Abstract
Herpesviruses or herpesviral sequences have been identified in various bat species. Here, we report the isolation, cell tropism, and complete genome sequence of a novel betaherpesvirus from the bat Miniopterus schreibersii (MsHV). In primary cell culture, MsHV causes cytopathic effects (CPE) and reaches peak virus production 2 weeks after infection. MsHV was found to infect and replicate less efficiently in a feline kidney cell, CRFK, and failed to replicate in 13 other cell lines tested. Sequencing of the MsHV genome using the 454 system, with a 224-fold coverage, revealed a genome size of 222,870 bp. The genome was extensively analyzed in comparison to those of related viruses. Of the 190 predicted open reading frames (ORFs), 40 were identified as herpesvirus core genes. Among 93 proteins with identifiable homologues in tree shrew herpesvirus (THV), human cytomegalovirus (HCMV), or rat cytomegalovirus (RCMV), most had highest sequence identities with THV counterparts. However, the MsHV genome organization is colinear with that of RCMV rather than that of THV. The following unique features were discovered in the MsHV genome. One predicted protein, B125, is similar to human herpesvirus 6 (HHV-6) U94, a homologue of the parvovirus Rep protein. For the unique ORFs, 7 are predicted to encode major histocompatibility complex (MHC)-related proteins, 2 to encode MHC class I homologues, and 3 to encode MHC class II homologues; 4 encode the homologues of C-type lectin- or natural killer cell lectin-like receptors;, and the products of a unique gene family, the b149 family, of 16 members, have no significant sequence identity with known proteins but exhibit immunoglobulin-like beta-sandwich domains revealed by three-dimensional (3D) structural prediction. To our knowledge, MsHV is the first virus genome known to encode MHC class II homologues.
INTRODUCTION
Cumulative data indicate that bats are important reservoirs of many emerging viruses (11), including some highly infectious and pathogenic viruses, such as henipaviruses (16, 58), severe acute respiratory syndrome (SARS) coronavirus (43), and filoviruses (41). The discovery of SARS-like coronavirus sequences in Chinese horseshoe bats intensified efforts to isolate novel viruses from different bats species, but to date virus isolation attempts have been largely unsuccessful, with a lack of permissive cell lines considered one of the contributing factors. We previously developed a method to generate primary cell lines from different bat organs (19). During the preparation of primary cells from Schreiber's long-fingered bat, Miniopterus schreibersii, we observed spontaneous cytopathic effects (CPE) in lymph node cells after two passages, which was subsequently identified as being caused by a betaherpesvirus.
Herpesviruses are highly disseminated in nature, infecting mammals, birds, reptiles, amphibians, fish, and oysters. The family Herpesviridae is divided into three subfamilies: Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. Phylogenetic analysis of the core genes yielded a tree with clear separation of the three subfamilies (51). Betaherpesvirinae is divided into five genera: Cytomegalovirus (CMV) (primate CMV), Muromegalovirus (murid CMV), Proboscivirus (elephantid herpesvirus), Roseolovirus (human herpesviruses 6 and 7 [HHV-6 and -7]), and unassigned members, which includes Tupaiid herpesvirus (tree shrew herpesvirus [THV]), Caviid herpesvirus 2 (guinea pig herpesvirus [GPCMV]), and Suid herpesvirus 2. Betaherpesviruses differ from alpha- and gammaherpesviruses in their restricted host range and long infection cycle (62).
Betaherpesvirus genome sizes range from 143 kbp for HHV-6 (31) to 241 kbp for chimpanzee CMV (CCMV) (21). The prototype strain of human CMV (HCMV) is estimated to encode 164 to 167 genes (21), of which about 70 are conserved in all betaherpesvirus genomes, including 40 core genes for all herpesviruses. The conserved genes are arranged largely colinearly in direction and position in betaherpesvirus genomes. It is known that betaherpesvirus genomes encode some proteins with homology to cellular counterparts, such as chemokine receptor, G protein-coupled receptor proteins, and major histocompatibility complex (MHC) class I molecules.
Alpha-, beta- and gammaherpesviruses have all been discovered in bats. Using PCR, Wibbelt et al. (94), Molnar et al. (56), and Watanabe et al. (93) discovered gamma- and betaherpesviruses in bats. Fifteen alphaherpesviruses were isolated from bats in Cambodia and Madagascar (65), and a betaherpesvirus (bat betaherpesvirus 2 [BHV-2]) was obtained from the spleen primary cell culture of Miniopterus fuliginosus (92).
In this study, we report the isolation, cell infection, and complete genome sequence of a novel bat betaherpesvirus from M. schreibersii primary cell culture (MsHV). Three groups of immune-related genes were found in the MsHV genome, including MHC class I and class II, C-type lectin, and a unique gene family of 16 members.
MATERIALS AND METHODS
Purification and propagation of MsHV.
M. schreibersii bat lymph node (MsLn) and kidney primary cells (MsKi) were prepared as previously described (19). The cells were cultured in Dulbecco's modified Eagle medium (DMEM)-F12-Hams (Sigma) supplemented with 10% fetal calf serum (FCS) (HyClone) and Antibiotic-Antimycotic antibiotics (Gibco) at 37°C with 5% CO2. To purify MsHV, MsKi cells in a 96-well tissue culture plate (4 × 104/well) were infected with 10-fold serial dilutions of MsHV and incubated for 15 days to allow CPE to develop. Supernatant was collected from wells with CPE at the dilution where less than 25% of wells showed CPE, and one of them was used for the second round of purification. A single clone with typical CPE was picked to propagate in MsKi cells grown in 4 150-cm2 flasks. The infected culture medium was subsequently clarified at 2,000 × g for 5 min and then ultracentrifuged at 55,000 rpm for 1 h in a Beckman SW 55Ti rotor. The resulting viral pellet was resuspended in phosphate-buffered saline (PBS), and aliquots were stored at −80°C. Virus was titrated using endpoint dilutions as described above.
Cell infection and real-time PCR.
Cells in Eagle minimal essential medium (EMEM) or DMEM with 10% fetal calf serum (FCS) were seeded in 35-mm plates and incubated overnight prior to infection with 1,000 IU (infectious units) in 0.5 ml total volume. After 2 h of incubation, 2 ml fresh medium with 5% FCS was added. At indicated days postinfection (dpi), cells were washed four times in PBS, collected by centrifugation, and stored at −20°C. Total DNA was extracted with Qiagen DNeasy blood and tissue kit. The same volume of total DNA for each cell species was used to detect MsHV genomic DNA by real-time PCR with the primers MsHV-F (5′-TCACC AGGAT AGGGC GAGAC A-3′) and MsHV-R (5′-CTTTT CAAAT TCCAG CTTCA CAGG-3′) using the following parameters: 95°C for 2 min and 40 cycles of 95°C for 15 s, 60°C for 30 s, 84°C for 30 s for signal collection, followed by melting-curve analysis from 60°C to 95°C.
Viral genomic DNA preparation and 454 sequencing.
The viral pellet was diluted with TE (10 mM Tris-Cl, 1 mM EDTA, pH 8.0) and treated with 0.2% SDS and 100 μg/ml proteinase K at 56°C for 2 h. DNA was extracted using phenol-chloroform and precipitated with 0.2 M NaCl and an equal volume of isopropanol. The DNA was separated on a 0.4% agarose gel, and the large and distinct band corresponding to viral genomic DNA was sliced out and recovered by electroelution into a dialysis tube (71), followed by filtering through a prewet 0.45-μm filter, phenol-chloroform extraction, and isopropanol precipitation. The DNA sequence of the purified MsHV viral genomic DNA was determined using the Roche 454 GS-FLX platform (454 Life Sciences, Branford, CT) with sample preparation as described in their Titanium series manuals, Rapid Library Preparation and emPCR Lib-L SV.
Genome sequence assembly.
De novo assemblies of the 454 sequencing output were performed independently with the Roche GS De Novo Assembler (Newbler) and CLC Genomics Workbench v4.8 (CLC Inc., Aarhus, Denmark) software programs, resulting in contigs ranging from 2 kb to 75 kb. Contigs generated from both next-generation-sequencing de novo algorithms were assembled using the Seqman module of the software program Lasergene DNASTAR 9 Core Suite (DNAStar, Madison, WI), resulting in a single large contiguous sequence of about 222.8 kb. To search for raw reads that had not been assembled at the ends of the large contig, 200 bp from each end was used as a template to find similarity with reads from the 454 raw reads library using the “scan for similarities” function in the program Clone Manager Professional 9 (Sci-Ed Software, Cary NC). This manual walking was repeated until no more sequences aligned. The final contig was then used as a reference to map back to the 454 raw reads using CLC Genomics Workbench, and the resulting consensus sequence was taken as the MsHV genome. PCR was used to confirm the sequence of regions with coverage lower than 50 reads and/or high GC content using primers designed according to the flanking sequences (L1-f, 5′-GTGGT TAAGT GTGGG TGTGT CTGG-3′; L1-r, GGAAG ACGGC GACAA CAGGC-3′; L2-f, 5′-GATTC GGTGG AATCA ATGAC CCTG-3′; L2-r, 5′-TTTCC TAACC AGTTA GCCCA ACGC-3′; L3-f, 5′-TTACA GCGTT AGGCG AATCA CACG-3′; L3-r, 5′-GTCAT CCTGC GCTTG GTGTA TCC-3′; L4-f, 5′-GTCTG CGACG AAGGC TCTCG C-3′; L4-r, 5′-TGTGG TTCTT GCAGG CAGCG-3′). 360 GC Enhancer buffer (ABI) was used for regions with high GC content according to the manufacturer's instructions.
Computer-assisted analysis.
Open reading frames (ORFs) were initially predicted with the software programs FgenesV (Softberry) and GeneMarkS (5). ORFs that contained canonical start and stop codons were BLASTed against the local protein database of betaherpesviruses, including THV (2), human CMV (HCMV) (24), murine CMV (MCMV) (64), rat CMV (RCMV) (89), GPCMV (38, 72), rhesus CMV (RhCMV) (33), chimpanzee CMV (CCMV) (21), HHV-6A (31), HHV-6B (25), and HHV-7 (53). Hits with an E-value of less than e−3 were used to annotate the MsHV genome, while ORFs with low-probability hits to betaherpesviruses were BLASTed against the GenBank nonredundant protein database; hits with E-values of less than e−3 were included in the annotation to obtain a draft annotated genome. The common strategy for herpesvirus' gene prediction, the presence of an ORF with a minimum length of 300 bp and less than 60% overlap with adjacent ORFs (64), was applied to other portions of the MsHV genome. The gaps between annotated ORFs were inspected again for ORFs with a minimum length of 150 bp. Analysis of the repeats in the genome was performed using the software program Tandem Repeats Finder (4). Protein sequences were subject to transmembrane and signal peptide prediction with the program Phobius (37). MsHV proteins were aligned pairwise with the homologues of THV, HCMV, and RCMV with the program ClustalW, and the pairwise identities are presented in Table 1. Multiple sequence alignment was conducted with ClustalW, and phylogenetic trees were constructed with the software program MEGA5 (82). Protein structure prediction was performed with Phyre2 (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index) (39).
Table 1.
