Abstract
BGLF4 of Epstein-Barr virus (EBV) encodes a serine/threonine protein kinase that phosphorylates multiple viral and cellular substrates to optimize the cellular environment for viral DNA replication and the nuclear egress of viral nucleocapsids. BGLF4 is expressed predominantly in the nucleus at early and late stages of virus replication, while a small portion of BGLF4 is distributed in the cytoplasm at the late stage of virus replication and packaged into the virion. Here, we analyzed systematically the functional domains crucial for nuclear localization of BGLF4 and found that both the N and C termini play important modulating roles. Analysis of amino acid substitution mutants revealed that the C terminus of BGLF4 does not contain a conventional nuclear localization signal (NLS). Additionally, deletion of the C-terminal putative helical regions at amino acids 386 to 393 and 410 to 419 diminished the nuclear translocation of BGLF4, indicating that the secondary structure of the C terminus is important for the localization of BGLF4. The green fluorescent protein-fused wild-type or C-terminal helical regions of BGLF4 associate with phenylalanine/glycine repeat-containing nucleoporins (Nups) in nuclear envelope fractionation. Both coimmunoprecipitation and in vitro pull-down assays further demonstrated that BGLF4 binds to Nup62 and Nup153. Remarkably, nuclear import assay with permeabilized HeLa cells demonstrated that BGLF4 translocated into nucleus independent of cytosolic factors. Data presented here suggest that BGLF4 employs a novel mechanism through direct interactions with nucleoporins for its nuclear targeting.
INTRODUCTION
Epstein-Barr virus (EBV) is a ubiquitous gammaherpesvirus that infects most of the human population worldwide (64). Upon stimulation of various kinds, the latent virus may be reactivated through expression of the viral immediate-early transactivators Zta and Rta, which then turn on the expression cascade of viral genes (39, 41). BGLF4 kinase is a virion-associated serine/threonine kinase expressed during the early and late stages of the lytic cycle (55). Several viral proteins expressed at different stages of virus replication have been shown to be phosphorylated by BGLF4, including EBNA2, EBNA-LP, BMRF1, BZLF1, viral DNA replication proteins, and structural components (3, 26, 27, 61, 65, 66). In cells replicating EBV, BGLF4 colocalizes with the viral DNA polymerase processivity factor BMRF1 in the viral DNA replication compartment and phosphorylates BMRF1 at multiple sites in vitro and in vivo (55, 61). BGLF4 phosphorylates the cellular replication origin binding complex MCM4-MCM6-MCM7, leading to inhibition of its helicase activities (31). Our study also indicated that BGLF4 recruits the cellular nucleotide excision repair protein XPC to the viral replication compartment to enhance viral DNA replication (40). Expression of BGLF4 alone in transfected cells induces premature chromosome condensation through the activation of condensin and topoisomerase II (33). Most importantly, small interfering RNA experiments demonstrated that BGLF4 regulates the nuclear egress of viral nucleocapsid through phosphorylation and disassembly of nuclear lamina (16, 35). BGLF4 also downregulates beta interferon production through phosphorylation and inhibition of interferon regulatory factor 3 (54).
BGLF4 is a member of the conserved herpesviral protein kinases (CHPKs) that have been identified in all human herpesviruses (7). CHPKs not only modulate multiple viral factors but also regulate the cellular machinery in the process of primary infection, nuclear egress, and tegumentation. Nevertheless, individual viral kinases display unique features, in addition to their conserved function, in the viral replication process (15, 34). Because BGLF4 and other CHPKs phosphorylate several proteins at cyclin-dependent kinase 1 (CDK1)-targeted sites, they are believed to at least in part function through a CDK mimicry mechanism (28, 33, 38). Unlike CDKs, whose nuclear translocations are regulated by cell cycle-specific cyclins, BGLF4 localizes predominantly in the nuclei of transiently transfected or virus-replicating cells, regardless of the cell cycle phase (14, 33).
