Abstract
Nonstructural protein 5A (NS5A) is essential for hepatitis C virus (HCV) replication and assembly and is a critical drug target. Biochemical data suggest large parts of NS5A are unfolded as an isolated protein, but little is known about its folded state in the cell. We used fluorescence resonance energy transfer (FRET) to probe whether or not different regions of NS5A are in close proximity within the cell. Twenty-three separate reporter constructs were created by inserting one or more fluorophores into different positions throughout the three domains of NS5A. FRET efficiency was maximal when donor and acceptor fluorophores were positioned next to each other but also could be observed when the two fluorophores flanked NS5A domain 1 or domain 3. Informatic and biochemical analysis suggests that large portions of the carboxy terminus of NS5A are in an unfolded and disordered state. Quercetin, a natural product known to disrupt NS5A function in cells, specifically disrupted a conformationally specific domain 3 FRET signal. Intermolecular FRET indicated that the NS5A amino termini, but not other regions, are in close proximity in multimeric complexes. Overall, this assay provides a new window on the intracellular conformation(s) of NS5A and how the conformation changes in response to cellular and viral components of the replication and assembly complex as well as antiviral drugs.
INTRODUCTION
About 170 million people are chronically infected with hepatitis C virus (HCV) worldwide, and more than 350,000 people die from hepatitis C-related liver diseases each year (37). While progress is being made on new antivirals for HCV, the cost, efficacy, and tolerability of the drugs are still not optimal. One target for drug therapy is the viral nonstructural protein 5A (NS5A), but the mechanisms of action through which antivirals disrupt NS5A function are unknown (6, 11).
NS5A is an ∼54-kDa phosphoprotein containing the distinct domains 1, 2, and 3, separated by repetitive low-complexity sequences (32). This organization is conserved across evolutionarily divergent NS5A genotypes. NS5A is an essential accessory protein that binds viral RNA and many different cellular partners. The first 213 amino acids of NS5A constitute domain 1, which contains an amino-terminal 34-amino-acid amphipathic helix that mediates membrane association (4, 26, 28) and plays a critical role in viral replication (33). Two different crystal structures have been obtained for domain 1. While the structures are similar in putative membrane association, secondary structure, and dimeric organization, the dimeric interface itself is dramatically different in each. It is unclear whether one, both, or neither is adopted in the cell (23, 34). Amino acids 250 to 342 of NS5A represent domain 2, which lacks detectable secondary structure as assessed by nuclear magnetic resonance (NMR) (15). Domain 3 comprises the region carboxy terminal to amino acid 356 and exhibits genotypic-length polymorphisms. Domain 3 from genotype 1b, which was used in the present study, is also suggested to be in an unfolded state by NMR (16). Domain 3 contains protein kinase CK2 phosphorylatable sites (2, 31) and plays a crucial role in viral assembly (3, 32).
Understanding the various roles of NS5A in the viral life cycle and disease pathogenesis has been hampered by the lack of structural models relevant to the cellular state, particularly for the carboxy terminus. Measurement of fluorescence resonance energy transfer (FRET) efficiency between a donor and an acceptor can provide information on proximity relationships between inter- and intramolecular regions of proteins (17). In this study, we have developed a FRET assay to examine the compactness of NS5A in hepatoma cells (Huh7.5). We used the biarsenical fluorescein derivative FlAsH (fluorescein arsenical hairpin) (14) as a FRET acceptor and enhanced cyan fluorescent protein (here referred to as CFP) as a FRET donor (13, 14, 18). FlAsH is advantageous because the tetracysteine domain required for FlAsH binding is small and less intrusive than other acceptors of choice such as yellow fluorescent protein (18). The technique has drawbacks, though, as even this small motif can be inaccessible or alter how the protein interacts with physiologic partners and there can be issues with background labeling in cells. In the present work, we have directly addressed these issues by verifying FlAsH load and FRET efficiency at fluorophore insertion sites with a CFP-CCPGCC directly sequential control. We detected a robust intramolecular and intermolecular FRET between two donor/acceptor positions within the HCV NS5A that was disrupted by quercetin, a food supplement with antiviral activity in cell culture (12).
MATERIALS AND METHODS
CFP/tetracysteine cDNAs.
A CFP-tetracysteine fusion protein was made and used to calibrate FRET measurements from FRET partners fused to NS5A. The 17-amino-acid FlAsH binding motif AEAAAREACCPGCCARA (here referred to as the CCPGCC motif), described previously (1), was introduced in frame in the C terminus of CFP by PCR techniques. Three additional tetracysteine constructs with genetic variants in the CCPGCC were made by the same strategy to test the binding efficiency of FlAsH. The PCR product was directly cloned into CFP-C1 vector (Clontech, CA) in each case. For determination of relative proximity within the NS5A, we inserted the FlAsH binding motif at four positions in the NS5A sequence and fused CFP to either the N terminus or C terminus of NS5A. Full-length HCV NS5A (amino acids 1 to 447) derived from the con1 genotype 1b sequence (GenBank accession no. AJ238799) was cloned in frame to both CFP-N1 and CFP-C1 vectors (Clontech, CA). We also made four different control constructs by inserting CFP-CCPGCC sequential motifs in the same NS5A positions as the CCPGCC motif (see Fig. 2) by a PCR strategy. Each construct was confirmed by sequencing and is shown in Table S1 in the supplemental material.
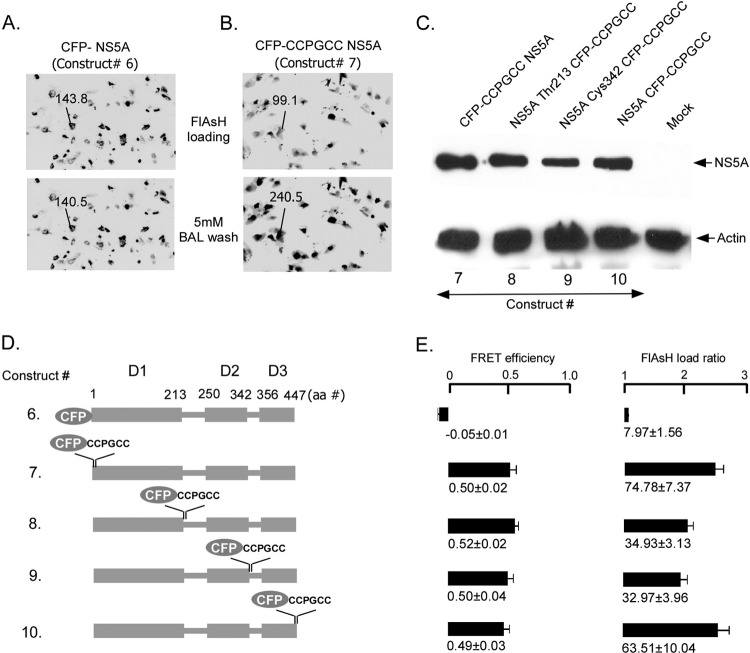
Fig 2.