Predicted ORFs in MsHV genome and identities with their homologs
| ORFa | Db | Location (nt) | Length (aa) | Homolog (% amino acid identity)c |
Comment(s)c | ||
|---|---|---|---|---|---|---|---|
| THV | HCMV | RCMV | |||||
| b1 | R | 493–675 | 60 | ||||
| b2 | L | 1825–2013 | 224 | ||||
| b3 | L | 2029–4008 | 62 | ||||
| b4 | R | 4134–4343 | 659 | Signal and transmembranee | |||
| b5 | R | 4295–4567 | 69 | Signal | |||
| b6 | R | 4650–4850 | 90 | ||||
| b7 | L | 5247–5846 | 66 | ||||
| b8 | R | 5509–6342 | 199 | Zinc carboxypeptidases, zinc-binding region 2 signature | |||
| b9 | R | 1052–1726 | 277 | Signal | |||
| B23 | L | 6516–7601 | 361 | T23 (35.8) | UL23 (28.6) | R23 (31.1) | US22 family |
| B24 | L | 7671–8978 | 435 | UL24 (30.5) | R24 (33.1) | US22 family; tegument protein | |
| b24.1 | L | 8819–9424 | 201 | ||||
| B25 | R | 9381–11501 | 706 | T25 (31.1) | UL25 (29.3) | R25 (27.3) | UL25 family |
| B26 | L | 11782–12423 | 213 | T26 (34.4) | UL26 (38.6) | R26 (36.3) | US22 family |
| B27 | L | 12618–14513 | 631 | T27 (33.0) | UL27 (30.6) | R27 (27.4) | |
| B28 | L | 14627–15628 | 333 | T28 (49.3) | UL43 (22.9) | R28 (33.5) | US22 family |
| B28.1 | L | 15838–16980 | 380 | T28 (23.6) | UL29 (19.3) | US22 family | |
| B28.2 | L | 17218–18261 | 347 | T28 (27.1) | UL29 (21.2) | R28 (26.0) | US22 family |
| B28.3 | L | 18350–19438 | 362 | T28 (26.8) | UL29 (21.0) | R28 (28.0) | US22 family |
| B28.4 | L | 19573–20652 | 359 | T28 (28.5) | UL29 (20.0) | R28 (26.0) | US22 family |
| B29E1 B29E2 | L | 21767–23089, 20633–21691 | 792 | T29 (36.2)d | UL29 (32.9) | R36 (22.7) | US22 family |
| B29.1 | L | 23131–23787 | 218 | T26 (25.4) | UL26 (25.7) | r25.1 (21.6) | US22 family |
| b29.2 | L | 23803–24030 | 75 | ||||
| b29.3 | L | 24051–24299 | 82 | UL95 (11.1) | |||
| B30 | L | 24394–25902 | 502 | T30 (38.3) | UL27 (23.0) | ||
| B31 | R | 26065–28245 | 726 | T31 (24.9) | UL31 (29.8) | R31 (26.2) | dUTPase family |
| B32 | L | 28262–29908 | 548 | T32 (26.1) | UL32 (23.3) | R32 (31.6) | Major tegument phosphoprotein |
| B33E1 B33E2 | R | 30013–30045, 30139–31224 | 371 | T33 (55.0) | UL33 (43.2) | R33 (47.4) | G protein-coupled receptor family |
| B34 | R | 31461–32792 | 443 | T34 (33.8) | UL34 (27.7) | R34 (26.9) | |
| B35 | R | 32951–34666 | 571 | T35 (38.6) | UL35 (30.8) | R35 (35.5) | UL25 family |
| b35.1 | L | 34651–35082 | 143 | Transmembrane | |||
| B36E1 | L | 36524–36745 | 445 | T36 (28.9) | UL36 (27.1) | R36 (30.2 | US22 family; immediate-early regulatory protein; inhibitor of caspase-8-induced apoptosis |
| B36E2 | 35175–36293 | ||||||
| b36.1 | L | 36319–36549 | 76 | Transmembrane | |||
| B37 | L | 36950–37927 | 325 | T37 (28.6) | UL37 (23.8) | R37 (29.0 | Immediate-early protein; mitochondrial inhibitor of apoptosis |
| b37.1 | R | 37946–38626 | 226 | ||||
| B38 | L | 38520–39533 | 337 | T38 (29.2) | UL38 (33.4) | R38 (30.0 | Virion envelope glycoprotein |
| b38.1 | L | 39543–39857 | 104 | Transmembrane | |||
| b39 | L | 39991–41148 | 385 | MHC class II; signal and transmembrane | |||
| b39.1 | L | 41206–42315 | 369 | Hereditary hemochromatosis protein (HFE); signal and transmembrane | |||
| 39.2 | L | 42384–43634 | 416 | MHC class II; signal and transmembrane | |||
| b39.3 | R | 43597–43923 | 108 | ||||
| b39.4 | L | 43831–45069 | 412 | MHC class I; signal and transmembrane | |||
| b39.5 | L | 45105–46190 | 361 | MHC class I; signal and transmembrane | |||
| b40 | L | 46237–47469 | 410 | 25.1% identity with HHV-7 U21; downregulates MHC class I; signal and transmembrane | |||
| b41 | R | 47767–48273 | 168 | ||||
| b42 | L | 48504–49352 | 282 | Two transmembrane | |||
| b42.1 | L | 49366–49683 | 105 | Transmembrane | |||
| b42.2 | L | 49871–50302 | 143 | Transmembrane | |||
| b42.3 | L | 50403–50735 | 110 | Transmembrane | |||
| B43 | L | 50826–51923 | 365 | UL43 (31.9) | R43 (26.8) | US22 family; tegument protein | |
| b43.1 | L | 51989–52948 | 319 | Seven transmembrane | |||
| B44 | L | 52942–54156 | 404 | T44 (58.9) | UL44 (56.5) | R44 (55.6) | DNA polymerase processivity subunit |
| b44.1 | L | 54353–54556 | 67 | ||||
| B45 | L | 54619–57585 | 988 | T45 (31.8) | UL45 (29.6) | R45 (31.4) | Large subunit ribonucleotide reductase homolog |
| B46 | L | 57623–58669 | 348 | T46 (39.4) | UL46 (39.0) | R46 (40.2) | Minor capsid binding protein |
| B47 | R | 58509–61448 | 979 | T47 (37.0) | UL47 (35.0) | R47 (33.6) | Tegument protein |
| B48 | R | 61456–67971 | 2171 | T48 (34.9) | UL48 (32.6) | R48 (33.6) | Largest tegument protein |
| B48A | L | 68059–68301 | 80 | T48A (48.4)d | UL48A (43.2) | R48A (43.3)d | Smallest capsid protein |
| B49 | L | 68291–69979 | 562 | T49 (49.0) | UL49 (42.2) | R49 (44.5) | Virion protein |
| B50 | L | 69963–70952 | 329 | T50 (49.2) | UL50 (43.2) | R50 (44.8) | Inner nuclear membrane protein |
| B51 | L | 71103–71432 | 109 | T51 (56.5) | UL51 (39.5) | R51 (50.8) | Terminase component |
| B52 | R | 71431–73248 | 605 | T52 (50.9) | UL52 (43.6) | R52 (44.0) | Virion protein |
| B53 | R | 73241–74125 | 294 | T53 (54.8) | UL53 (44.5) | R53 (52.8) | Tegument protein |
| B54 | L | 74246–77491 | 1081 | T54 (53.6) | UL54 (50.3) | R54 (50.2) | DNA polymerase |
| B55 | L | 77496–80264 | 922 | T55 (50.7) | UL55 (46.9) | R55 (47.9) | Virion envelope glycoprotein B (gB) |
| B56 | L | 80152–82623 | 823 | T56 (58.1) | UL56 (54.9) | R56 (54.7) | Terminase component |
| b56.1 | R | 82568–82921 | 117 | Arg-rich profile | |||
| b56.2 | R | 83019–83180 | 53 | ||||
| B57 | L | 83144–86770 | 1208 | T57 (59.4) | UL57 (53.9) | R57 (48.5) | Major DNA binding protein |
| b58 | L | 86936–87109 | 57 | ||||
| b59 | L | 87087–87362 | 91 | ||||
| b60 | L | 87316–87699 | 127 | ||||
| b61 | L | 87826–88029 | 67 | ||||
| b62 | L | 88096–88320 | 74 | ||||
| b63 | L | 88785–89102 | 105 | ||||
| b64 | L | 89111–89317 | 68 | ||||
| b65 | L | 89326–89496 | 56 | ||||
| b66 | R | 89558–89761 | 67 | ||||
| b67 | L | 90040–90204 | 54 | ||||
| b68 | L | 90265–90570 | 101 | ||||
| b68.1 | L | 90972–91124 | 50 | ||||
| b68.2 | R | 91840–92019 | 59 | ||||
| b68.3 | R | 92322–92501 | 59 | ||||
| B69 | L | 92543–94642 | 699 | T69 (31.2) | UL69 (28.4) | R69 (29.5) | Multiple regulatory protein; tegument protein |
| b69.1 | L | 94746–95030 | 94 | ||||
| b69.2 | R | 95252–95572 | 106 | ||||
| B70 | L | 95483–98368 | 961 | T70 (46.2) | UL70 (44.5) | R70 (43.9) | DNA helicase primase subunit |
| B71 | R | 98603–99289 | 228 | T71 (33.5) | UL71 (31.8) | Tegument protein; envelopment | |
| B72 | L | 99403–100461 | 352 | T72 (26.3) | UL72 (25.4) | R72 (27.3) | dUTPase family |
| B73 | R | 100460–100843 | 127 | T73 (41.9) | UL73 (34.0) | R73 (40.0) | Virion envelope glycoprotein N (gN) |
| B74 | L | 100829–101893 | 354 | T74 (28.3) | UL74 (28.3) | r74 (26.1) | Virion envelope glycoprotein O (gO) |
| B74A | R | 101853–102152 | 99 | T74A (27.2)d | UL74A (30.2) | R74A (25.9)d | |
| B75 | L | 102237–104381 | 714 | T75 (38.6) | UL75 (34.2) | R75 (35.0) | Virion envelope glycoprotein H (gH) |
| B76 | R | 104522–105394 | 290 | T76 (41.3) | UL76 (40.2) | R76 (43.7) | Inducers of cell cycle arrest and apoptosis |
| B77 | R | 105084–106838 | 584 | T77 (52.2) | UL77 (50.4) | R77 (45.5) | Portal capping protein |
| B78 | R | 106930–108150 | 406 | T78 (27.3) | UL78 (27.9) | R78 (25.1) | G protein-coupled receptor family |
| B79 | L | 108218–109204 | 328 | T79 (42.6) | UL79 (39.8 | R79 (44.9) | |
| B80 | R | 109065–111251 | 728 | T80 (40.0) | UL80 (35.9) | R80 (38.0) | Protease; assembly protein; |
| B80.5 | R | 109932–111251 | 439 | Assembly protein, C-terminal part of B80 | |||
| B82 | L | 111320–112888 | 522 | T82 (33.1) | UL82 (30.7) | R82 (29.1) | dUTPase family; upper matrix protein |
| B84 | L | 113062–114576 | 504 | T84 (35.2) | UL84 (27.4) | R84 (11.0) | Lytic DNA replication initiation |
| B85 | L | 114731–115618 | 295 | T85 (57.0) | UL85 (59.7) | R85 (59.5) | Minor capsid protein |
| B86 | L | 115834–119883 | 1349 | T86 (59.4) | UL86 (59.0) | R86 (61.2) | Major capsid protein |
| B87 | R | 119976–122621 | 881 | T87 (53.8) | UL87 (48.4) | R87 (54.3) | |