The transport of material between the cytoplasm and nucleus of eukaryotic cells is controlled via channel-like nuclear pore complexes (NPCs) embedded in the nuclear envelope. The NPC is assembled from multiple copies of about 30 different nucleoporins (Nups) (10). Hydrophobic interactions between phenylalanine/glycine (FG) repeats on the NPC and nuclear transport receptors (NTRs) are critical for the translocation step of nuclear transport through NPC. Transport of most macromolecules (those with molecular masses of >40 kDa) into the nucleus is controlled by so-called nucleus-targeting sequences or nuclear localization signals (NLSs) that are recognized by specific members of the importin receptor family. The proteins containing a bipartite motif or a stretch of basic amino acids (classical or conventional NLSs) bind to a heterodimeric receptor complex composed of importin-α and -β (52). For example, the NLS of simian virus 40 (SV40) large T antigen is PKKKRK, and the NLS of nucleoplasmin consists of two clusters of positively charged residues with a space of 10 amino acids (aa) between them, KR-10 aa-KKKL (53). In the transportation process, importin-α binds to the NLS-bearing protein and importin-β mediates the binding of the transport complex to the NPC (18, 57). The importin-bound protein complexes can pass through the NPC by interacting with specific FG repeat-containing nucleoporins (FG-Nups). The small GTPase Ran regulates the association and dissociation of the importin-cargo complex between GTP- and GDP-bound forms (53, 58).
The precise nucleus-targeting mechanism remains unclear for BGLF4 and other herpesviral homologs. For example, a putative NLS was mapped in a green fluorescent protein (GFP)-UL97 fusion protein at aa 48 to 110 by immunofluorescence analysis (46). However, it was recently reported that human cytomegalovirus UL97 can be translated from two alternative ATGs, which results in isoforms of 90 kDa and 100 kDa (56). In a GFP–β-galactosidase (β-Gal) fusion cassette, the N-terminal region from aa 6 to 35 is a functional bipartite NLS, while aa 164 to 198 and 190 to 213 are required to coordinate for optimal nuclear translocation. The small isoform containing aa 74 to 707 showed nuclear/nucleocytoplasmic staining, coupling with up to 15% cytoplasmic localization. Whether the small UL97 isoform translocates to the nucleus through interaction with other molecules is not clear (56). In previous studies, truncation of the C-terminal BGLF4 beyond the conserved kinase domain changed its distribution to the cytoplasm (14, 17). A basic motif RSLKKRFK at aa 386 to 393 was suggested to be the NLS of BGLF4, but its precise contribution was not fully explored. Recently, two SUMO binding motifs (SIMs) were identified in BGLF4 at aa 36 to 40 and 344 to 350. Both SIMs are crucial for BGLF4 function in many aspects, including its proper nuclear localization. A nuclear export signal (NES) at aa 342 to 359 overlapping with the second SIM was identified, suggesting that the nuclear export of BGLF4 may be regulated by its SUMO-interacting abilities (37). Thus, the subcellular distribution of herpesviral kinases may be more complicated than previously thought.
In this study, we aimed to identity the functional domains and the mechanism involved in the nuclear localization of BGLF4. By analyzing mutants with various deletion and point mutations, we show that aa 386 to 393 and multiple helical regions are important for the nuclear targeting of BGLF4. The helix domains of BGLF4 interact with FG repeat-containing nucleoporins in coimmunoprecipitation and glutathione S-transferase (GST)–BGLF4 pull-down assays, suggesting a novel nucleus-targeting mechanism of BGLF4.
MATERIALS AND METHODS
Cell culture and transfection.
The HeLa cell line was derived from a human cervical carcinoma. The NA cell line was derived from NPC-TW01 infected with recombinant Akata EBV (6). All cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C with 5% CO2. DNA transfection was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For EBV lytic cycle induction, NA cells were treated with 40 ng/ml 12-O-tetradecanoylphorbol-13-acetate (TPA; Sigma) and 3 mM sodium butyrate (Sigma) for the indicated times.
Plasmids.