FRET signals from NS5A incorporating CFP tags or CFP-CCPGCC sequential tags in different domains expressed in Huh7.5 cells. (A) Huh7.5 cells were transfected with CFP-NS5A. Incubation with 1 μM FlAsH followed by 5 mM BAL had no effect on CFP fluorescence or FRET efficiency. (B) Huh7.5 cells transfected with CFP-CCPGCC tagged on the N terminus of NS5A. Fluorescence was measured after incubating cells with 1 μM FlAsH and again after incubation with 5 mM BAL. (C) Western blot of CFP-CCPGCC sequential tags, tagging protein in four different positions of NS5A using an antibody to NS5A. (D) Schematic diagrams of tagged NS5A constructs that were used to assess nonspecific binding of FlAsH (construct 6) or to assess accessibility of the FlAsH compound to tetracysteine tags placed in each domain of NS5A (constructs 7 to 10). (E) Comparison of the FRET efficiencies and FlAsH loading in Huh7.5 cells for the different NS5A constructs shown in panel D. Mean FRET efficiencies ± standard errors are shown.
FlAsH labeling and fluorescence microscopy.
We used modifications of a previously described protocol (13). Approximately 50,000 Huh7.5 cells were transfected with the indicated DNA construct and incubated at 37°C in 5% CO2 for 48 h for expression. Transfected plates were thoroughly rinsed with 1× Hanks balanced salt saline solution (HBSS) (Cellgro, VA) with calcium and magnesium and without phenol red and equilibrated for 30 min by adding HBSS in the dark. Each plate was covered with 750 μl of a freshly made loading solution consisting of 10 μM EDT (1,2-ethanedithiol) (Fluka Analytical, CA) and 1 μM FlAsH-EDT2 (Lumio Green from Invitrogen, CA, in this paper referred to as FlAsH) in HBSS. Dishes were incubated at 37°C in a CO2 incubator for 1 h. The FlAsH loading solution was thoroughly rinsed with HBSS, and nonspecific labeling of the cells was reduced out by incubating with 250 μM EDT in HBSS for 10 min in the dark at room temperature. The plates were again washed with HBSS and covered with 1 ml HBSS before imaging. For FlAsH removal, a freshly made stock solution of 2,3-dimercapto-1-propanol (British Anti-Lewisite [BAL] from Sigma, CA) in HBSS was added to yield a final BAL concentration of 5 mM. Dimming of the fluorescein intensity below visual levels occurred within 10 min in the majority of cases. The epifluorescence of the cell was captured with a cooled charge-coupled device (CCD) Photometrics CoolSNAP HQ camera (Roper Scientific, Inc., NJ) using a 10×/0.30 numerical aperture (NA) objective in an inverted Nikon microscope. Excitation was provided by a 100-W Hg lamp filtered with standard band-pass excitation (ex)/emission (em) filter cubes. Chroma 31036 (ex, 436/20 nm; em, 480/30 nm; dichroic, 455) for CFP and Chroma 41028 (ex, 500/20 nm; em, 535/30 nm; dichroic, 515) for fluorescein were used for imaging. Images of 680 × 512 pixels were acquired at 8-bit resolution without compression of the A/D conversion scale. Integration time (500 ms) and camera sensitivity settings were set as constant. Images were averaged 5 times and analyzed with an online ImageJ program (http://imagej.nih.gov/ij/download.html). Mean pixel intensities were measured in regions of interest including 100 to 600 pixels (0.04 μm per pixel). Background intensities were measured near the regions of interest and were subtracted. Mean pixel intensities for CFP and FlAsH emissions were in the range of 70 to 110 on a scale of 0 to 255. The FRET ratio was defined as (CFP emission after FlAsH loading and after BAL wash)/(CFP emission after FlAsH loading and before BAL wash). Using this definition, FRET efficiency was calculated as 1 − [1/(FRET ratio)] (24). We also determined the FlAsH load ratio by calculating FlAsH emission after FlAsH loading of individual cells corrected by background subtraction of transfected cells near the measured area outside the cell. In each experiment 30 to 50 cells whose expression was low enough to be in the quantifiable range for ImageJ software were randomly chosen because in pilot experiments with constructs that yielded FRET, measuring this number of cells provided standard errors of <10% of the total FRET signal (21).
Purified CFP/tetracysteine fusion protein.
cDNA was amplified by PCR from a CFP-CCPGCC construct with introduction of the BamHI site at the 5′ ends and the HindIII site at 3′ ends. The PCR products were then cloned directly to pQE-30 N terminus 6×His prokaryotic expression vector (Qiagen, CA) in frame at BamHI/HindIII sites. The pQE-30 6×His tag CFP-CCPGCC clone was transformed in M15(pREP4) Escherichia coli (Qiagen, CA). The cells were grown at 37°C in LB to an optical density at 600 nm (OD600) of 0.7, and expression was induced at 28°C by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 7 h. The recombinant protein was then bound to Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, CA) and purified according to the manufacturer's protocol. The purified recombinant protein was incubated with FlAsH-EDT2 in labeling buffer (50 mM HEPES, pH 7.1, 140 mM NaCl, 1 mM β-mercaptoethanol, 1 mM Tris[2-carboxyethyl]phosphine hydrochloride [TCEP]) in a 1:1 ratio for 2 h at 4°C in the dark. For SDS-PAGE, FlAsH-labeled protein was added to Laemmli sample buffer (Bio-Rad, CA) with 50 mM TCEP (Sigma, CA) instead of β-mercaptoethanol or dithiothreitol (DTT). The mixture was incubated for 30 min at room temperature, loaded, and run in the dark. The fluorescent bands in unstained gel were revealed by a UV-transluminator and photographed. The same gel was then analyzed by Western blotting with anti-CFP polyclonal antibody (BioVision, CA). The excitation and emission spectra of purified CFP/tetracysteine protein in the presence and absence of FlAsH were measured with an AB2 luminescence spectrometer (Thermo Electron Corporation, WI). CFP and FlAsH emissions were measured in the range of 450 nm to 600 nm using 430-nm excitation. Donor quenching due to FRET was measured after incubation of the sample with 5 mM BAL for 10 min in the dark at room temperature. The spectra were analyzed by AB2 luminescence spectrometer analysis software (Thermo Electron Corporation, WI).
Intermolecular NS5A FRET.
For intermolecular FRET, CFP and the tetracysteine FlAsH binding motif were inserted either at the N terminus or at the C terminus in different NS5A plasmids. Three different types of cotransfection were done in Huh7.5 cells: NS5A-CFP + NS5A-CCPGCC (cotransfection I), CFP-NS5A + CCPGCC-NS5A (cotransfection II), and CFP-NS5A + NS5A-CCPGCC (cotransfection III). FlAsH labeling and FRET measurement were done as described above at 48 h posttransfection. For quercetin experiments, at 8 h posttransfection, cells were incubated with indicated amounts of quercetin (Sigma-Aldrich, MO), and FRET was measured 40 h later (48 h posttransfection).
Coimmunoprecipitation assay.