| B88 | R | 122652–123794 | 380 | T88 (34.4) | UL88 (37.0) | R88 (35.4) | Tegument protein |
| B89E1 B89E2 | L | 129510–130388, 123791–124918 | 668 | T89 (72.5) | UL89 (66.5) | R89 (68.7) | Terminase component |
| b90 | L | 124947–125747 | 266 | Signal and transmembrane | |||
| B91 | R | 125737–125985 | 82 | T91 (37.9) | UL91 (38.4) | R91 (20.8) | |
| B92 | R | 126019–126735 | 238 | T92 (64.9) | UL92 (47.5) | R92 (60.0) | |
| B93 | R | 126707–128305 | 532 | T93 (35.1) | UL93 (31.5) | R93 (33.5) | Tegument protein |
| B94 | R | 128259–129284 | 341 | T94 (42.7) | UL94 (42.7) | R94 (39.8) | Tegument protein |
| B95 | R | 130387–131511 | 374 | UL95 (37.7) | R95 (48.8) | Tegument protein | |
| B96 | R | 131540–131905 | 121 | T96 (43.2) | UL96 (39.7) | R96 (32.6) | Tegument protein |
| B97 | R | 131931–133835 | 634 | T97 (38.7) | UL97 (36.4) | R97 (35.9) | Phosphotransferase |
| B98 | R | 133867–135546 | 559 | T98 (41.4) | UL98 (44.9) | R98 (38.2) | Exonuclease |
| B99 | R | 135483–135833 | 116 | T99 (34.4) | UL99 (26.6) | R99 (30.5) | Tegument phosphoprotein |
| B100 | L | 135965–137017 | 350 | T100 (59.4) | UL100 (52.8) | R100 (49.9) | Virion envelope glycoprotein M (gM) |
| b101 | R | 137240–137695 | 151 | ||||
| b101.1 | L | 137577–137903 | 108 | ||||
| B102 | R | 137945–140254 | 769 | T102 (38.3) | UL102 (32.1) | R102 (27.2) | DNA helicase primase subunit |
| B103 | L | 140308–141042 | 244 | T103 (34.9) | UL103 (34.6) | R103 (28.3) | Tegument protein |
| B104 | L | 140996–143125 | 709 | T104 (46.6) | UL104 (48.9) | R104 (39.3) | Portal protein; DNA encapsidation |
| B105 | R | 142848–145409 | 853 | T105 (55.8) | UL105 (54.0) | R105 (50.2) | DNA helicase primase subunit |
| b106 | L | 148805–148975 | 56 | ||||
| b107 | L | 149051–149365 | 104 | ||||
| B112 | R | 150046–150690 | 214 | T112 (30.7) | UL112 (29.5) | R112EX1 (27.8) | Lytic DNA replication initiation |
| B113 | R | 150767–151993 | 408 | UL113 (26.2) | R113 (28.4) | Lytic DNA replication initiation | |
| B114 | L | 152078–152863 | 261 | T114 (57.9) | UL114 (51.9) | R114 (51.8) | Uracil-DNA glycosylase |
| B115 | L | 152878–153597 | 239 | T115 (36.6) | UL115 (36.8) | R115 (33.8) | Virion envelope glycoprotein L (gL) |
| B116 | L | 153600–154454 | 284 | t116 (35.2) | UL116 (25.0) | ||
| B117 | L | 154489–155982 | 497 | T117 (21.2) | UL117 (27.5) | ||
| B118 | L | 156019–156330 | 103 | t119 (19.6) | Transmembrane | ||
| b119 | R | 156499–156933 | 144 | Arg-rich profile; Cys-rich profile; signal | |||
| b120 | L | 156945–157220 | 91 | Transmembrane | |||
| b121 | L | 157264–157911 | 215 | Signal | |||
| b121.1 | L | 157914–158213 | 99 | Signal and transmembrane | |||
| b121.2 | L | 158214–158879 | 221 | Transmembrane | |||
| b121.3 | L | 159021–159638 | 205 | ||||
| B121.4 | L | 159901–160752 | 283 | t121.4 (29.6) | r119.5 (24.8) | ||
| B121.5 | L | 160886–161383 | 165 | t121.5 (22.3) | Signal and transmembrane | ||
| B122 | L | 161567–162817 | 416 | T122 (43.5) | UL122 (32.5) | R122 (31.9) | Immediate-early 2 transactivator |
| b123 | L | 163205–164347 | 380 | 17.7% identity with γHHV-4 LMP-1; Asp-rich profile | |||
| b124 | L | 164409–164651 | 80 | ||||
| b124.1 | R | 164947–165189 | 80 | Signal and transmembrane | |||
| b124.2 | L | 165660–166349 | 229 | Molecular chaperone DnaK; Gly-rich profile | |||
| B125 | L | 170815–172374 | 519 | r127 (22.1) | 26.5% identity with HHV-6B U94; parvovirus Rep protein | ||
| B126 | R | 172473–173663 | 396 | T2 (17.2) | US22 (22.9) | r128 (28.5) | US22 family |
| b127 | L | 173650–174042 | 130 | ||||
| B128 | L | 173996–174172 | 58 | UL128 (22.0) | |||
| b129 | L | 174398–174757 | 119 | ||||
| B130 | L | 174807–175070 | 87 | T13.1 (33.0) | UL130 (23.4) | ||
| b131 | L | 175905–176333 | 142 | Transmembrane | |||
| B132 | L | 176434–177150 | 238 | T9 (24.9) | Signal | ||
| b133 | L | 177144–178052 | 302 | G-protein coupled receptors family 1 signature; signal and transmembrane | |||
| B148 | L | 178191–179156 | 321 | t8 (25.3) | UL148 (25.4) | ||
| b148.1 | L | 179536–179844 | 102 | Transmembrane | |||
| b148.2 | L | 180067–180888 | 273 | Transmembrane | |||
| b149 | L | 180944–181603 | 219 | b149 family; signal and transmembrane | |||
| b149.1 | L | 182401–183387 | 328 | b149 family; two transmembrane | |||
| b149.2 | L | 183972–184309 | 112 | b149 family | |||
| b149.3 | L | 184351–185343 | 330 | b149 family; signal and transmembrane | |||
| b149.4 | L | 185404–186390 | 328 | b149 family; signal and transmembrane | |||
| b149.5 | L | 186441–187400 | 319 | b149 family; signal and transmembrane | |||
| b149.6 | L | 187504–188469 | 321 | b149 family; transmembrane | |||
| b149.7 | L | 188938–189903 | 321 | b149 family; signal and transmembrane | |||
| b149.8 | L | 189996–190964 | 322 | b149 family; signal and transmembrane | |||
| b149.9 | L | 191080–192051 | 323 | b149 family; signal and transmembrane | |||
| b149.10 | L | 192610–193578 | 322 | b149 family; signal and transmembrane | |||
| b149.11 | L | 193664–194596 | 310 | b149 family; signal and transmembrane | |||
| b149.12 | L | 194656–195630 | 324 | b149 family; signal and transmembrane | |||
| b149.13 | L | 195830–196507 | 225 | b149 family; signal and transmembrane | |||
| b149.14 | L | 196701–197552 | 283 | b149 family; two transmembrane | |||
| b149.15 | L | 198108–198974 | 288 | b149 family | |||
| b149.20 | L | 199874–200674 | 266 | Signal | |||
| b149.21 | R | 200539–200973 | 144 | Transmembrane | |||
| b149.22 | L | 202217–203680 | 487 | Ig-like domain profile | |||
| B150 | L | 203690–205828 | 712 | T5 (32.4) | US22 (28.9) | r139 (22.8) | US22 family |
| B151 | L | 206171–208063 | 630 | T4 (36.2 | US23 (36.9) | r140 (32.2) | US22 family |
| B152 | L | 208606–210156 | 516 | T3 (39.5) | US24 (36.5) | r141 (30.5) | US22 family |
| B153 | L | 210361–212190 | 609 | T2 (31.6) | US26 (30.4) | r142 (25.4) | US22 family |
| B154 | R | 212886–214421 | 511 | T5 (30.2) | US22 (29.9) | r140 (28.6) | US22 family |
| B155 | L | 214594–216507 | 637 | T1 (27.4) | US23 (23.0) | r143 (26.5) | US22 family |
| b156 | L | 216904–217818 | 304 | C-type lectin family; transmembrane; 38.8% identity with b163 | |||
| b157 | R | 217842–218021 | 59 | ||||
| b158 | L | 218031–218639 | 202 | b158 family; transmembrane; 27.9% identity with b160 | |||
| b159 | R | 218744–219106 | 120 | ||||
| b160 | L | 219697–220251 | 184 | b158 family; transmembrane; 27.9% identity with b158 | |||
| b161 | L | 220223–221023 | 266 | C-type lectin family; transmembrane; 29.2% identity with b162 | |||
| b162 | L | 221017–221658 | 213 | Natural killer cell lectin-like receptor; transmembrane; 29.2% identity with b161 | |||
| b163 | L | 221777–222604 | 276 | C-type lectin family; transmembrane; 38.8% identity with b156 | |||
ORFs with homologs in THV, HCMV or RCMV are named with uppercase prefixes and unique ORFs with lowercase prefixes. The herpesvirus core genes are underlined.
D, direction. “L” indicates the ORFs toward left, and “R” indicates those toward the right.
Bold indicates the homolog with highest identity.
T48A, R48A, T74A, and R74A were not identified previously; T29 was repredicted.
“Signal” stands for a signal peptide predicted by Phobius (37), and “transmembrane” stands for a transmembrane domain predicted.
Nomenclature.
All the predicted ORFs are listed in Table 1. The nomenclature used for MsHV ORFs is based on a similar strategy used for other betaherpesviruses (2, 64, 72, 89), where ORFs are numbered in the order of appearance in the genome from left (5′) to right (3′); ORFs with homologues in THV, HCMV, or RCMV were named with uppercase prefixes (e.g., B23). ORFs with no obvious homologues in those viruses were designated using lowercase prefixes (e.g., b9). Decimal points were introduced when necessary, but the decimal suffixes do not necessarily indicate a relationship between these genes, e.g., B24 and b24.1 are not related, while B28 and B28.1 to B28.4 are all members of one gene family.
Nucleotide sequence accession numbers.
