhibitory functions of PARP13 were demonstrated on murine leukemia virus and two alphaviruses, Sindbis virus and Semliki Forest virus (16, 34). Interference of PARP13 with virus replication appears to be based on its ability to bind and degrade virus-specific RNAs (56). PARP12 protein demonstrates a number of similarities with PARP13 in terms of domain structure (Fig. 10A), such as a PARP domain at the carboxy terminus and five Zn finger domains. Additionally, PARP13 and PARP12 are expressed in two splice variants, with the short variants lacking the carboxy-terminal PARP-encoding fragment. However, unlike PARP13, PARP12 has not been previously shown to possess virus-inhibitory functions. In our experiments, the long isoform of PARP12, PARP12L, was capable of downregulating replication of a broad range of evolutionarily distinct alphaviruses and other RNA viruses that do not belong to the alphavirus genus at all (Fig. 8). The mechanism of this broad inhibitory effect needs to be further investigated. The previously published data suggested that PARP13 mediates RNA degradation (56) and, thus, indirectly inhibits virus replication. We can speculate that based on its similarities to PARP13, PARP12 may have a similar function. However, our preliminary data indicate that it inhibits translation by a different mechanism (data not shown), which is now under detailed investigation. Another possibility comes from the recent finding that a conserved domain of the alphavirus nonstructural protein nsP3 is structurally similar to a PARP domain (35). The exact function of nsP3 in virus replication is not completely understood, but regardless of nsP3's activity, overexpression of PARP12L might interfere with the function of this alphavirus nonstructural protein. However, this hypothesis needs additional experimental support and does not explain the negative effect of PARP12L expression on other RNA viruses. Experiments with other PARPs have indicated that in the PARP superfamily, PARP12 and PARP13 are not the only proteins exhibiting anti-alphavirus activities. PARP7 and PARP10 also interfere with VEEV replication. Thus, the importance of the PARP superfamily in the development of an antiviral response is likely to be greater than we currently know. In an additional experiment, we decreased PARP12 expression almost 10-fold using RNA interference (RNAi), but, as expected, this failed to affect virus clearance significantly. The ability of other PARPs to inhibit VEEV replication and activation of a very wide variety of other ISGs during VEEV/GFP/C1 replication explains the lack of an observable effect of PARP12L knockdown on the rates of virus clearance from NIH 3T3 cells (data not shown).
The results of this study suggest that PARP genes are members of a growing group of IFN-inducible genes whose products exhibit antiviral effects (7, 30, 33, 39, 43, 46, 47, 55). So far, all of these proteins, expressed individually, demonstrate low but detectable antiviral activities. Thus, despite these antiviral effects, overexpression of these individual proteins is insufficient for complete protection of cells against viral infections. Additionally, knockout of the individual genes usually does not have a deleterious effect on the development of the innate immune response to viral infections (38). Thus, it appears that there is a highly redundant system involving numerous gene families, as well as multiple genes within these families, in the development of the antiviral response. This redundancy and lack of a dominant antiviral gene product make evolution of resistant virus variants an almost impossible task. This also complicates analysis of antiviral proteins, because the need to dissect the small effects of numerous individual gene products makes accurate attribution of their particular antiviral functions very difficult. As a result, we are probably still in the early stages of understanding how hundreds of innate immune response genes interact in a coordinated fashion to regulate and thwart viral infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Niall J. Foy for helpful discussions and for critical reading and editing of the manuscript.
This work was supported by Public Health Service grants AI070207 and R01AI073301.