NS5A dimerization was studied by cotransfecting NS5A-CFP and NS5A (with no tag) plasmids in Huh7.5 cells. For background controls four cotransfection/transfection assays were also done in parallel: CFP + NS5A, NS5A, NS5A-CFP, and a mock transfection. After 48 h of transfection, the cells were lysed on ice in a lysis buffer (25 mM Tris-Cl, pH 7.6, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% NP-40) with EDTA-free 1× halt protease inhibitor cocktail (Thermo Scientific, IL). The expression of each protein was confirmed by Western blotting of total cell lysate with anti-NS5A monoclonal antibody (Meridian Life Science, Inc., ME). The detergent from the cell lysates was removed by a detergent removal spin column (Thermo Scientific, IL) according to the manufacturer's protocol. The wash/equilibration buffer for this column was the same as the lysis buffer with protease inhibitor cocktail without the detergents. The detergent-free lysates were treated with 5 μl/ml of an RNase cocktail (Ambion, NY) enzyme mix overnight at 4°C as described previously (19). The detergent-free and RNase-treated total cell lysates were coimmunoprecipitated with an anti-CFP polyclonal antibody (BioVision, CA), and Western blotting was performed with anti-NS5A monoclonal antibody.
Cells, replicon, HCV infectious clone, and luciferase assay.
Huh7.5 cells with or without the con1b replicon were maintained as described before (10). For quercetin experiments in the infectious clone, the cells were either untreated or treated with 25 μM and 50 μM quercetin for 15 days. Approximately 2 million cells from each group (0 μM, 25 μM, and 50 μM) were electroporated with 6.0 μg of in vitro-transcribed RNA derived from JFH-luciferase-wild type (JFH-Luc-WT) cDNA clone and resuspended in 5.0 ml complete media. An aliquot corresponding to 50,000 cells was plated separately in 24-well plates, and the rest of the suspension was plated in T25 flasks and further incubated at 37°C. Following 4 h of incubation the media were replaced with complete growth media containing 0 μM, 25 μM, or 50 μM quercetin. The luciferase activity was monitored in cells plated in 24 wells at 72 h. The supernatants were collected from T25 flasks at 72 h, filtered through 0.45-μm filters, and passed onto naïve Huh7.5 cells that had been plated in 24 wells a day before. Following virus adsorption, the virus inoculum was replaced with complete medium and further incubated for 48 h at 37°C. The cells were lysed, and luciferase activity was monitored as described before (10). The average results of two independent experiments, each done in triplicate, were calculated.
Confocal microscopy.
Huh7.5 cells were grown on 12-mm glass coverslips and transfected with NS5A(Cys342-CCPGCC)-CFP plasmid DNA. After 4 h the transfection media were replaced with fresh complete media containing 0 μM, 5 μM, or 50 μM quercetin. Forty-eight hours posttransfection the cells were fixed with 4% paraformaldehyde for 30 min at room temperature. The cells were washed with phosphate-buffered saline (PBS)-Tween and treated with absolute ethanol for 5 min on ice followed by incubation with 5% bovine serum albumin (BSA) in PBS-Tween for 30 min at room temperature. Cells were incubated with anti-CFP polyclonal antibodies and anti-protein disulfide isomerase (anti-PDI) monoclonal antibodies (Santa Cruz Biotechnology, CA) diluted in PBS-Tween containing 3% BSA for 1 h. Cells were then washed three times and further incubated with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 594 goat anti-mouse IgG (Invitrogen, CA) for 1 h at room temperature. Cells were also stained with DAPI (4′,6-diamidino-2-phenylindole) (0.5 mg ml-1) to reveal nuclei. After the final wash the cells were mounted upside down on a glass cover slide in the presence of fluoromount (Sigma, CA) and visualized with a motorized Zeiss Axioplan IIi equipped with a rear-mounted excitation filter wheel (0.2-μm Z-increment, 40× PlanApo, oil immersion lens), a triple-pass (DAPI/fluorescein isothiocyanate [FITC]/Texas Red) emission cube, and an Orca AG CCD camera (Hamamatsu, NJ). The images were collected using OpenLabs 5.0 software (Improvision, MA), and further analysis was performed using Volocity 5.0 software (Improvision, MA) to get the merge images.
Statistics.
Data are presented as means ± standard errors of the means. Comparisons between two groups were done using Student's t test. Statistical significance was defined at values of P of <0.05.
RESULTS
Assessment of FlAsH-mediated FRET biochemically, as well as in Huh7.5 cells.
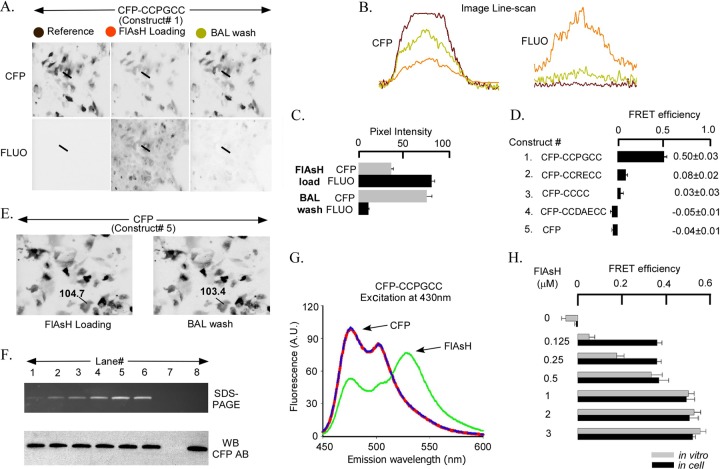
To validate the FlAsH-based FRET strategy in Huh7.5 cells, we first created fusion constructs of CFP and the tetracysteine motifs designed to bind FlAsH (1). Different CFP-CCXXCC sequential fusion proteins (constructs 1 to 5) were expressed in Huh7.5 cells for 48 h followed by 1 h of labeling with 1 μM FlAsH-EDT2 in FlAsH loading solution. Following FlAsH labeling, cells were incubated with 5 mM BAL to track the increase in CFP emission (decrease in FRET) due to the removal of bound FlAsH. Figure 1A shows the changes in donor and acceptor fluorescence in Huh7.5 cells expressing the CFP-CCPGCC sequential fusion protein during the FlAsH labeling protocol. CFP emission decreases after incubation with FlAsH (compare Fig. 1A left and middle panels in CFP channel) and recovers following FlAsH removal with BAL (compare Fig. 1A middle and right panels in CFP channel). FlAsH emission is drastically reduced after BAL treatment (Fig. 1A in FLUO channel). The line scans in Fig. 1B show pixel intensity across the width of a cell in the donor and acceptor channels. To ensure reproducibility and to conform to the definition of FRET efficiency (24), we focused only on cells in which FlAsH removal was substantial (less than 10% residual FlAsH emission after BAL incubation). In the cells with substantial FlAsH unloading by BAL, the CFP emission increased ∼2-fold (Fig. 1C). The increase in donor emission with concomitant decrease in acceptor emission is a characteristic sign of FRET (24). Robust FRET, with an efficiency of 0.50 ± 0.03, was measured in the CFP-CCPGCC sequential fusion protein expressed in Huh7.5 cells (Fig. 1D, construct 1).
Fig 1.