The MsHV genome sequence has been submitted to the GenBank database with the accession number JQ805139. Other herpesviruses used in this study are bat BHV-2 (accession no. AB517983), THV (NC_002794.1), HCMV Merlin (NC_006273.2), RCMV (Maastricht strain; NC_002512.2), GPCMV (ATCC-P5 strain; AB592928.1), MCMV (Smith strain; NC_004065.1), RhCMV (strain 68-1; AY186194.1), CCMV (AF480884.1), cynomolgus macaque CMV (CyCMV) (JN227533.1), HHV-6A (NC_001664.2), HHV-6B (NC_000898.1), HHV-7 (strain RK; AF037218.1), (herpes simplex virus 1 (HSV) (NC_001806.1), and Epstein-Barr virus (EBV) (NC_007605.1).
RESULTS
Identification and cell infection of a betaherpesvirus from M. schreibersii primary cell culture.
During establishment of primary cell cultures from different organs from the microbat M. schreibersii, CPE was observed in confluent cultures of primary lymph node cells. Initial PCR with family-specific degenerate primers revealed a betaherpesvirus. Since the original primary cell cultures were made from an organ pool from different individuals, we purified the virus with two rounds of endpoint dilution in MsKi primary cells.
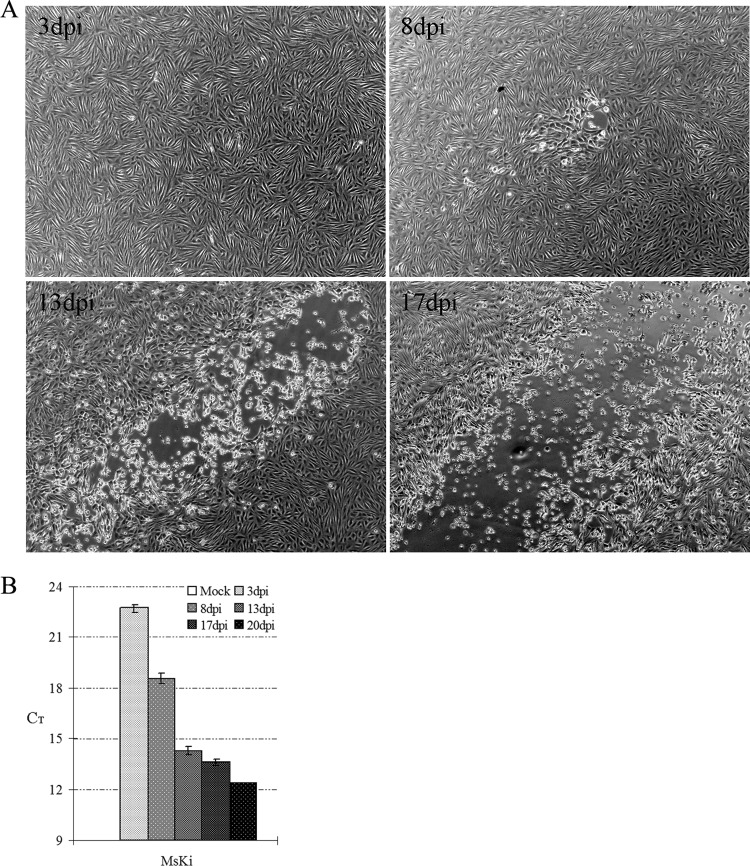
MsKi cell monolayers were infected with MsHV at a multiplicity of infection (MOI) of 0.1 and incubated for up to 20 days. On 7 to 8 dpi, CPE started to form as some cells swelled, rounded up, and then detached from the culture surface; the viral CPE spread to neighboring cells until most of the cells in the monolayer died (Fig. 1A). Real-time PCR revealed that the amount of viral DNA detected increased with time (Fig. 1B).
Fig 1.
Infection of MsKi primary cells. (A) CPE development in infected MsKi cells. Cells were infected with MsHV at an MOI of 0.1. The same CPE locus is shown at 8, 13, and 17 dpi. (B) Real-time PCR quantification of viral DNA. Total DNA was extracted from infected cells, and viral DNA was detected with real-time PCR. Error bars represent standard deviations (n = 3).
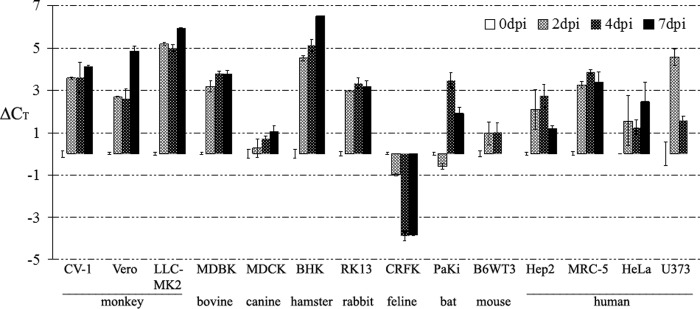
To evaluate the species specificity of MsHV infection, cells from different species, including monkey (CV-1, Vero, and LLC-MK2), human (Hep2, MRC-5, HeLa, and U373), and seven other species (MDBK, MDCK, BHK, RK13, CRFK, PaKi, and B6WT3), were infected, and viral replication was monitored with real-time PCR at 0, 2, 4, and 7 dpi. As shown in Fig. 2, the amount of viral DNA decreased after infection in all the cell lines, as indicated by increasing threshold cycle (CT) values compared to the CT at 0 dpi, with the only exception being the feline kidney cell line, CRFK. In the CRFK cells, the CT value decreased by 4 cycles at 4 and 7 dpi, compared to 0 dpi, and by 6 to 10 cycles compared to results for other cells. PaKi, a bat kidney primary cell from the fruit bat Pteropus alecto (19), is nonpermissive for MsHV.
Fig 2.
Infection of different cell lines by MsHV. Cells in 35-mm plates were infected with 1,000 IU and sampled at 0, 2, 4, and 7 dpi. MsHV viral DNA in the cells was detected using real-time PCR. The CT values at 0 dpi were set as zero. U373 and B6WT3 were not determined at 7 dpi due to poor cell viability. Error bars represent standard deviations (n = 2).
454 sequencing of the MsHV genome.
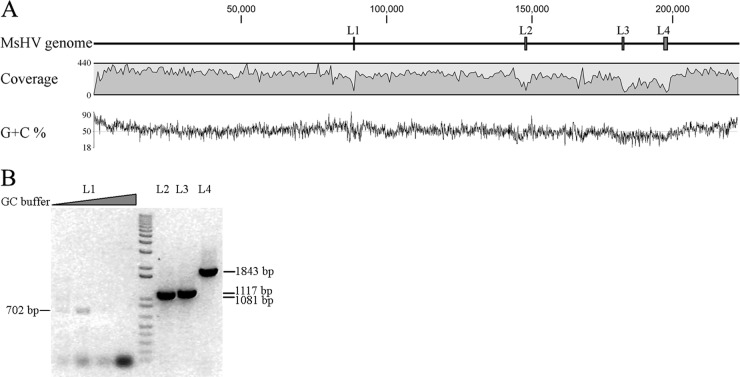
Next-generation 454 sequencing was used to sequence the genome of MsHV. We obtained 135,774 raw reads, with an average length of 428 bp. Two different NGS software programs were used for de novo assembly (Newbler and CLC Genomics Workbench) to obtain the full-length viral genome, but neither resulted in a single full-length contig. However, when the contigs from the different assembly approaches were pooled, a draft single large contig was obtained. This contig was then used as a reference sequence to map back to the raw reads, resulting in a consensus sequence of 222,870 bp, which we regard as the MsHV genome. The MsHV genome is close in size to the primate (14, 21, 24, 33) and murid (64, 89) CMV genomes and about 27 kbp longer than that of THV (2). The 222,870-bp genome incorporated 114,047 reads (84.0%). The read coverage ranged from 5- to 440-fold, with an average of 224-fold (Fig. 3A). The regions with less than 50-fold coverage were manually confirmed before being included in the final contig. Four regions, labeled L1, L2, L3, and L4 in Fig. 3A, had the lowest coverage, 5-, 34-, 11- and 14-fold, respectively. The sequence assembly across these regions was further confirmed by PCR with flanking primers, and the L1 region could be amplified only with an appropriate amount of GC enhancer buffer (Fig. 3B).
Fig 3.
Assembly of the MsHV genome. The MsHV consensus genome was assembled from, and mapped back to, the 454 sequencing raw reads. (A) Coverage and G+C content along the genome. Bars labeled L1 to L4 indicate the low-coverage regions (<50). (B) PCR confirmation of low-coverage regions.
Features of MsHV genome sequence.
The overall G+C content of the MsHV genome is 51.9%, less than those of THV (66.5%), HCMV (57.5%), RCMV (61%), and MCMV (58.7%), respectively. As in THV (2), HCMV (24), and RhCMV (33), the termini of the genome have the highest G+C content (Fig. 3A). In contrast, there is a region of ∼18 kbp, between L3 and L4, that has a significantly lower G+C content of 39.5% and a significantly lower sequence coverage than other regions of the genome. This was also true for L2, with a G+C content of 37.9%. L1, however, has a very high G+C content of 72% in 450 bp and up to 88% in 146 bp, which explains the difficulty in amplification of this region.
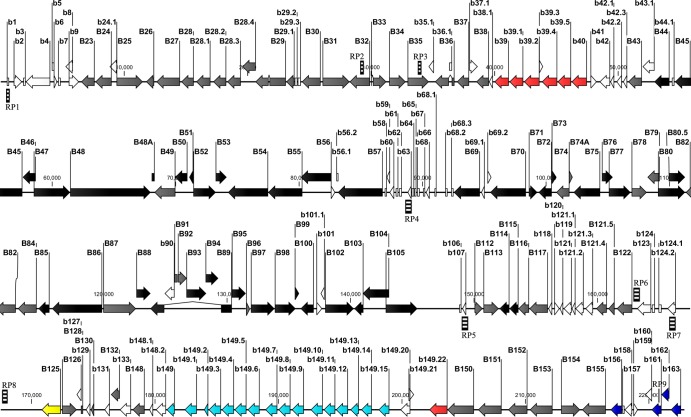
The MsHV genome was analyzed to identify repetitive sequences. No repeats were found at either terminus, but nine regions (RP1 to RP9) were found to contain tandem repeat sequences (Table 2). RP1, located at nucleotides 431 to 734, contains 4 repeat units, 3 of which overlap each other, while RP6, RP7, and RP8 have 2 overlapping units. RP5 has a single unit of 205 bp repeated 2.1 times. Of note is RP4, in the low-coverage area of L1, which includes two physically close units of 13 bp and 21 bp, repeated 4.8 and 6.8 times, respectively. RP4 is located between the genes B57 and B69, the conserved position of the betaherpesvirus origin of lytic replication (OriLyt). Repetitive and GC-rich sequences were found in the THV (2), HCMV (49), RCMV (88), and MCMV (64) genomes at the same location and were also difficult to sequence, as was CyCMV, by both next-generation and Sanger sequencing (48). RP1 to RP9 are annotated in Fig. 4.
Table 2.