Plasmids expressing wild-type (WT) BGLF4 kinase (pYPW17), kinase-dead mutant K102I (pYPW20), and BMRF1 (pYPW26) were generated in the background of the pSG5 vector as described previously (55). GFP-BGLF4-expressing plasmid (pCPL4) was generated in the pEGFP-C1 vector in a previous study (33). All mutants with deletion and point mutations of BGLF4 or GFP-BGLF4 were generated using a single-primer mutagenesis protocol (42). All the plasmid DNA templates and primers used for generating BGLF4 mutants are listed elsewhere (data not shown; https://www.space.ntu.edu.tw/navigate/share/GLQQYJUGVN). N-terminal deletion mutants d(1-26) (a mutant with a deletion from aa 1 to 26), d(1-70), and d(27-70), C-terminal deletion mutants d(294-429) and d(327-429), putative NLS-deletion and point mutation mutants d(386-393), R386A, K393A, and 3A, C-terminal helix deletion mutants d(290-300), d(303-313), d(378-389), and d(410-419), mutants with point mutations in the helix region (S300A, Q299P, and Q299A), and mutants with kinase domain mutations D195A, N200K, and D219E were generated using pYPW17 as the template. The templates for mutants with putative NLS point mutations 2A and 5A were pHYH8 (K393A) and pHYH10 (3A), respectively. For generating kinase domain double mutation mutants D195A/D219E and D195A/N200K, pJTW34 (D195A) was used as the template, and for N200K/D219E, pJTW36 (N200K) was used as the template. In addition, the spontaneous S300P mutant was identified in a PCR cloning for wild-type pSG5-BGLF4 using pCF4 (8) as the template. pCPL4(GFP-BGLF4) was used to generate a GFP-BGLF4 mutant truncated from aa 289 to 429 [GFP-B(289-429)] and GFP-B(369-429). For nuclear import assay, the yeast expression plasmid of GST-BGLF4 (pEG-BGLF4) was a gift from Diane S. Hayward (Johns Hopkins University School of Medicine). pEG-K102I (GST-K102I) was generated using a single-primer mutagenesis protocol with pEG-BGLF4 as the template (data not shown, https://www.space.ntu.edu.tw/navigate/share/GLQQYJUGVN). To generate pEG-LazZ-NLS (GST-LacZ-NLS), a SalI-BglII DNA fragment containing LacZ-NLS was digested from pEYFP-LacZ-NLS (kindly provided by Mitsuhiro Kawata, Kyoto Prefectural University of Medicine, Japan [25]) and cloned into the SalI and BamHI sites of yeast expression vector pEG(KT) (47). For the GST pull-down assay, recombinant GST-BGLF4 and GST-K102I expression plasmids were generated in an Escherichia coli expression vector, pGEX3X (40). pGEX-2T-HA-PTAC97 (GST–importin-β) was a gift from Y. Yoneda (Osaka University, Japan) (22). For in vitro transcription-coupled translation, Nup62 and Nup153 expression plasmids (pCWC13 and pCWC14) were generated by cloning the PCR-amplified human Nup62 cDNA (BC003663; a gift from J. Lu, Academia Sinica, Taiwan) or Nup153 cDNA (BC052965; Origene) into the XhoI site of the pCDNA3.1(+) vector (Invitrogen).
Antibodies and immunoblotting.
After transfection, cells were lysed in NP-40 lysis buffer (1% NP-40, 50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1 mM Na3VO4), resolved by electrophoresis in SDS-polyacrylamide gels, and transferred onto Hybond-C membranes (Amersham). The blots were blocked with washing buffer (100 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.2% Tween 20) containing 5% skim milk at room temperature (RT) for 1 h and incubated with primary antibodies at 4°C overnight. After washing three times with TBST (10 mM Tris-HCl, pH 7.4, 24 mM NaCl, 0.2% Tween 20), the blots were incubated with horseradish peroxidase-conjugated secondary antibodies at RT for 1 h. Finally, the blots were developed using enhanced chemiluminescence (Amersham) and exposed to X-ray films. For immunoblotting, BGLF4 monoclonal antibodies (MAbs) 2616 and 2224, recognizing epitopes within aa 327 to 429 and 27 to 70, respectively, were used (55). Other primary antibodies used were rabbit BGLF4 antiserum (55), β-actin (Sigma), poly-ADP-ribose polymerase (PARP; clone F-2; Santa Cruz), α-tubulin (Calbiochem), anti-lamin A/C (Santa Cruz), and anti-GFP (Clontech). To detect the nuclear pore complex, mAb414 (Abcam) was used to recognize the nucleoporins that contain conserved FXFG repeats, including Nup62, Nup153, Nup214, and Nup358 (11).
Immunofluorescence assay.