In-cell and in vitro FRET signals from CFP-CCXXCC sequential tags and WT-CFP. (A) CFP-CCPGCC-transfected Huh7.5 cells show a high level of CFP expression as indicated by fluorescence in the CFP channel. Before FlAsH loading, there is no fluorescence in the FLUO channel for the reference condition (gray circle, left panels). After 1 h of incubation in 1 μM FlAsH to load the tetracysteine motif, fluorescence in the CFP channel decreased and in the FLUO channel increased (red circle, middle panel). Incubation with 5 mM BAL for 10 min to displace FlAsH from the tetracysteine tag reversed the fluorescence changes back toward the reference conditions (yellow circle, right panels). (B) Pixel intensity of a line scan across the Huh7.5 cell (straight line indicated in each part of panel A in the CFP versus FLUO channels). Colors of traces correspond to the colors of filled circles in panel A. (C) Summary of mean pixel intensities for the experiment represented in panels A and B. (D) Summary of FRET efficiencies for a series of putative FlAsH binding sequences. (E) Wild-type CFP (WT-CFP)-transfected Huh7.5 cells (without the tetracysteine tag) show no change in CFP fluorescence after 1 μM FlAsH loading or after 5 mM BAL wash. (F) The upper part shows bacterially purified CFP-CCPGCC sequential tag protein incubated with increasing concentrations of FlAsH and then subjected to SDS-PAGE. Purified CFP-CCPGCC protein (1 μM) plus FlAsH (in μM): 0.125 (lane 1), 0.25 (lane 2), 0.5 (lane 3), 1 (lane 4), 2 (lane 5), and 3 (lane 6). Fluorescence was saturated in 1 μM FlAsH incubation (lane 4). Fluorescence was not detected in control lane 7 (3 μM FlAsH, no protein) or lane 8 (1 μM protein without FlAsH). In the lower part, a Western blot of the same SDS-PAGE gel using an antibody to CFP shows equal loading. (G) Fluorescence emission spectra of purified CFP-CCPGCC protein excited at 430 nm before (red line) and after (green line) 1 μM FlAsH loading and then 5 mM BAL incubation (blue line). (H) Summary of FRET efficiencies for CFP-CCPGCC evaluated in Huh7.5 cells (black) and in vitro (gray) at different FlAsH concentrations.
To confirm that the FRET signal involved the engineered tetracysteine and not spurious FlAsH acceptors at nonspecific sites, we tested CCXXCC tags with different affinities for FlAsH (1), as well as CFP without the tetracysteine tag (Fig. 1D, constructs 2 to 5). The CCPGCC motif yielded the highest FRET efficiency, whereas CCDEACC demonstrated only background levels. The measured FRET efficiency correlated well with the reported affinities of FlAsH for tetracysteine binding sites, which are 4 pM for CCPGCC, 70 pM for CCRECC, 1,800 pM for CCCC, and 92,000 pM for CCDEACC (1). Furthermore, FRET was undetectable in an untagged CFP (Fig. 1D, construct 5, and E). The last result confirmed that CFP bleaching during image acquisition and incubation with BAL was insignificant. Thus, the FRET signal involves only the donor and FlAsH acceptor bound to the tetracysteine motif.
Determination of saturation of FlAsH sites in cell-free and cell culture assays.
To test for saturation of FlAsH sites (measurement of donor with acceptor at 1:1 ratio), we compared spectrofluorometric FRET measurements using recombinant CFP-CCPGCC purified from recombinant E. coli with FRET measurements made in Huh7.5 cells. The spectral properties of recombinant, purified CFP and FlAsH incubation procedures in vitro are well described (13). The goal was to obtain FRET versus FlAsH concentration curves and empirical evidence of FlAsH binding to CFP-CCPGCC at a 1:1 donor/acceptor ratio using equilibrium binding analysis. Different concentrations (0 μM to 3 μM) of FlAsH were incubated with 1 μM bacterially expressed recombinant protein carrying the tetracysteine tag (Fig. 1H). Labeling of the recombinant protein by FlAsH was detected in fluorograms of unstained SDS-PAGE gels under broad-wavelength UV illumination (Fig. 1F, upper panel) and confirmed by Western blotting with an anti-CFP antibody (Fig. 1F, lower panel). FRET efficiency of the recombinant protein increased with increasing amounts of FlAsH (Fig. 1G and H). FRET efficiency reached a maximum at 1 μM FlAsH with the recombinant purified protein at a FRET efficiency of ∼0.52 (Fig. 1H). The “in cell” FlAsH binding showed a comparable saturation point (1 μM) with a similar maximal FRET efficiency of ∼0.50 (Fig. 1H). Thus, maximal FRET efficiencies for the sequential fusion protein were similar when measured in cells and in cell-free experiments when enough FlAsH was used.
Intramolecular FRET of HCV NS5A in Huh7.5 cells.
After validating the FlAsH-based FRET system using the CFP-CCPGCC sequential motif in Huh 7.5 cells, we sought to evaluate FRET in tagged HCV NS5A. To determine if accessibility of FlAsH to the CCPGCC motif is a limitation in expressed NS5A, we inserted the CFP-CCPGCC sequential motif in four different positions: the amino terminus, after Thr 213 (junction of domain 1 and domain 1-2 linker), after Cys 342 (junction of domain 2 and domain 2-3 linker), and the carboxy terminus (Fig. 2D, constructs 7 to 10). FRET efficiency at these four positions was similar (Fig. 2E) to that of the sequential motifs without fusion to NS5A (Fig. 1, construct 1). Thus, the tertiary structure of NS5A does not hinder access of the FlAsH reagent to the tetracysteine sites. Furthermore, all four of these NS5A constructs with CFP-CCPGCCs (constructs 7 to 10) are actively expressed recombinant proteins in Huh7.5 cells (Fig. 2C). Cells with low- to midlevel expression were analyzed because ImageJ software has a limitation to read maximal pixel intensity of 255 in each transfected group. Control experiments revealed that FRET was not detected in the CFP-NS5A fusion that lacked the tetracysteine tag (Fig. 2A and D, construct 6, and E), demonstrating that nonspecific binding of FlAsH does not create spurious FRET.
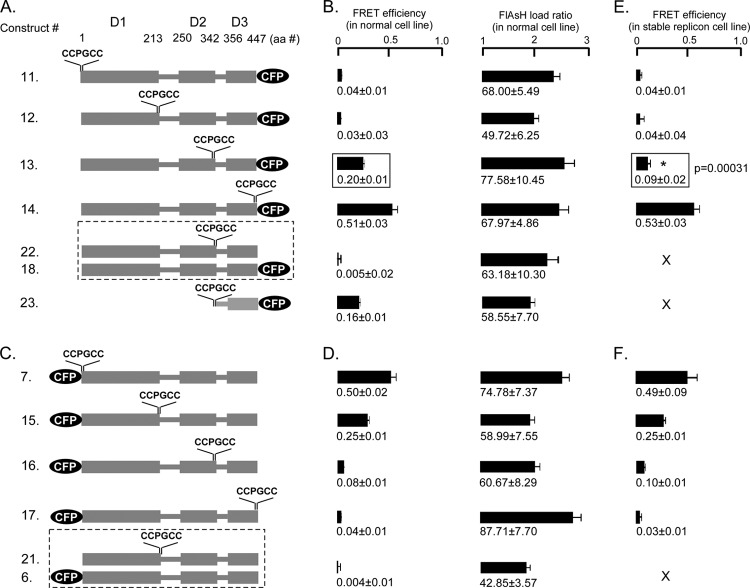
To map the compactness of the domains of NS5A by FRET, we fixed the donor (CFP) position at either the carboxy terminus (Fig. 3A, constructs 11 to 14) or the amino terminus (Fig. 3C, constructs 7 and 15 to 17) and introduced the acceptor (CCPGCC motif) at the same four critical positions as those described above in the NS5A molecule (Fig. 2). FRET efficiency measurements were conducted in Huh7.5 cells as well as the Con1b stable replicon cell line at 48 h after transfection. In general, comparatively weak FRET or in some cases almost no FRET was detected when CFP and the tetracysteine tag were separated by an intervening NS5A sequence compared to an inserted CFP-CCPGCC sequential motif; however, in two cases [construct 13, NS5A(Cys342-CCPGCC)-CFP, and construct 15, CFP NS5A(Thr213-CCPGCC)], FRET was observed despite over 100 amino acids separating the two fluorophores (Fig. 3). The FlAsH load ratio was also determined in each case to confirm that a significant amount of FlAsH was bound when we calculated the FRET efficiency.