Repeat regions in MsHV genomea
| Repeat region | Location (nt) | Consensus unit | Length (nt) | No. of copies | Match (%) | Mismatch (%) | In-del (%) |
|---|---|---|---|---|---|---|---|
| RP1 | 431–472 | GCGGCGGCGATGACGGCGCCG | 21 | 2 | 92.9 | 7.1 | 0 |
| 563–666 | TCGGAGAGAGCGTCATTCCCG | 21 | 4.9 | 89.4 | 12.5 | 1.0 | |
| 635–682 | GCGTCGTTCCAATCGGAGAG | 21 | 2.3 | 93.8 | 2.1 | 8.3 | |
| 668–734 | TCGGAAAAGGCGCCGCTCCCG | 20 | 3.5 | 92.5 | 3.0 | 13.4 | |
| RP2 | 28886–28957 | GCCAACGCCGCC | 12 | 6 | 97.2 | 2.8 | 0 |
| RP3 | 34459–34507 | CAGGCGGGAACGCGACAGAACGGC | 24 | 2 | 91.8 | 6.1 | 2.0 |
| RP4 | 88639–88701 | TAGAAAGCTTATA | 13 | 4.8 | 98.4 | 3.2 | 1.6 |
| 88960–89100 | CCCGGCTCGCCCCGGCGCTCC | 21 | 6.8 | 91.5 | 6.4 | 2.8 | |
| RP5 | 149151–149571 | ATTTCCACACATGGGTCAGATATTATGCCGTATAAGGTCACCTTATGCGATCTTGGACTTCCCCGCGCGGTAGTCATCTGAAAAGTCCAGGCGACCGGCCAATTTCTCTCAACCGTGTCATTTCTTCTAAATTGATGAAGCGTTGGAAAGTACCCGGACCTCTGCCAGTGGAACATTGTCGGTGAGTCACGGTCGACGCAGGGGG | 205 | 2.1 | 97.1 | 1.7 | 1.0 |
| RP6 | 163038–163100 | ATATAAAAGTTGTGGATATACCATCG | 26 | 2.4 | 92.1 | 7.9 | 0 |
| 163364–163441 | ATCGTCATCATCCTCGTCCCCATT | 24 | 3.3 | 92.3 | 7.7 | 0 | |
| RP7 | 165780–166261 | CCCAAGCCTGCCTTCCACCCCAAGCCTGCCTCCCCAAG | 38 | 11.7 | 88.4 | 3.1 | 7.9 |
| 165961–166302 | GCCTGCCTCCCCAAGCCCAAGCCTGCCTTCCACACCAAGCCTGCCTGCCTCACCAAGCCCAAGCCTGCCTTCCACCCCAAGCCCAAGCCT | 90 | 3.8 | 93.9 | 5.0 | 4.7 | |
| RP8 | 167292–167601 | CAGACCTTAAAAGGCCATCCGGAAGGGTGGAATAGGTCAAACGAGTCACATTCCCCCTAACAGATGGGTGAATAAGCCCAGTAAATCCCCCGAATGTCATGGGAAAGGTCCCGCGGGTTGACTCACAGTCCCTGCCCTTCTCAGAAAGGAAGGGTGAGTGGGCCTCC | 167 | 1.8 | 96.5 | 2.6 | 0.6 |
| 167630–167960 | GTTGAGTCACTCCATTGTGCTTTTAAGAAAAAAAAGATAGATGGGACCCTAAGACCTTAAAAGGCCGTCCGGAAAGGACCGGGGGGTCAGACGCGTCTCATCATCACCCAAGAGATGCGTGAATAAGTCCGACAAACCCAACCAAGGTCACGCGAAAAGTCCATCCC | 167 | 2 | 93.1 | 5.1 | 1.5 | |
| RP9 | 220796–220862 | CGGATCGCGGACCGGATCGCGCCT | 24 | 2.8 | 86.6 | 11.9 | 1.5 |
Statistics refer to the percentages of match, mismatch, and insertion and deletion (in-del) between the repeat region and the consensus unit over the length of the repeat region.
Fig 4.
Map of the MsHV genome. ORFs that code for proteins with homology to herpesvirus core genes are depicted as black arrows, and those with homology to proteins of THV, HCMV, or RCMV are shown as gray arrows. Both of these types of ORFs are named using uppercase prefixes. Unique ORFs are named using lowercase prefixes. MHC-related proteins are in red, the b149 family in blue, the C-type lectin family in green, and parvovirus Rep protein homolog in yellow. Zebra-crossing bars represent repeat regions (RP) but not exactly proportional in length to the real sequences.
ORFs of the MsHV genome.
Initial ORF analysis found 658 ORFs of 50 amino acid residues or greater. However, when analyzed by protein BLAST search against a protein data set of fully sequenced and well-annotated betaherpesviruses (a common approach used for herpesvirus gene annotation), 190 ORFs were predicted with high confidence to be true coding ORFs, and they are depicted in the genome map (Fig. 4). MsHV proteins were individually analyzed in comparison with those of THV, HCMV, and RCMV. Of the 190 ORFs, 93 encode proteins that have homologues in THV, HCMV, or RCMV, with 60 having the highest similarity with homologues of THV, 20 with HCMV, and 15 with RCMV (Table 2). All the core genes of herpesviruses were found in the MsHV genome; they are conserved in both position and orientation and are indicated by black arrows in Fig. 4, while other genes conserved in betaherpesviruses are indicated by gray arrows. In general, the core genes share higher identities, from 30 to 60%, than the noncore genes, from 20 to 50%, with other betaherpesvirus' counterparts (P < 0.001).
Analysis of individual genes and gene families. (i) Proteins essential for DNA replication and nucleotide metabolism.
Like other betaherpesviruses (52, 59, 77, 78), MsHV encodes the homologues of the six genes of the DNA replication, including UL44 (DNA polymerase processivity subunit), UL54 (DNA polymerase [DPOL]), UL57 (single-stranded DNA binding protein), and the heterotrimeric helicase-primase consisting of UL70 (primase), UL102 (helicase-primase complex-associated factor), and UL105 (helicase). B44, B54, B57, and B105 are among the genes with highest identities among THV, HCMV and RCMV, ranging from 48% to 60% (Table 1), while B70 and B102, encoding the primase subunit and complex-associated factor, share about 45% and 33% identities, respectively. B98 and B114 encode the homologues of HCMV alkaline nuclease UL98 (74) and uracil DNA glycosylase UL114, both of which influence viral DNA replication (18). The large subunit ribonucleotide reductase homologue (B45) and the dUTPase homologue (B72) share 31% to 26% identity with their counterparts, which for HCMV are enzymatically inactive and dispensable for cell culture (12, 32) but in MCMV function as a cell death suppressor (10). HCMV UL84 (98) and UL112-113 (60) contribute to the initiation of lytic DNA replication, and the latter produces four products by alternative splicing. B112 and B113 were found to have less than 30% identity with UL112 and UL113.
(ii) Regulatory genes.
HCMV UL36 and UL37 (17), UL69 (97), UL122 and UL123 (IE1 and IE2) (59), TRS1/IRS1 (80), and US3 (17) are reported to be immediate-early proteins that play a key role in initiation of viral replication via the regulation of gene expression. UL36 (viral inhibitor of caspase-8 activation [vICA]) and UL37 exon 1 (UL37x1) (viral mitochondrion-localized inhibitor of apoptosis [vMIA]) are two important proteins that inhibit cell death from apoptosis; the former prevents cleavage by binding to the prodomain of procaspase-8 (75), while the latter inhibits apoptosis by binding Bax and sequestering it at the mitochondrial membrane (1). All betaherpesviruses characterized to date carry homologues of UL36, but only primate CMVs retain a complete form of UL37 (UL37x1); MCMV, RCMV, and other betaherpesviruses, including MsHV, lack a homologue of UL37x1. Like THV, MsHV encodes the homologues of UL69 and UL122 but not UL123 or US3. TRS1/IRS1 belong to US22 family; MsHV has two loci at both ends of the genome encoding multiple US22 family members, with those at the right end sharing more similarity with TRS1/IRS1 than those at the left end. A UL82 (pp71) homologue was also found, which is the recently recognized viral transcriptional activator that activates viral immediate-early gene (IE1) expression in HCMV-infected cells through degradation of Daxx (70).
(iii) Structural proteins.
The structure of the HCMV virion has been divided into three components: nucleocapsid, tegument, and envelope. The nucleocapsid is composed of at least five proteins: UL86 (major capsid protein), UL85 (minor capsid protein), UL48A (the smallest capsid protein), UL46 (minor capsid binding protein), and UL80 (assembly protein) (8). As core genes, UL86 and UL85 are conserved with about 60% identity among MsHV, THV, HCMV, and RCMV, but B46 and B80 share less than 40% identity with UL46 and UL80. The homologue of UL48A, which is less than 100 amino acids (aa) in size but comprises 12.6% of virion mass (87), was also identified in MsHV, THV, and RCMV at the congruent position in the genome (2, 89). UL104, the portal protein for viral DNA encapsidation, reportedly interacting with the large terminase subunit UL56 and together with UL89, is involved in the resistance of HCMV to benzimidazole ribonucleosides (23).
An amorphous tegument is located between the virion capsid and envelope and comprises 50% of virion total mass (87). More than 27 HCMV proteins were reported to be tegument proteins, and of these the most abundant are the product of UL83 (pp65, lower matrix protein), with 15% of virion mass, and 9% (each) for UL82 (pp71, upper matrix protein and virion transactivator), UL32 (pp150, large matrix phosphoprotein), and UL48 (largest tegument protein). Although UL83 is the most abundant protein of the virion, it shares very low identity among HCMV, RCMV, and MCMV and was reported to be dispensable for growth in tissue culture (73). Neither MsHV nor THV carries the homologue of UL83. It will be interesting to determine which protein is instead the predominant protein in the virion of both viruses. In addition to their structural functions in virion morphogenesis, some tegument proteins have been shown to regulate viral gene expression or modify host cell responses to HCMV infection (44, 47, 79).
On the HCMV viral envelope, glycoproteins form three complexes: gCI, gCII and gCIII, or gB (glycoprotein B; UL55), gM-gN (UL100 and UL73), and gH-gL-gO (UL75, UL115, and UL74, respectively) (8). A study of the HCMV proteome using mass spectrometry revealed that the abundances of these glycoproteins relative to the total virion protein content are ordered as follows (from greatest to least): UL100 (9.2%), UL55 (1.4%), UL75/UL115 (0.6%/0.5%), UL73 (0.1%),and UL74 (<0.1%). Interestingly, the identities between the counterparts of MsHV and HCMV follow the same order: 52.8% (B100), 46.9% (B55), 34.2%/36.8% (B75/B115), 34% (B73), and 28.3% (B74).
(iv) MHC-related proteins.