To detect the distribution of BGLF4 in EBV-replicating cells, slide-cultured NA cells were treated with 40 ng/ml TPA and 3 mM sodium butyrate for the indicated times. Slide-cultured HeLa cells were transfected with WT, kinase-dead (K102I), or BGLF4 mutant-expressing plasmids. At 24 h posttransfection, slides were fixed with methanol at −20°C for β-actin or 4% paraformaldehyde for NPC staining at RT for 20 min, washed with phosphate-buffered saline (PBS; 145 mM NaCl, 1.56 mM Na2HPO4, 1 mM KH2PO4, pH 7.2) at RT for 5 min, and permeabilized with 0.1% TNX-100 at RT for 5 min. BGLF4 was stained with rabbit BGLF4 antiserum. For NPC staining, cells were incubated with MAb 414 (Abcam) at RT for 1.5 h, washed with PBS, and incubated with rhodamine-conjugated antimouse antibody for another 1 h. DNA was stained with Hoechst 33258 at RT for 30 s and covered with mounting medium (H-1000; Vector).
Subcellular fractionation.
HeLa cells (1 × 107) were transfected with plasmid-expressing wild-type or mutant BGLF4. At 24 h posttransfection, cells were harvested and treated with CE buffer (10 mM HEPES, 60 mM KCl, 1 mM EDTA, 0.075% NP-40, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], pH 7.2) on ice for 3 min. After centrifugation at 200 × g for 5 min, the supernatant was harvested as the cytosolic fraction. The pellet, referred to as the nuclear fraction, was then washed twice with NE buffer (CE buffer without NP-40) at 4°C and resuspended in sample lysis buffer (200 mM Tris-HCl, pH 6.8, 8% SDS, 0.4% bromphenol blue, 40% glycerol, 400 mM DTT). The nuclear fraction was sonicated before it was applied to SDS-polyacrylamide gels for immunoblotting analysis. PARP and α-tubulin were detected as nuclear and cytosolic markers, respectively (4).
Nuclear envelope fractionation.
Nuclear envelope fractionation was performed as described previously (30, 45), with slight modification. For isolation of nuclei, HeLa cells (3 × 106) transfected with BGLF4, K102I, or vector were incubated with 700 μl hypotonic buffer (5 mM Tris-HCl, pH 7.4, 5 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 1 mM PMSF) on ice for 1 h, harvested by scraping, homogenized by passage through 27-gauge needles 15 times, and centrifuged at 500 × g for 5 min at 4°C. The resulting pellet with intact nuclei was washed once with hypotonic buffer, resuspended, and frozen rapidly at −80°C overnight.
The frozen nuclei were rapidly thawed by placing tubes in a 30°C water bath and then centrifuged at 500 × g for 1 min at 4°C. The resulting nuclear pellet was resuspended in 1 ml of buffer A with DNase and RNase (0.1 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 1× protease inhibitor, 5 μg/ml DNase I, 5 μg/ml RNase A) at RT to disrupt the nuclei and remove the nuclear membrane-associated DNA or RNA. The reaction mixtures were then incubated with 4 ml of buffer B (0.1 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 10% sucrose, 20 mM triethanolamine, pH 8.5, 1× protease inhibitor) at RT for 15 min and 4 ml of buffer C (sucrose cushion solution, 0.1 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 30% sucrose, 20 mM triethanolamine, pH 7.5, 1× protease inhibitor) and centrifuged at 4,000 × g for 15 min at 4°C. The supernatant (S1 fraction) contained nucleoplasm, and the pellet (P1 fraction) contained crude nuclear envelope.
The P1 fraction was then sequentially incubated with 1 ml of buffer D (0.1 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 10% sucrose, 20 mM triethanolamine, pH 7.5, 1× protease inhibitor), treated with 0.5 ml of buffer D with 0.3 mg/ml heparin and 4 ml of buffer C, and centrifuged at 4,000 × g for 15 min at 4°C. The resulting supernatant (S2 fraction) contained membrane-associated ribosomes and chromatin, and the pellet (P2 fraction) contained enriched nuclear envelope. The P2 fraction was sequentially incubated with 1 ml of buffer D, treated with 0.5 ml of buffer D containing 3% TNX-100 and 0.075% SDS, treated with 4 ml of buffer C, and then centrifuged at 4,000 × g for 15 min at 4°C. The resulting supernatant (S3 fraction) contained lipid bilayers, and the pellet contained the nuclear pore complex and lamina. To separate the nuclear pore complex from the lamina, the P3 fraction was incubated with 1 ml of buffer D containing 0.3% Empigen BB detergent on ice for 10 min and centrifuged at 15,000 × g for 15 min. The supernatant (S4 fraction) contained solubilized nucleoporins, and the pellet fraction (P4) contained insoluble nuclear lamina.