Fig 3.
Intramolecular FRET in HCV1B NS5A depends upon donor-acceptor position and can be altered by Con1b replicon. (A) Schematic of NS5A constructs when CFP was located at the carboxy terminus (constructs 11 to 14) and the CCPGCC acceptor site was inserted at four different positions surrounding the three domains (at S1, T213, C342, and C447), doubly transfected constructs (constructs 18 and 22), and isolated NS5A domain3 (construct 23). (B) FRET efficiencies and FlAsH load ratios for the NS5A-CFP constructs in Huh7.5 cells. (C) Schematic of NS5A constructs when the CFP donor was located at the amino terminus (constructs 7 and 15 to 17) and the CCPGCC acceptor site was inserted at the same positions as shown in panel A and doubly transfected constructs (constructs 6 and 21). (D) FRET efficiencies and FlAsH load ratios for the CFP-NS5A constructs in Huh7.5 cells. (E) FRET efficiencies for NS5A-CFP constructs shown in panel A that were transfected in Con1b replicon stable cell line. (F) The FRET efficiencies for NS5A-CFP constructs shown in panel C that were transfected in Con1b replicon stable cell line. Mean FRET efficiencies ± standard errors are shown. Dashed-line boxes, cotransfected constructs; solid-line boxes, t test between the FRET efficiencies; X, not determined in the present study.
The results were indistinguishable between experiments in which NS5A was expressed alone or coexpressed with the Con1b replicon for all constructs except in the case of NS5A(Cys342-CCPGCC)-CFP (construct 13) (Fig. 3A, B, and E). The FRET efficiency at this location was 0.20 ± 0.01 in NS5A alone and was reduced to 0.09 ± 0.02. Reduction of the FRET of this construct could be due to binding of other nonstructural proteins, changes in the subcellular environment of NS5A, or the conformation of NS5A in a replication complex.
To ascertain whether the observed FRET for construct 13 (domain 3 flanked by fluorophores) and construct 15 (domain 1 flanked by fluorophores) occurred through intra- or intermolecular interactions, each fluorophore was individually engineered into separate NS5A proteins and the pair was cotransfected: constructs 18 and 22 to assess domain 3 FRET and constructs 6 and 21 to assess domain 1 FRET. Neither cotransfection of construct 18 with 22 nor cotransfection of construct 6 with 21 yielded any FRET, suggesting that the FRET observed for probes flanking domain 1 or 3 occurs intramolecularly.
Since studies of purified domain 3 had suggested an extended confirmation that potentially might not show FRET (16), we also expressed domain 3 flanked by fluorophores without an endoplasmic reticulum (ER) membrane anchor or domain 1/2 (construct 23). FRET was observed for isolated domain 3 despite lacking a membrane anchor, reinforcing that FRET likely occurs intramolecularly and is not exclusively a result of concentration on membranes.
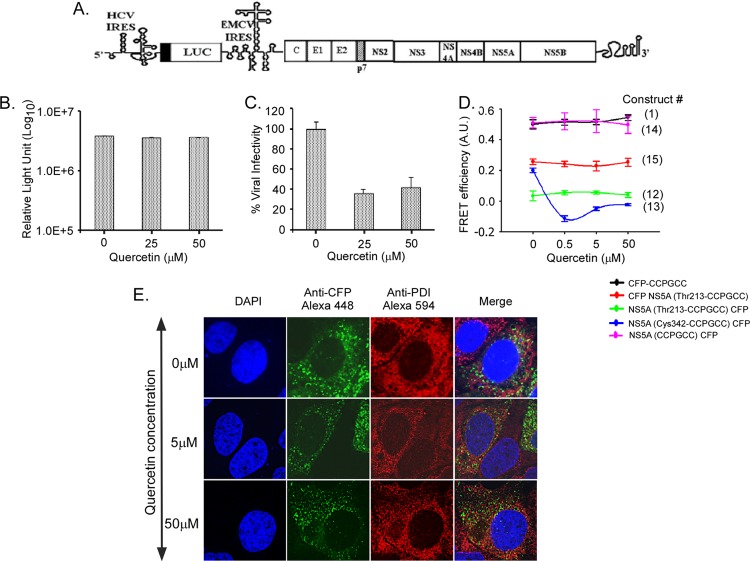
Quercetin reduces infectious HCV particle production and disrupts domain 3 FRET.
Quercetin decreases HCV particle production (12) and has been suggested to act at least in part through blocking the ability of NS5A to facilitate viral cap independent translation (20). We utilized a bicistronic RNA construct to measure the effects of quercetin on both translation mediated by the HCV 5′ untranslated region (UTR) and particle production. This construct uses the HCV 5′ UTR to drive luciferase, but the HCV open reading frame itself is driven by the 5′ UTR of encephalomyocarditis virus (EMCV) (Fig. 4A). Transfection of this construct into cells treated with quercetin resulted in a lower production of luciferase in the first 24 h than in cells untreated with quercetin (data not shown), but by 72 h the overall intracellular levels of luciferase were similar (Fig. 4B). We confirmed that quercetin has no cytopathic effect on Huh7.5 cells when cells are treated with up to 75 μM quercetin (data not shown). The supernatants from cells treated with various amounts of quercetin were then used to infect new Huh7.5 cells that had no quercetin in order to quantitate the amount of infectious HCV particles produced from cells treated with quercetin. As shown in Fig. 4C, quercetin resulted in a significantly lower production of infectious HCV particles, suggesting an effect on particle assembly/release independent of any effect on the HCV 5′ UTR-directed translation.
Fig 4.
Quercetin reduces production of infectious HCV particles and specifically alters FRET of NS5A domain 3 tags. (A) Schematic of JFH-Luc-WT (luciferase-wild type) replicon transfected into Huh7.5 cells, in which replication is monitored by independent expression of luciferase (10). (B) The in vitro-transcribed RNA from JFH1-Luc-WT was electroporated into naïve Huh7.5 cells, and the cells were treated with 25 μM or 50 μM quercetin for 15 days. The viral genome replication was monitored by measuring luciferase activity in the cells at 72 h postelectroporation as described before (10). (C) Production of infectious virions was monitored by passing the supernatant from panel B to naïve Huh7.5 cells, and luciferase activity was measured 48 h postinfection. The numbers of infectious virions obtained from quercetin-treated cells were presented relative to those in untreated cells, which were plotted as 100%. (D) Effect of quercetin on FRET efficiency in CFP-CCPGCC sequential tags (construct 1) and four different NS5A constructs (constructs 12 to 15). Mean FRET efficiencies ± standard errors are shown. (E) Confocal microscopy shows colocalization of NS5A in Huh7.5 cells. The transfected cells were stained with anti-CFP followed by Alexa Fluor 488 to show NS5 and with anti-PDI followed by Alexa Fluor 594 to show ER. The merged images are shown on the right. The cells were also stained with DAPI to show nuclei. The concentration of quercetin is indicated on the left.