Some betaherpesviruses encode multiple MHC class I homologues, such as UL18 and UL142 of HCMV (24) and CCMV (21), m144 of MCMV (64), R144 of RCMV (89), and gp147, gp148, and gp149 of GPCMV (72), and most recently, a rodent gammaherpesvirus, rodent herpesvirus Peru (RHVP), was reported to encode three ORFs, each of which was similar to a different region of the rat MHC class I protein (46). Although the scores are very low when BLASTed in GenBank, MsHV b39.5 is most similar to chimpanzee MHC class I chain-related protein, b39.4 to the MHC class I chain-related protein of crab-eating macaque, b39 and b39.2 to the MHC class II antigens of bovine and crab-eating macaque, respectively, and b39.1 to the hereditary hemochromatosis protein (HFE), which is a nonclassical MHC-related protein that associates with the transferrin receptor and regulates iron metabolism (68). Consistent with known MHC class I molecules, b39, b39.1, b39.2, b39.4, and b39.5 have a signal peptide at the N terminus, a transmembrane domain, and a cytoplasmic tail at the C terminus. Another MsHV protein, b149.22, contains an Ig-like domain profile according to a Prosite search but produces no significant hits in GenBank BLAST search. Hence, it most likely represents a new protein with no homologue in the known betaherpesvirus. MsHV b149.22 was predicted to be a type II transmembrane protein. The functions of these betaherpesvirus proteins were not fully characterized. For example, HCMV UL18 and UL142 and MCMV m144 have been reported to modulate natural killer cells (20, 63, 96), while RCMVΔr144 shows restricted replication in salivary glands and spleen of neonatal rats (3, 40). These proteins have no significant homology to any mammalian proteins (55) and are dispensable for virus replication in fibroblasts (7, 9, 28).
In addition to MHC class I homologues, betaherpesviruses also encode other genes which interfere with the MHC class I antigen-processing pathway, such as the HCMV U2 and U3 family members (45), MCMV m04, m06, and m152 (91), and RhCMV rh178 (67). Here we find that another protein in MsHV, b40, has 25% amino acid sequence identity with HHV-7 U21, which was recently reported to downregulate the MHC class I complex on the cell surface (50).
(v) C-type lectin-like proteins.
At the 3′ proximity of the MsHV genome is a cluster of four ORFs: b156, b161, b162, and b163. Three of them, b156, b161, and b163, contain C-type lectin family domains, while b162 does not. Instead, b162 it is more related to the natural killer cell lectin-like receptors than to C-type lectin proteins. Pairwise comparison showed that b156 and b163 share an identity of 39% whereas b162 and b163 have a sequence identity of 29% (Table 2). A transmembrane domain was predicted at the N-terminal end for all of these C-type lectin homologues, which is consistent with the type II transmembrane structure of C-type lectin proteins (100). The MsHV C-type lectin family shows a 24% sequence identity with its homologue in the English isolate of RCMV, which carries a spliced form of C-type lectin, and its interruption results in no change in virus replication in cell culture (90).
(vi) Parvovirus Rep protein homologue.
RCMV r127 (89) and HHV-6 U94 (83) were reported to be the only herpesvirus homologues of the parvovirus NS1 or Rep protein. Remarkably, situated at the congruent position in the MsHV genome, B125 has 22% and 27% identities with r127 and U94, respectively. The RCMV r127 protein is dispensable for virus replication both in vitro and in vivo (86); however, HHV-6 U94 was shown to inhibit the replication of not only HHV-6A and HHV-6B but HHV-7 and HCMV, suggesting a role in the regulation of replication and latency of human betaherpesviruses (13). The conservation in direction and position of the parvovirus Rep homologues in the genomes of MsHV, RCMV, and HHV-6 indicates a likely common origin.
(vii) The b149 family.
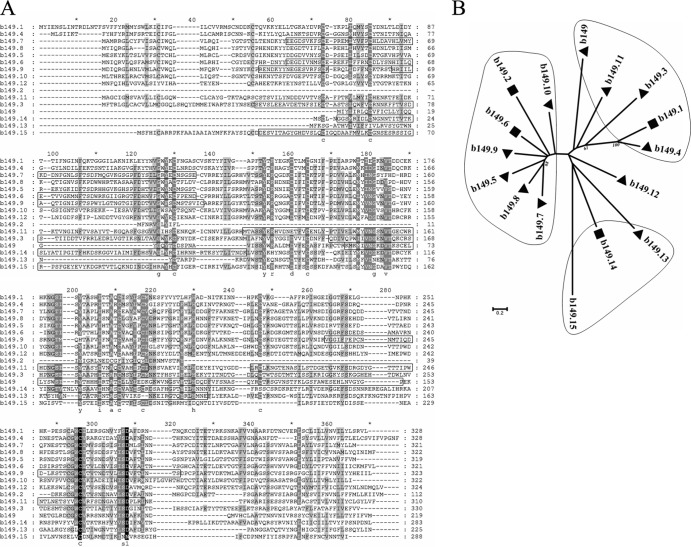
There is an ∼18-kbp region which has significantly lower G+C content than other parts of the MsHV genome. Interestingly, toward the left-hand end of this region, there is a unique gene family of 16 tandemly arranged members (Fig. 4). The proteins encoded by this gene family vary in length from 219 to 330 amino acid residues, except for b139.2, which is only 112 amino acid residues long with a truncated N terminus. Most of them have a potential signal peptide at the N terminus and a transmembrane domain at the C terminus. A sequence alignment revealed more than 10 conserved cysteine residues (Fig. 5A). An initial sequence BLAST search in GenBank resulted in no significant hits; however, 9 of the 16 proteins were predicted to have one or two immunoglobulin-like beta-sandwich domains or other folds related to proteins functioning in the immune system. As shown in Fig. 5A, the predicted domains cover most of the aligned sequence except for the two ends. Phylogenetic analysis indicates that the b149 family forms 4 clades and vicinal members tend to cluster together (Fig. 5B). The sequence identities range from 33% to 48% among the b149.2 clade, 19% to 48% among the b149.1 clade, and 16% to 28% among the b149.13 clade.
Fig 5.
Analysis of the MsHV b149 family. (A) Alignment of the b149 family using ClustalW. The sequences in boxes were predicted to fold like immunoglobulin-like beta-sandwich domains by the software program Phyre2. (B) Neighbor-joining tree of the b149 family, based on the sequence alignment. The tree was reconstructed using MEGA5, with the parameter setting of the Jones-Taylor-Thornton model, gaps/missing-data treatment of pairwise deletion, and 1,000 bootstraps. Bootstrap percentages of more than 50% are shown. Triangles indicate that both the signal peptide at the N terminus and the transmembrane domain at the C terminus were predicted, while rectangles indicate that only the transmembrane domain at the C terminus was predicted.
(viii) Other gene families.
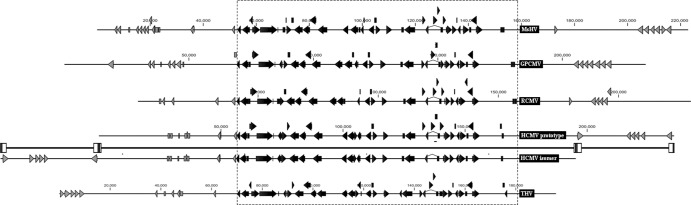
The US22 family is a large gene family present in all betaherpesviruses. Different betaherpesviruses carry different numbers of US22 family members, but the genomic arrangement is similar, with two clusters of tandemly repeated homologues at each end of the genome and some isolated homologues located in between. The exception is the THV genome, where the two clusters of US22 family tandem repeats are both located at the left end (2). The MsHV genome encodes 18 members of the US22 family. Although most of the members have highest sequence identities with those of THV, the distribution of the MsHV US22 family members more resembles that of GPCMV (38) (Fig. 6). B154, which is positioned in a direction opposite to that of its neighboring genes, is an exception. This gene organization has not been observed in any other betaherpesviruses.
Fig 6.
Comparison of the genome arrangement of MsHV, GPCMV, RCMV, HCMV, and THV. The genome maps of MsHV, GPCMV, RCMV, and THV were adjusted in order that their core gene blocks (black arrows in the box) line up with that of HCMV. US22 family genes are shown as gray arrows. The prototype of the HCMV genome and one isomer with a reverse US at the other end are represented. HCMV terminal repeats are shown as blocks on the line between the prototype and the isomer.
MsHV B33 and B78 belong to the G-protein coupled receptor (GCR) family, which is characterized by seven transmembrane domains. While HCMV UL33 is genetically diverse and UL78 is highly conserved among HCMV clinical isolates, both are dispensable for replication in fibroblasts (22, 54). MsHV b132 is another protein with a GCR signature but with only one transmembrane domain instead of seven, and it is not homologous to any known proteins.
There are several predicted proteins in MsHV which have no homologue in GenBank. One of these, protein b43.1, is a protein predicted to span the membrane seven times. Two others, proteins b158 and b160, have 28% identity with each other but have no homologue found in GenBank.
Phylogenetic analysis.
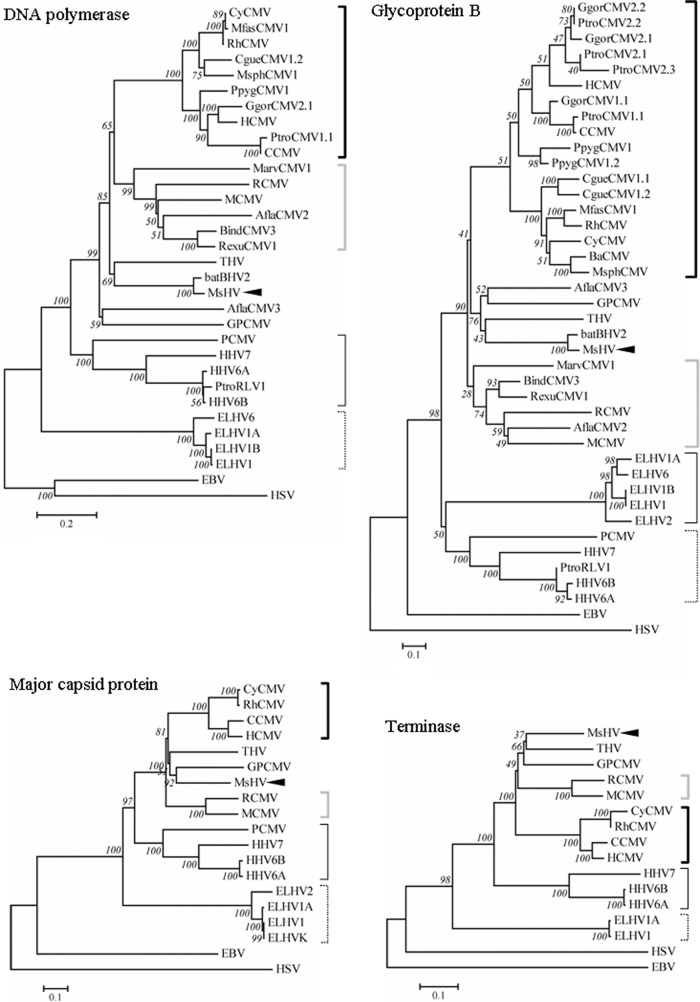
The DPOL and gB sequences of MsHV were compared with those of two other betaherpesviruses from bats, bat BHV-2 (92) and bat BHV-1 (94). As predicted, DPOL and gB of bat BHV-2, which was isolated from Miniopterus fuliginosus, have high sequence identities to their MsHV counterparts, 86% and 91%, respectively. Bat BHV-1 was detected in two relatively distant species, Myotis nattereri and Pipistrellus pipistrellus. It was not included in the phylogenetic analysis because of the limited sequence available: only a 58-aa region of DPOL, with 51% identity to its counterparts in MsHV and bat BHV-2. Phylogenetic trees were constructed based on protein sequences of DPOL, gB, major capsid protein, and terminase, respectively (Fig. 7). The topologies of the four trees are very consistent with the taxonomy of betaherpesviruses, since the trees generally divide into 4 subfamilies, cytomegalovirus, muromegalovirus, roseolovirus, and proboscivirus. As expected, MsHV and bat BHV2 are closely related to THV according to these phylogenetic trees. Interestingly, GPCMV from guinea pig (38) together with Apodemus flavicollis cytomegalovirus 3 (AflaCMV3) from yellow-necked mouse (27), both within the Rodentia, were clustered closer to MsHV and THV than the muromegaloviruses. They form a branch distinct from but related to cytomegalovirus and muromegalovirus. Similarly, porcine cytomegalovirus (PCMV) consistently groups with the roseoloviruses rather than cytomegalovirus (30, 95).