Expression and purification of GST-fusion protein.
E. coli-expressed recombinant GST-BGLF4, GST-K102I, and GST–importin-β were purified as previously described (40), with minor modifications. Plasmid-transformed E. coli BL21(DE3) was cultured in ampicillin (100 μg/ml) containing Luria broth medium until the optical density at 600 nm (OD600) was 0.6, and the cultures were kept on ice for 30 min. Isopropyl-β-d-thiogalactoside was then added to a final concentration of 0.5 mM, and cultures were incubated at 24°C for 18 h (GST-BGLF4 and GST-K102I) or 20°C for 14 h (GST–importin-β). The bacteria were harvested and resuspended in ice-cold PBS containing a protease inhibitor cocktail (Roche) and 1 μg/ml lysozyme, incubated on ice for 1 h, and sonicated at 4°C. The extracts were incubated overnight with glutathione-agarose at 4°C. After they were washed 3 times with PBST (1× PBS, 1% TNX-100), GST fusion proteins were eluted in elution buffer (50 mM Tris, pH 8.0, 10 mM reduced glutathione).
To purify recombinant GST-BGLF4 and GST-LacZ-NLS from Saccharomyces cerevisiae yeast for in vitro import assays, yeast cultures were grown in SC-URA dropout medium at 30°C to an OD600 of 0.8 and induced with 2% galactose for 4 h for GST fusion protein expression. The yeast cells were harvested and lysed with glass beads in lysis buffer (40 mM triethanolamine, pH 7.2, 2 mM EDTA, 150 mM NaCl, 2 mM DTT, 1× protease inhibitor). The yeast lysates were incubated overnight with glutathione-agarose at 4°C and washed 5 times with wash buffer (1× PBS, 1% TNX-100, 300 mM NaCl). GST fusion proteins were eluted in elution buffer and dialyzed against 20 mM HEPES, pH 7.3–110 mM potassium acetate at 4°C for 1 day (48). The concentrations of the purified GST proteins were estimated relative to the Coomassie blue staining signals of bovine serum albumin standards on SDS-polyacrylamide gels.
Coimmunoprecipitation assay.
HeLa cells (2 × 106) were transfected with plasmid expressing wild-type or mutant BGLF4. At 24 h posttransfection, cells were harvested and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS) containing a protease inhibitor cocktail. The cell lysates were separated by centrifugation for 10 min at 16,000 × g, and supernatant was precleared by incubation with protein A-Sepharose beads (Amersham Biosciences) and rotation at 4°C for 1 h and then reacted with BGLF4 monoclonal antibody 2224 at 4°C for 2 h. Protein A-Sepharose beads (100 μl at 20%) were added to pull down the immunocomplexes with rotation for 1 h at 4°C. Immunocomplexes were collected by centrifugation, washed extensively with NP-40 buffer, and detected by immunoblotting with mAb414 antibody for FG-Nups. GFP fusion proteins were immunoprecipitated with rabbit polyclonal anti-GFP (Clontech), and the immunocomplexes were detected using BGLF4 MAb 2616 or mAb414.
In vitro transcription-coupled translation and GST pull-down assays.
[35S]Met-labeled Nup62 and [35S]Met-labeled Nup153 were generated by adding pCWC13 (pCDNA3.1-Nup62) and pCWC14 (pCDNA3.1-Nup153) into a TNT T7 quick-coupled transcription/translation system according to the manufacturer's protocol (Promega). After gel analysis for translation products, 10 μl lysates was incubated with 2 μg of bacterially expressed GST, GST-BGLF4, GST-K102I, or GST–importin-β in a final volume of 500 μl in a mixture containing binding buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10% glycerol, 1% NP-40) at 4°C for 2 h. Proteins that interacted with the GST-fusion protein were selected by 20 μl glutathione-agarose beads for 2 h, displayed in 10% SDS-polyacrylamide gels, and detected by autoradiography.