NS5A domain 3 has previously been implicated in viral assembly (3, 32). Our data indicated that the conformation of domain 3 could be monitored by FRET (Fig. 3). Therefore, cells were transfected with FRET constructs, treated with three different concentrations of quercetin, and monitored for FRET activity. The CFP-CCPGCC sequential motif (construct 1) did not change in FRET efficiency, indicating that the CFP-CCPGCC motif alone is unaffected by quercetin (Fig. 4D). In contrast, quercetin treatment significantly disrupted FRET for NS5A(Cys342-CCPGCC)-CFP, where the fluorophore pair flanks domain 3. The FRET efficiency at this location was reduced from 0.20 ± 0.01 to −0.11 ± 0.01 upon exposure to as little as 0.5 μM quercetin (Fig. 4D). FRET was unaltered for all of the other tested constructs. Thus, these data raise the possibility that quercetin treatment induced a conformational change within domain 3 and/or the carboxy terminus of NS5A. This ablation of FRET could result from a number of different possibilities: direct interaction with quercetin, indirect effects resulting from suppression of HSP70 expression (12), or other uncharacterized effects of quercetin. We performed colocalization of the NS5A(Cys342-CCPGCC)-CFP construct with protein disulfide isomerase (PDI), an ER marker, in the presence and absence of quercetin (Fig. 4E). Our results show no gross change in the distribution of NS5A by quercetin. NS5A still forms puncta in treated cells and associates with PDI, although the size of the puncta and their association with PDI are slightly less than in untreated cells. Importantly, quercetin treatment did not cause NS5A(Cys342-CCPGCC)-CFP to become diffusely cytoplasmic, eliminating this as a possible reason for the decrease in FRET.
Intracellular multimerization of NS5A.
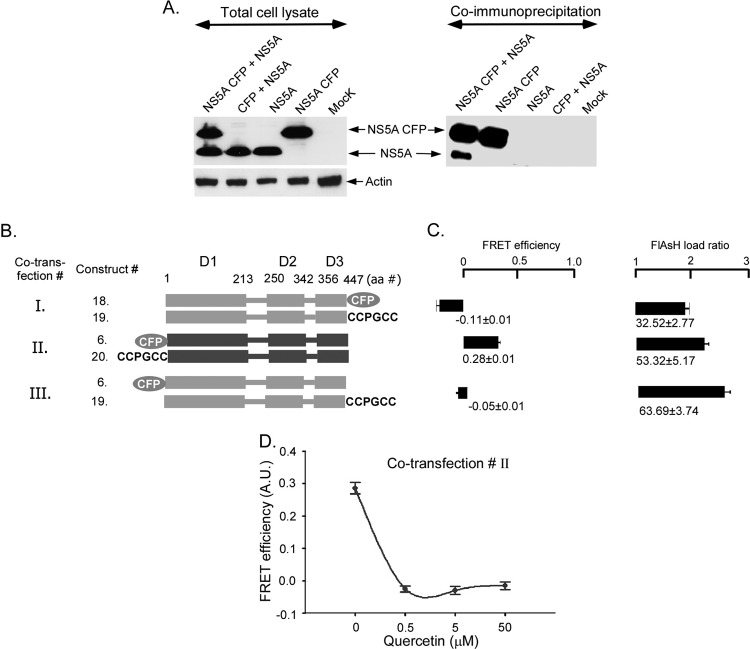
The two crystal structures of NS5A domain 1 reveal two different homodimers, yet whether NS5A multimerizes in vivo is unknown (24, 33). We tested the ability of NS5A protein expressed from different constructs to associate closely in the cell. We performed immunoprecipitation experiments of NS5A and NS5A fused to CFP. The NS5A-CFP fusion protein was coexpressed with untagged NS5A and subject to immunoprecipitation with an anti-CFP antibody (Fig. 5A). Lysates were treated to extract NS5A from membranes, and then detergent and RNA were removed to reduce nonspecific clustering of NS5A aggregates (see Materials and Methods). Western blotting revealed that untagged NS5A was coprecipitated by NS5A-CFP, suggesting that two NS5A proteins expressed from different genetic constructs associate in cells.
Fig 5.
Dimerization of HCV1B NS5A altered by quercetin in Huh7.5 cells. (A) NS5A, NS5A fused to CFP, and CFP were expressed in Huh7.5 cells in the indicated combinations for 48-h cell lysates. Expression was confirmed by Western blotting of total cell lysate with an anti-NS5A antibody (left panel). Lysates were subjected to immunoprecipitation with an anti-CFP antibody, and coprecipitated proteins were detected by Western blotting with an anti-NS5A antibody (right panel). (B) Schematic of NS5A constructs and the three conditions for cotransfection of Huh 7.5 cells. NS5A was fused to either CFP or the CCPGCC motif at either the N or the C terminus (constructs 6, 18, 19, and 20). FRET efficiencies were measured after 48 h of transfection. (C) FRET efficiencies and FlAsH loading for the different cotransfection conditions shown in panel B. (D) FRET efficiency in cotransfection II upon treatment with increasing concentrations of quercetin. Mean FRET efficiencies ± standard errors are shown.
Having established that NS5As expressed from different plasmids coimmunoprecipitate, we then used our FlAsH methodology to study NS5A multimerization in living cells. Four different tagged constructs (constructs 6, CFP-NS5A; 18, NS5A-CFP; 19, NS5A-CCPGCC; and 20, CCPGCC NS5A) were created by introducing CFP or the CCPGCC motif to either the N or the C terminus of NS5A (Fig. 5B). Three different cotransfections were performed (Fig. 5B, cotransfection conditions I, II, and III) in which FRET could theoretically occur between multimers with the carboxy termini of a multimer in close proximity (condition I), the amino termini in close proximity (condition II), or between the amino terminus of one subunit and the carboxy terminus of another (condition III). FRET was detected only when the N termini of NS5A were differentially tagged (condition II), suggesting that the amino termini are within 100 Å of each other, and probably much closer. FRET was not detected when the extreme C termini were tagged. This is in contrast to our earlier results, wherein FRET was observed when domain 3 at the C terminus was flanked by fluorophores in cis (Fig. 3C and D). Thus, FRET from probes flanking domain 3 likely occurs due to intramolecular interaction.
We then tested if quercetin treatment of cells ablated the FRET that occurred between the amino termini of a NS5A multimer. Cells transfected with both CFP-NS5A and CCPGCC-NS5A and subsequently treated with quercetin displayed significantly lower FRET efficiency (−0.02 ± 0.009) than that of the untreated control cells (0.23 ± 0.01) (Fig. 4D). Together, these data suggest that NS5A assembles in the cell as a dimer or higher-order multimer, that NS5As have N termini that are in close proximity to each other and to the C terminus of domain 1 (Fig. 3C and D, construct 15), and that quercetin treatment ablates intra- and intermolecular FRET, possibly due to changes in the structure of organization or the multimer.