Fig 7.
Phylogenetic analysis of betaherpesviruses. Protein sequences were aligned with ClustalW, and then neighbor-joining phylogenetic trees were reconstructed with MEGA5, with the Jones-Taylor-Thornton model, gaps/missing-data treatment of pairwise deletion, and 1,000 bootstraps. MsHV is pointed out with black arrows; thick black brackets indicate cytomegalovirus, gray ones indicate muromegalovirus, thin black ones indicate roseolovirus, and thin dotted ones indicate proboscivirus; alphaherpesvirus HSV and gammaherpesvirus EBV are used as outgroups.
DISCUSSION
Here we report the isolation and characterization of a new betaherpesvirus from the bat M. schreibersii. Similar to previously described betaherpesviruses, MsHV has a low rate of replication, reaching a peak 2 weeks after infection, which is similar to that of HCMV (61). MsHV has a restricted species tropism in vitro and is able to infect only one cell line of nonbat origin from a total of 14 cell lines representing 10 different animal species. The genome of MsHV was sequenced using 454 next generation sequencing and is 222,870 bp in length.
MsHV was initially identified in lymph node primary cells and then subjected to two rounds of purification and propagation with multiple passages in kidney primary cells. The number of passages was less than 10. Genetic stability of betaherpesviruses varies during in vitro passaging. HCMV and GPCMV have been shown to undergo genetic alterations, including gene loss (24, 38), while MCMV was reported to be stable (15). The genetic stability of MsHV is unknown. Given that our approach to identifying MsHV genes is based on finding homologues with the genes of other betaherpesviruses and the common criteria used for unique gene prediction, we acknowledge that data presented in this paper may not be a complete representation of MsHV genome content and that some of the predicted ORFs, especially those with splicing, may need revision in the future when we have transcription data to help better annotation.
Many herpesviruses have terminal repeat (TR) sequences, and according to the presence and location, herpesvirus genomes can be divided into six groups (61). The pattern of TR sequences in betaherpesviruses varies significantly. HCMV and CCMV have large TR at both termini and the junction of UL and US (21); RCMV (89) and MCMV (64) have 504-bp and 31-bp TR, respectively, and GPCMV has TR at the left terminus only (72). A number of herpesviruses have no TR, including THV, RhCMV, CyCMV (48), and the gammaherpesvirus RHVP (46). MsHV also has no TR. We cannot exclude the possibility that only one copy of the TR unit was incorporated into the assembled genome, but taking the 428-bp average length of the sequencing reads into consideration, we think it is unlikely. MsHV, RCMV, MCMV, GPCMV, RhCMV, and CyCMV share a similar genome organization, which is colinear with the prototype isomer of the HCMV genome, while the THV genome is colinear with another isomer of the HCMV genome because its US22 family locus reversely translocates from one end to the other (Fig. 6). So far, of all the betaherpesvirus genomes sequenced, only HCMV and CCMV have a large TR and form four genomic isomers; none of the others, including RhCMV and CyCMV from primates in the Cercopithecidae, share these genomic features. The emergence of large TR and genomic isomers may have occurred very recently in betaherpesvirus evolutionary history, but this hypothesis needs to be validated with more genome sequences from different species.
One of the most significant findings in this study is that MsHV encodes multiple families of gene homologues implicated in immunity. One family encodes MHC class I homologues. Virus-encoded MHC class I homologues have previously been identified in some betaherpesviruses (21, 64, 72, 89) and a gammaherpesvirus RHVP (46, 99). HCMV evades T-cell recognition by downregulating expression of MHC class I molecules on the surface of infected cells, an effect mediated by the products of the virus-encoded genes US2, US3, US6, and US11 (85). Although MsHV doesn't carry any homologues of US2, US3, US6, or US11, MsHV b40 is a homologue of HHV-7 U21, which downregulates classical and nonclassical MHC class I complexes from the cell surface (50). The downregulation of endogenous MHC class I would expose infected cells to attack by natural kill (NK) cells.
MsHV also encodes two MHC class II homologues and one homologue of the HFE protein which is closely related to MHC class II. To our knowledge, virus-encoded MHC class II homologues have not previously been reported. However, HCMC US2 and US3 not only inhibit the MHC class I antigen presentation pathway, which allows infected cells to evade recognition by CD8+ T lymphocytes, but also inhibit presentation of exogenous protein antigens to CD4+ T lymphocytes, which is part of the MHC class II antigen presentation pathway (84). Other viruses were also reported to target the MHC class II processing pathway for immune evasion (34). Epstein-Barr virus, a gammaherpesvirus, downregulates MHC class II expression during its reactivation (42). Future functional studies of MsHV MHC class II homologues are required to fully understand the significance of the roles they may play in potentially novel virus immune evasion pathways.
The term C-type lectin was originally introduced to distinguish between Ca2+-dependent and Ca2+-independent carbohydrate-binding lectins, but the C-type lectin superfamily now includes a large number of proteins that are homologous to the C type but don't bind carbohydrates (100). Recent studies have identified C-type lectins as an important family of pattern recognition receptors (PRRs) that are involved in the induction of specific gene expression profiles in response to specific pathogens (29). The human NK receptor complex includes a group of type II transmembrane proteins resembling C-type lectins, for which there is accumulating evidence to support crucial roles in the innate immune system (36, 69). MsHV b156, b161, b162, and b163 are related to C-type lectin or natural killer cell lectin-like receptors. The English strain of RCMV was reported to express a C-type lectin protein with high similarity with host C-type lectins (90), but a genome-wide comparative analysis between this virus and MsHV was not possible because of the limited sequence available to RCMV. C-type lectin homologues have also been found in the genomes of other viruses, including gammaherpesviruses (46, 57), poxviruses (6, 26, 81), and African swine fever virus (35). While some of them are dispensable or play some role in virus infection, the EBV C-type lectin homologue gp42 was reported to bind to the MHC class II receptor HLA-DR1 (57).
A gene family of 16 members, the b149 family was identified at the 3′ end of the MsHV genome and before the US22 family. The b149 family gene products have no significant homologues in GenBank, but three-dimensional (3D) structure predication analysis indicated they all contain immunoglobulin-like beta-sandwich folds. Interestingly, in the MCMV genome, the m145 family, a unique gene family of 12 ORFs, was predicted with MHC class I-like folds with a confidence of 70% to 90% (76). Remarkably, in both the b149 and m145 families, the members located in the center of the gene cluster are more conserved with each other (66). As reviewed by Revilleza et al. (66), the m145 family plays an important role in disrupting NK-cell recognition. For example, m138, m145, m152, and m155 downregulate different ligands of NKG2D, and m157 binds both the inhibitory NK receptor Ly49I and the activating receptor Ly49H. And interestingly, the surface molecules of NK cells, NKG2D, Ly49I, and Ly49H, belong to the C-type lectin family. The prediction confidence of the b149 family is low, ranging from less than 10% to 35%, which could be due to the fact that there is little information available in the public database for bat immune molecules. If we can experimentally prove their functionality in immune evasion, these molecules may represent an important new class of immune modulators yet to be fully characterized in other mammalian viruses.
ACKNOWLEDGMENTS
We thank Jackie Pallister and Chris Cowled for critical readings of the manuscript, Chris Cowled, Michelle Baker, and Justin Ng for useful discussions about MHC and other immune-relevant molecules, and Hume Field, Craig Smith, and Carol De Jong for providing bats in the original primary cell line work. The technical help from Kaylene Selleck and the AAHL sequencing group is highly appreciated.
This study is supported in part by an NHMRC Australia-China Postdoctoral Exchange Fellowship (to H.Z.) and an OCE Science Leader Award from the CSIRO Office of the Chief Executive (to L.-F.W.).