Footnotes
Published ahead of print 23 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aguilar PV, Leung LW, Wang E, Weaver SC, Basler CF. 2008. A five-amino-acid deletion of the eastern equine encephalitis virus capsid protein attenuates replication in mammalian systems but not in mosquito cells. J. Virol. 82:6972–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguilar PV, Weaver SC, Basler CF. 2007. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 81:3866–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atasheva S, Fish A, Fornerod M, Frolova EI. 2010. Venezuelan equine encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin alpha/beta that obstructs nuclear pore complex function. J. Virol. 84:4158–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atasheva S, Garmashova N, Frolov I, Frolova E. 2008. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in mammalian but not in mosquito cells. J. Virol. 82:4028–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atasheva S, Krendelchtchikova V, Liopo A, Frolova E, Frolov I. 2010. Interplay of acute and persistent infections caused by Venezuelan equine encephalitis virus encoding mutated capsid protein. J. Virol. 84:10004–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burdeinick-Kerr R, Wind J, Griffin DE. 2007. Synergistic roles of antibody and interferon in noncytolytic clearance of Sindbis virus from different regions of the central nervous system. J. Virol. 81:5628–5636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Der SD, Yang YL, Weissmann C, Williams BR. 1997. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 94:3279–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dryga SA, Dryga OA, Schlesinger S. 1997. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: The importance of a mutation in the nsP2 gene. Virology 228:72–83 [DOI] [PubMed] [Google Scholar]
- 9. Fazakerley JK, Boyd A, Mikkola ML, Kaariainen L. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76:392–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frolov I, Schlesinger S. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68:1721–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frolov I, Schlesinger S. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frolov I, Schlesinger S. 1994. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 68:8111–8117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frolova EI, et al. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 84:11679–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Froshauer S, Kartenbeck J, Helenius A. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706 [DOI] [PubMed] [Google Scholar]
- 17. Garmashova N, et al. 2007. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J. Virol. 81:13552–13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garmashova N, Gorchakov R, Frolova E, Frolov I. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garmashova N, et al. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 81:2472–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gorchakov R, Frolova E, Frolov I. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397–9409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gorchakov R, et al. 2008. A new role for ns polyprotein cleavage in Sindbis virus replication. J. Virol. 82:6218–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gorchakov R, Frolova E, Williams BR, Rice CM, Frolov I. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 78:8455–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in sindbis virus-infected cells. J. Virol. 82:10088–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griffin DE. 1986. Alphavirus pathogenesis and immunity, p 209–250 In Schlesinger SS, Schlesinger MJ. (ed), The Togaviridae and Flaviviridae. Plenum Press, New York, NY [Google Scholar]
- 25. Griffin DE. 2010. Recovery from viral encephalomyelitis: immune-mediated noncytolytic virus clearance from neurons. Immunol. Res. 47:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kerns JA, Emerman M, Malik HS. 2008. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 4:e21 doi:10.1371/journal.pgen.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim DY, et al. 2011. Design of chimeric alphaviruses with a programmed, attenuated, cell type-restricted phenotype. J. Virol. 85:4363–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klimstra WB, et al. 1999. Infection of neonatal mice with Sindbis virus results in a systemic inflammatory response syndrome. J. Virol. 73:10387–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lemm JA, Durbin RK, Stollar V, Rice CM. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lenschow DJ, et al. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levine B, et al. 1993. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature 361:739–742 [DOI] [PubMed] [Google Scholar]
- 32. Liljeström P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu SY, Sanchez DJ, Cheng G. 2011. New developments in the induction and antiviral effectors of type I interferon. Curr. Opin. Immunol. 23:57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. MacDonald MR, Machlin ES, Albin OR, Levy DE. 2007. The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J. Virol. 81:13509–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malet H, et al. 2009. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 83:6534–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petrakova O, et al. 2005. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J. Virol. 79:7597–7608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rice CM, Levis R, Strauss JH, Huang HV. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: Mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryman KD, White LJ, Johnston RE, Klimstra WB. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 15:53–76 [DOI] [PubMed] [Google Scholar]
- 39. Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sauter D, et al. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sauter D, Specht A, Kirchhoff F. 2010. Tetherin: holding on and letting go. Cell 141:392–398 [DOI] [PubMed] [Google Scholar]
- 42. Schreiber V, Dantzer F, Ame JC, de Murcia G. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7:517–528 [DOI] [PubMed] [Google Scholar]
- 43. Silverman RH. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sjöberg EM, Garoff H. 1996. The tranlation-enhancing region of the Semliki Forest virus subgenome is only functional in the virus-infected cell. J. Gen. Virol. 77:1323–1327 [DOI] [PubMed] [Google Scholar]
- 45. Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Szretter KJ, et al. 2011. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J. Virol. 85:11557–11566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thakur CS, et al. 2007. Small-molecule activators of RNase L with broad-spectrum antiviral activity. Proc. Natl. Acad. Sci. U. S. A. 104:9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trgovcich J, Aronson JF, Johnston RE. 1996. Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis. Virology 224:73–83 [DOI] [PubMed] [Google Scholar]
- 49. Trgovcich J, et al. 1997. Sindbis virus infection of neonatal mice results in a severe stress response. Virology 227:234–238 [DOI] [PubMed] [Google Scholar]
- 50. Ventoso I, et al. 2006. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 20:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Volkova E, Gorchakov R, Frolov I. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology 344:315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang E, Kim DY, Weaver SC, Frolov I. 2011. Chimeric Chikungunya viruses are nonpathogenic in highly sensitive mouse models but efficiently induce a protective immune response. J. Virol. 85:9249–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang E, et al. 2007. Chimeric Sindbis/eastern equine encephalitis vaccine candidates are highly attenuated and immunogenic in mice. Vaccine 25:7573–7581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weaver SC, Frolov I. 2005. Togaviruses, p 1010–1024 In Mahy BWJ, ter Meulen V. (ed), Virology, vol 2 IRL Press, Salisbury, United Kingdom [Google Scholar]
- 55. Zhang Y, Burke CW, Ryman KD, Klimstra WB. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81:11246–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu Y, Gao G. 2008. ZAP-mediated mRNA degradation. RNA Biol. 5:65–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.