DISCUSSION
In the present study, we have demonstrated that FlAsH-based FRET can report proximity relationships for donor/acceptor pairs fused to NS5A in Huh7.5 cells under appropriate experimental conditions. FRET is dependent on the distance separating the donor and acceptor, their relative orientation, and the refractive index between FRET partners, so it is not possible to calculate absolute changes in distances between donor and acceptor without verifying the orientations (25). Nevertheless, given the steep dependence (sixth power) of FRET on the distance separating donor and acceptor, it is possible to calculate that the upper limit estimates of distance separating two potential FRET partners is not more than ∼100 Å (8) with maximal FRET typically occurring at 16 to 70 Å depending upon the specific experiment (29). Some potential limitations for the use of FlAsH as a FRET acceptor, including nonspecific binding and limited access to tetracysteine binding sites, were overcome here. Of six possible FRET pairings where CFP and the CCPGCC motif were inserted and flanked between domains in NS5A in one molecule, two specific donor/acceptor pairs (Fig. 3, constructs 13 and 15) demonstrated FRET and presumably are in close proximity (<100 Å). While intermolecular FRET could theoretically occur with these constructs, no FRET occurred for constructs 11, 12, 16, and 17, nor did it occur when the fluorophores were expressed on separate molecules (cotransfections of constructs 18 and 22 or constructs 6 and 21 [Fig. 3]), which suggests intermolecular FRET likely occurred under only one of the cotransfection conditions (Fig. 5) and that intermolecular FRET is an unlikely explanation for FRET from single genetic constructs. Our data support NS5A existing as a multimer in the cell, so other intermolecular FRET cannot be entirely excluded, but our data are most consistent with a model of intramolecular FRET occurring for probes flanking domain 1 or 3.
FRET substrates for high-throughput screening (30, 38) and FlAsH dye detection of HCV core in cells assembling particles (7) have been used in several innovative studies of HCV recently, but to our knowledge they have not been used to study intracellular proximity relationships of any HCV protein. Now that specific FRET-accessible positions in NS5A are identified, future studies in which NS5A contains one FRET partner tag and the other FRET partner is in an assembly-competent core, for example (7), may shed new light on how NS5A functions during the life cycle.
Over 12 years ago, variation in the sequence of NS5A was linked to interferon response (9), and ever since then, the functions and forms of NS5A have been aggressively studied and vigorously debated. The high glycine, proline, serine, and acidic residue content of NS5A, along with disorder prediction programs (22, 27) and the wide assortment of cellular and viral partners that NS5A likely interacts with, have suggested that at least some portion of the protein was unfolded or disordered. Biochemical and NMR studies of isolated domain 2 (15) and domain 3 of genotype 1b (36) give further evidence of the loose unfolded state of the purified protein including (i) a larger size than expected by weight gel filtration elution, (ii) circular dichroism devoid of secondary structure signatures, and (iii) limited dispersion of amide proton NMR resonances. Our FRET data argue for the fact that in some situations in the cell domain 3 can be relatively compact. Whereas a loose, extended random coil of 100 residues, if such a state existed in the cell, would likely exceed the 100-Å limit of FRET (35), we detected FRET when fluorophores were placed on opposite sides of domain 3, indicating significantly closer proximity. This agrees with theoretical measurements whereby domain 3 is assumed to be in a compact, globular rigid sphere, with a predicted volume of over 13,000 Å3 and a diameter of ∼30 Å (http://www.basic.northwestern.edu/biotools/proteincalc; predictions made assuming the CFP fluorophore had no width). Supporting these conclusions are recent NMR data of domain 3 from JFH1 NS5A (36). These studies suggest that domain 3 may have some helical content, particularly at residues corresponding to Ser364-Gly381 of JFH1 genotype 2a and Ser441-Val445 in genotype 1b. While JFH1 might have more helical propensity than the Con1 isolate used here, similar regions are nevertheless present in both variants. Isolate domain 3 (construct 23) retained most of the FRET of the domain 3 expressed in the context of the entire protein (Fig. 3, construct 13), even though it is not membrane localized (data not shown). To our knowledge, our data are the first assessment of NS5A structure in the cellular environment, and this is consistent with at least portions of domain 3 spending some time in a compact configuration. In the presence of the replicon this domain 3 FRET conformation was reduced but not eliminated, perhaps arguing that the FRET-positive conformation is not present in the replication complex or even relevant to this stage of the viral life cycle, but the loss of FRET could also be seen as evidence of relevant binding of other proteins that change the refractive index between the fluorophores (5).
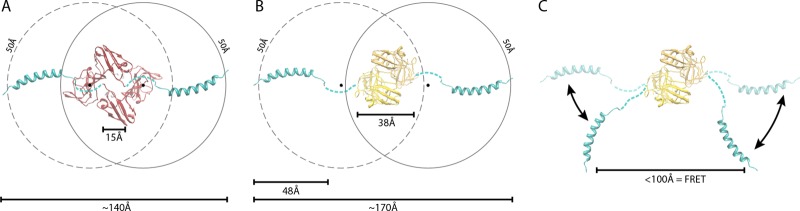
The X-ray crystallographic structures of recombinantly produced NS5A domain I lacking the N-terminal amphipathic helix yielded NS5A dimers in two distinct conformations (23, 34). In both structures, the monomeric subunits are nearly identical (∼1-Å root mean square deviation [RMSD] for Cα atoms). The N termini are located at the same end of the dimer in a region that has been proposed to facilitate interactions with host membranes. The N-terminal amphipathic helix is also thought to associate with membrane and can be modeled onto these structures in the same area (Fig. 6A and B) (26). However, the mode of dimerization is dramatically different. In one orientation, dimerization is mediated primarily by the N-terminal halves of the crystallized subunits and the C-terminal halves are spread to reveal a basic groove (Fig. 6A) (34). In the other, a totally separate interface is utilized to assemble subunits in an almost completely parallel orientation with no obvious groove in the structure. For each structure, when the amphipathic helices are modeled at their maximal distance from each other, as has been suggested (34), they exceed the FRET radius of 100 Å. This does not account for any additional distance derived from the fluorophores themselves but does assume the connecting segment between the amphipathic helix and domain 1 is unfolded. Despite the differences between these two conformations, both can readily assume a FRET-competent state assuming rotation of the amphipathic helix within the plane of the membrane (Fig. 6C). This is in agreement with our data showing significant FRET efficiency when the N termini of NS5A monomers are differentially labeled with CFP or CCPGCC. The amino acids between the amphipathic helix and the crystallized core of domain 1 are disordered (23, 34), supporting our putative model whereby their flexibility permits movement of the N termini close to each other without clashing with the body of the domain. Strikingly, treatment of cells with quercetin disrupted FRET between the N-terminal pairs. Thus, quercetin likely disrupts both intra- and intermolecular FRET, suggesting conformational rearrangements within NS5A that may disrupt its function, possibly causing the observed decrease in the production of infectious virus. Models in which FRET occurs for fluorophore pairs between rather than within dimers (as in Fig. 6) are certainly possible, but quercetin in that case would still be altering intermolecular FRET.
Fig 6.