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Andoniou CE, Degli-Esposti MA. 2006. Insights into the mechanisms of CMV-mediated interference with cellular apoptosis. Immunol. Cell Biol. 84:99–106 [DOI] [PubMed] [Google Scholar]
- 2. Bahr U, Darai G. 2001. Analysis and characterization of the complete genome of tupaia (tree shrew) herpesvirus. J. Virol. 75:4854–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beisser PS, et al. 2000. The r144 major histocompatibility complex class I-like gene of rat cytomegalovirus is dispensable for both acute and long-term infection in the immunocompromised host. J. Virol. 74:1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acid Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blasco R, Sisler JR, Moss B. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 67:3319–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bradley AJ, et al. 2009. High-throughput sequence analysis of variants of human cytomegalovirus strains Towne and AD169. J. Gen. Virol. 90:2375–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Britt WJ, Boppana S. 2004. Human cytomegalovirus virion proteins. Hum. Immunol. 65:395–402 [DOI] [PubMed] [Google Scholar]
- 9. Browne H, Churcher M, Minson T. 1992. Construction and characterization of a human cytomegalovirus mutant with the UL18 (class I homolog) gene deleted. J. Virol. 66:6784–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brune W, Menard C, Heesemann J, Koszinowski UH. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303–305 [DOI] [PubMed] [Google Scholar]
- 11. Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19:531–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caposio P, Riera L, Hahn G, Landolfo S, Gribaudo G. 2004. Evidence that the human cytomegalovirus 46-kDa UL72 protein is not an active dUTPase but a late protein dispensable for replication in fibroblasts. Virology 325:264–276 [DOI] [PubMed] [Google Scholar]
- 13. Caselli E, et al. 2006. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology 346:402–414 [DOI] [PubMed] [Google Scholar]
- 14. Chee MS, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169 [DOI] [PubMed] [Google Scholar]
- 15. Cheng TP, Valentine MC, Gao J, Pingel JT, Yokoyama WM. 2010. Stability of murine cytomegalovirus genome after in vitro and in vivo passage. J. Virol. 84:2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chua KB, et al. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435 [DOI] [PubMed] [Google Scholar]
- 17. Colberg-Poley AM, Santomenna LD, Harlow PP, Benfield PA, Tenney DJ. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 66:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Courcelle CT, Courcelle J, Prichard MN, Mocarski ES. 2001. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J. Virol. 75:7592–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crameri G, et al. 2009. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One 4:e8266 doi:10.1371/journal.pone.0008266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cretney E, et al. 1999. m144, a murine cytomegalovirus (MCMV)-encoded major histocompatibility complex class I homologue, confers tumor resistance to natural killer cell-mediated rejection. J. Exp. Med. 190:435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davison AJ, et al. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17–28 [DOI] [PubMed] [Google Scholar]
- 22. Deckers M, et al. 2009. High genotypic diversity and a novel variant of human cytomegalovirus revealed by combined UL33/UL55 genotyping with broad-range PCR. Virol. J. 6:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dittmer A, Drach JC, Townsend LB, Fischer A, Bogner E. 2005. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides. J. Virol. 79:14660–14667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dolan A, et al. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301–1312 [DOI] [PubMed] [Google Scholar]
- 25. Dominguez G, et al. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duncan SA, Smith GL. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 66:1610–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ehlers B, et al. 2007. Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J. Virol. 81:8091–8100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Farrell HE, et al. 1997. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature 386:510–514 [DOI] [PubMed] [Google Scholar]
- 29. Geijtenbeek TB, Gringhuis SI. 2009. Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 9:465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goltz M, Widen F, Banks M, Belak S, Ehlers B. 2000. Characterization of the DNA polymerase loci of porcine cytomegaloviruses from diverse geographic origins. Virus Genes 21:249–255 [DOI] [PubMed] [Google Scholar]
- 31. Gompels UA, et al. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29–51 [DOI] [PubMed] [Google Scholar]
- 32. Hahn G, et al. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551–9555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J. Virol. 77:6620–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hegde NR, Chevalier MS, Johnson DC. 2003. Viral inhibition of MHC class II antigen presentation. Trends Immunol. 24:278–285 [DOI] [PubMed] [Google Scholar]
- 35. Hurtado C, et al. 2004. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology 326:160–170 [DOI] [PubMed] [Google Scholar]
- 36. Huysamen C, Brown GD. 2009. The fungal pattern recognition receptor, Dectin-1, and the associated cluster of C-type lectin-like receptors. FEMS Microbiol. Lett. 290:121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kall L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027–1036 [DOI] [PubMed] [Google Scholar]
- 38. Kanai K, et al. 2011. Re-evaluation of the genome sequence of guinea pig cytomegalovirus. J. Gen. Virol. 92:1005–1020 [DOI] [PubMed] [Google Scholar]
- 39. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 40. Kloover JS, Grauls GE, Blok MJ, Vink C, Bruggeman CA. 2002. A rat cytomegalovirus strain with a disruption of the r144 MHC class I-like gene is attenuated in the acute phase of infection in neonatal rats. Arch. Virol. 147:813–824 [DOI] [PubMed] [Google Scholar]
- 41. Leroy EM, et al. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576 [DOI] [PubMed] [Google Scholar]
- 42. Li D, et al. 2009. Down-regulation of MHC class II expression through inhibition of CIITA transcription by lytic transactivator Zta during Epstein-Barr virus reactivation. J. Immunol. 182:1799–1809 [DOI] [PubMed] [Google Scholar]
- 43. Li W, et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 44. Lischka P, Rauh C, Mueller R, Stamminger T. 2006. Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm. J. Virol. 80:10274–10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loenen WA, Bruggeman CA, Wiertz EJ. 2001. Immune evasion by human cytomegalovirus: lessons in immunology and cell biology. Semin. Immunol. 13:41–49 [DOI] [PubMed] [Google Scholar]
- 46. Loh J, et al. 2011. Identification and sequencing of a novel rodent gammaherpesvirus that establishes acute and latent infection in laboratory mice. J. Virol. 85:2642–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu M, Shenk T. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marsh AK, et al. 2011. Genomic sequencing and characterization of cynomolgus macaque cytomegalovirus. J. Virol. 85:12995–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Masse MJ, Karlin S, Schachtel GA, Mocarski ES. 1992. Human cytomegalovirus origin of DNA replication (oriLyt) resides within a highly complex repetitive region. Proc. Natl. Acad. Sci. U. S. A. 89:5246–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. May NA, Glosson NL, Hudson AW. 2010. Human herpesvirus 7 u21 downregulates classical and nonclassical class I major histocompatibility complex molecules from the cell surface. J. Virol. 84:3738–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McGeoch DJ, Rixon FJ, Davison AJ. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90–104 [DOI] [PubMed] [Google Scholar]
- 52. McMahon TP, Anders DG. 2002. Interactions between human cytomegalovirus helicase-primase proteins. Virus Res. 86:39–52 [DOI] [PubMed] [Google Scholar]
- 53. Megaw AG, Rapaport D, Avidor B, Frenkel N, Davison AJ. 1998. The DNA sequence of the RK strain of human herpesvirus 7. Virology 244:119–132 [DOI] [PubMed] [Google Scholar]
- 54. Michel D, et al. 2005. The human cytomegalovirus UL78 gene is highly conserved among clinical isolates, but is dispensable for replication in fibroblasts and a renal artery organ-culture system. J. Gen. Virol. 86:297–306 [DOI] [PubMed] [Google Scholar]
- 55. Mocarski ES, Jr, Shenk T, Pass RF. 2006. Cytomegaloviruses, p 2703–2773 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 56. Molnar V, et al. 2008. Detection of a novel bat gammaherpesvirus in Hungary. Acta Vet. Hung. 56:529–538 [DOI] [PubMed] [Google Scholar]
- 57. Mullen MM, Haan KM, Longnecker R, Jardetzky TS. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375–385 [DOI] [PubMed] [Google Scholar]
- 58. Murray K, et al. 1995. A novel morbillivirus pneumonia of horses and its transmission to humans. Emerg. Infect. Dis. 1:31–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pari GS, Anders DG. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park MY, et al. 2006. Interactions among four proteins encoded by the human cytomegalovirus UL112-113 region regulate their intranuclear targeting and the recruitment of UL44 to prereplication foci. J. Virol. 80:2718–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pellett PE, Roizman B. 2006. The family Herpesviridae: a brief introduction, p 2703–2773 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 62. Pellett PE, Roizman B. 2006. The family Herpesviridae: a brief introduction, p 2480–2502 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 63. Prod'homme V, et al. 2007. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1− NK cells. J. Immunol. 178:4473–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rawlinson WD, Farrell HE, Barrell BG. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Razafindratsimandresy R, et al. 2009. Partial molecular characterization of alphaherpesviruses isolated from tropical bats. J. Gen. Virol. 90:44–47 [DOI] [PubMed] [Google Scholar]
- 66. Revilleza MJ, et al. 2011. How the virus outsmarts the host: function and structure of cytomegalovirus MHC-I-like molecules in the evasion of natural killer cell surveillance. J. Biomed. Biotechnol. 2011:724607 doi:10.1155/2011/724607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Richards R, Scholz I, Powers C, Skach WR, Fruh K. 2011. The cytoplasmic domain of rhesus cytomegalovirus Rh178 interrupts translation of major histocompatibility class I leader peptide-containing proteins prior to translocation. J. Virol. 85:8766–8776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roy CN, Penny DM, Feder JN, Enns CA. 1999. The hereditary hemochromatosis protein, HFE, specifically regulates transferrin-mediated iron uptake in HeLa cells. J. Biol. Chem. 274:9022–9028 [DOI] [PubMed] [Google Scholar]
- 69. Ryan JC, Naper C, Hayashi S, Daws MR. 2001. Physiologic functions of activating natural killer (NK) complex-encoded receptors on NK cells. Immunol. Rev. 181:126–137 [DOI] [PubMed] [Google Scholar]
- 70. Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 72. Schleiss MR, et al. 2008. Analysis of the nucleotide sequence of the guinea pig cytomegalovirus (GPCMV) genome. Virol. J. 5:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schmolke S, Kern HF, Drescher P, Jahn G, Plachter B. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sheaffer AK, Weinheimer SP, Tenney DJ. 1997. The human cytomegalovirus UL98 gene encodes the conserved herpesvirus alkaline nuclease. J. Gen. Virol. 78(Pt 11):2953–2961 [DOI] [PubMed] [Google Scholar]
- 75. Skaletskaya A, et al. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U. S. A. 98:7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Smith HR, et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. U. S. A. 99:8826–8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smith JA, Jairath S, Crute JJ, Pari GS. 1996. Characterization of the human cytomegalovirus UL105 gene and identification of the putative helicase protein. Virology 220:251–255 [DOI] [PubMed] [Google Scholar]
- 78. Smith JA, Pari GS. 1995. Human cytomegalovirus UL102 gene. J. Virol. 69:1734–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stamminger T, et al. 2002. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J. Virol. 76:4836–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stasiak PC, Mocarski ES. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Su HP, Singh K, Gittis AG, Garboczi DN. 2010. The structure of the poxvirus A33 protein reveals a dimer of unique C-type lectin-like domains. J. Virol. 84:2502–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2713–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thomson BJ, Efstathiou S, Honess RW. 1991. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nature 351:78–80 [DOI] [PubMed] [Google Scholar]
- 84. Tomazin R, et al. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039–1043 [DOI] [PubMed] [Google Scholar]
- 85. Tortorella D, Gewurz B, Schust D, Furman M, Ploegh H. 2000. Down-regulation of MHC class I antigen presentation by HCMV; lessons for tumor immunology. Immunol. Invest. 29:97–100 [DOI] [PubMed] [Google Scholar]
- 86. van Cleef KW, et al. 2004. The rat cytomegalovirus homologue of parvoviral rep genes, r127, encodes a nuclear protein with single- and double-stranded DNA-binding activity that is dispensable for virus replication. J. Gen. Virol. 85:2001–2013 [DOI] [PubMed] [Google Scholar]
- 87. Varnum SM, et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vink C, Beuken E, Bruggeman CA. 1997. Cloning and functional characterization of the origin of lytic-phase DNA replication of rat cytomegalovirus. J. Gen. Virol. 78(Pt 11):2963–2973 [DOI] [PubMed] [Google Scholar]
- 89. Vink C, Beuken E, Bruggeman CA. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Voigt S, Sandford GR, Ding L, Burns WH. 2001. Identification and characterization of a spliced C-type lectin-like gene encoded by rat cytomegalovirus. J. Virol. 75:603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wagner M, Gutermann A, Podlech J, Reddehase MJ, Koszinowski UH. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Watanabe S, et al. 2010. Novel betaherpesvirus in bats. Emerg. Infect. Dis. 16:986–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Watanabe S, et al. 2009. Detection of a new bat gammaherpesvirus in the Philippines. Virus Genes 39:90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wibbelt G, et al. 2007. Discovery of herpesviruses in bats. J. Gen. Virol. 88:2651–2655 [DOI] [PubMed] [Google Scholar]
- 95. Widen F, et al. 2001. Identification and sequence analysis of the glycoprotein B gene of porcine cytomegalovirus. Virus Genes 23:339–346 [DOI] [PubMed] [Google Scholar]
- 96. Wills MR, et al. 2005. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 175:7457–7465 [DOI] [PubMed] [Google Scholar]
- 97. Winkler M, Rice SA, Stamminger T. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xu Y, Cei SA, Rodriguez Huete A, Colletti KS, Pari GS. 2004. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J. Virol. 78:11664–11677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yang Z, West AP, Jr, Bjorkman PJ. 2009. Crystal structure of TNFalpha complexed with a poxvirus MHC-related TNF binding protein. Nat. Struct. Mol. Biol. 16:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zelensky AN, Gready JE. 2005. The C-type lectin-like domain superfamily. FEBS J. 272:6179–6217 [DOI] [PubMed] [Google Scholar]