Model of NS5A domain 1 dimer in a FRET-competent state. The solution structure of the N-terminal amphipathic helix PDB 1R7E (26) was modeled onto the crystal structures of two NS5A domain 1 dimer configurations. Structures are oriented to view the proposed membrane-binding surface of NS5A. Models and distance measurement were created with PDB 1ZH1 (33) (A) and PDB 3FQM (B) (23). The models were constructed with the N termini separated by a maximal distance assuming a flexible linker between the N-terminal helix and domain 1, consistent with the disorder in this region of the crystal structures. Shown are the distances between the N termini of the crystal structure monomers for the amphipathic helix, the maximal length between the modeled termini, the 48-Å length of the amphipathic helix, and a hypothetical 50-Å radius assuming rotation of the helix around the flexible linker. The midpoint of this linker is indicated by a black spot. In this configuration, the distance between N termini exceeds the range compatible with FRET. (C) Potential rotation of the N-terminal helix around the flexible linker in the plane of the membrane readily positions the termini in a FRET-competent state without causing clashes with the core of domain 1 for PDB 3FQM and 1ZH1 (not shown).
In conclusion, we have provided novel tools to investigate the conformation and spatial constraints of an important drug target, HCV NS5A. Although there is no crystal structural information for domains 2 and 3 of NS5A, FRET studies such as this one can suggest some distance constraints on the tertiary and quaternary organization of these regions. Our studies offer the first intracellular examination of NS5A interactions and show that in at least one configuration the amino termini are in close proximity (Fig. 6). Furthermore, our studies provide data from cells supportive of the emerging view that domain 3 in some configurations may be more compact than previously thought, but consistent with some NMR data suggesting some helical content (33). The data here provide only some constraints on how close different parts of NS5A are in the cell but nonetheless have value, since the phosphorylation state and binding partners of NS5A in the cell are different from those of the purified NS5A used in other studies. Given the number of different antiviral compounds that act on one or more forms of NS5A and may soon be in the clinic, we feel this assay may shed new insight into the mechanistic details of how these compounds alter NS5A structure and function. Intracellular FRET measurements and X-ray crystallography data can provide complementary data on the cellular structural arrangement of a protein. As our results show, while the spatial resolution and overall structural detail of FRET measurements are less precise than those obtained by X-ray diffraction, FRET assays include the unique advantages of making measurements in living cells and being able to detect changes in conformation in response to acute interventions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the American Cancer Society Research Scholar Grant (07-077-01) to R.S., by the Office of Research and Development, Biomedical Laboratory R&D Service, Department of Veterans Affairs, and by R00GM088484 to A.M.
Footnotes
Published ahead of print 23 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adams SR, et al. 2002. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124:6063–6076 [DOI] [PubMed] [Google Scholar]
- 2. Appel N, Pietschmann T, Bartenschlager R. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Appel N, et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035 doi:10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brass V, et al. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130–8139 [DOI] [PubMed] [Google Scholar]
- 5. Brzostowski JA, Meckel T, Hong J, Chen A, Jin T. 2009. Imaging protein-protein interactions by Forster resonance energy transfer (FRET) microscopy in live cells. Curr. Protoc. Protein Sci. Chapter 19:Unit19.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coelmont L., et al. 2010. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS One 5:e13687 doi:10.1371/journal.pone.0013687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Counihan NA, Rawlinson SM, Lindenbach BD. 2011. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog. 7:e1002302 doi:10.1371/journal.ppat.1002302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. dos Remedios CG, Moens PD. 1995. Fluorescence resonance energy transfer spectroscopy is a reliable “ruler” for measuring structural changes in proteins. Dispelling the problem of the unknown orientation factor. J. Struct. Biol. 115:175–185 [DOI] [PubMed] [Google Scholar]
- 9. Enomoto N, et al. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77–81 [DOI] [PubMed] [Google Scholar]
- 10. Fernandes F, Ansari IU, Striker R. 2010. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One 5:e9815 doi:10.1371/journal.pone.0009815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao M, et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gonzalez O, et al. 2009. The heat shock protein inhibitor Quercetin attenuates hepatitis C virus production. Hepatology 50:1756–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griffin BA, Adams SR, Jones J, Tsien RY. 2000. Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 327:565–578 [DOI] [PubMed] [Google Scholar]
- 14. Griffin BA, Adams SR, Tsien RY. 1998. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281:269–272 [DOI] [PubMed] [Google Scholar]
- 15. Hanoulle X, et al. 2009. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanoulle X, et al. 2009. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 381:634–638 [DOI] [PubMed] [Google Scholar]
- 17. Heidecker M, Yan-Marriott Y, Marriott G. 1995. Proximity relationships and structural dynamics of the phalloidin binding site of actin filaments in solution and on single actin filaments on heavy meromyosin. Biochemistry 34:11017–11025 [DOI] [PubMed] [Google Scholar]
- 18. Hoffmann C, et al. 2005. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat. Methods 2:171–176 [DOI] [PubMed] [Google Scholar]
- 19. Kang J, Lee MS, Gorenstein DG. 2007. Application of RNase in the purification of RNA-binding proteins. Anal. Biochem. 365:147–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khachatoorian R, et al. 20 December 2011. A cell-permeable hairpin peptide inhibits hepatitis C viral nonstructural protein 5A-mediated translation and virus production. Hepatology [Epub ahead of print.] doi:10.1002/hep.25533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lakowicz JR. 2009. Principles of fluorescence spectroscopy, 3rd ed, p 443–475 Springer Science, New York, NY [Google Scholar]
- 22. Li X, Romero P, Rani M, Dunker AK, Obradovic Z. 1999. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 10:30–40 [PubMed] [Google Scholar]
- 23. Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83:4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miyawaki A, Tsien RY. 2000. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 327:472–500 [DOI] [PubMed] [Google Scholar]
- 25. Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. 2004. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 101:10554–10559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Penin F, et al. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279:40835–40843 [DOI] [PubMed] [Google Scholar]
- 27. Romero P, et al. 2001. Sequence complexity of disordered protein. Proteins 42:38–48 [DOI] [PubMed] [Google Scholar]
- 28. Sapay N, et al. 2006. NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus. Biochemistry 45:2221–2233 [DOI] [PubMed] [Google Scholar]
- 29. Stankova K, et al. 2012. Conformational transitions of proteins engaged in DNA double strand break repair, analyzed by tryptophan fluorescence emission and FRET. Biochem. J. 443:701–709 [DOI] [PubMed] [Google Scholar]
- 30. Sudo K, et al. 2005. High-throughput screening of low molecular weight NS3-NS4A protease inhibitors using a fluorescence resonance energy transfer substrate. Antivir. Chem. Chemother. 16:385–392 [DOI] [PubMed] [Google Scholar]
- 31. Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032 doi:10.1371/journal.ppat.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 34. Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uversky VN. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11:739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Verdegem D, et al. 2011. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J. Biol. Chem. 286:20441–20454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. World Health Organization 2011. Fact sheet, p N 166 World Health Organization, Geneva, Switzerland [Google Scholar]
- 38. Yu X, Sainz B, Jr, Uprichard SL. 2009. Development of a cell-based hepatitis C virus infection fluorescent resonance energy transfer assay for high-throughput antiviral compound screening. Antimicrob. Agents Chemother. 53:4311–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.