Cell permeabilization and in vitro nuclear import assay.
Nuclear import assay was performed on the basis of a previous protocol with slight modification (1). HeLa cells grown on slides were permeabilized at RT with 40 μg/ml digitonin (Sigma-Aldrich) in transport buffer (TB; which contains 20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium diacetate, 1 mM EGTA, 2 mM DTT, 1× protease inhibitor) for 5 min, followed by placement on ice for 5 min, and washed with ice-cold TB twice. Nuclear import assay was performed with the import substrate (0.5 to 1 μM yeast recombinant GST-BGLF4, GST-K102I, or GST-LacZ-NLS) alone or mixed with 75% (vol/vol) cytosol factor (10% rabbit reticulocyte lysate, 0.5 mM ATP, 0.5 mM GTP, 10 mM creatine phosphate, 30 U/ml creatine phosphokinase) at RT for 1 h. For wheat germ agglutinin (WGA) treatment, 250 μg/ml WGA was added for 15 min before washing and addition of import substrates. After they were washed with TB twice to stop the reaction, cells were fixed with 4% paraformaldehyde and subjected to immunofluorescence assay with BGLF4 MAb 2224 or anti-GST antibody.
RESULTS
Mapping the regulatory domains for nuclear localization of BGLF4.
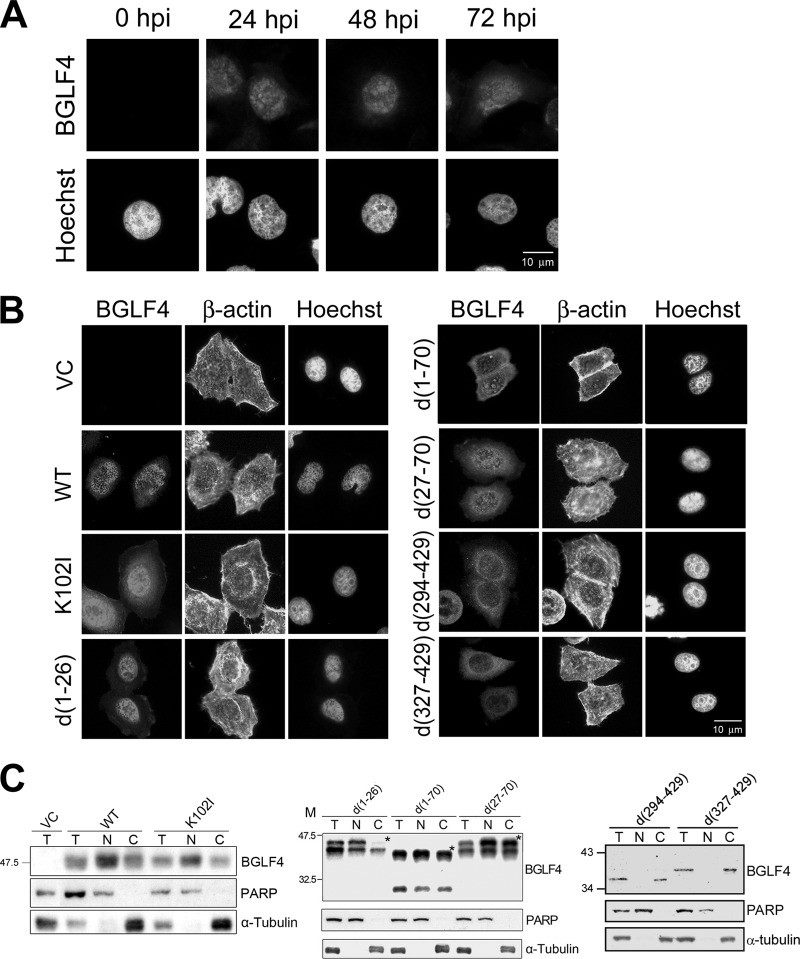
Through the sequence alignment of BGLF4 with cellular protein kinases, seven conserved kinase motifs with additional long N- and C-terminal regions were identified (Fig. 1A). Before the analysis of BGLF4 subcellular localization, we first detected the BGLF4 expression patterns in chemically induced EBV-positive NA cells. As indicated in our previous study, BGLF4 was predominantly detected in the nucleus of NA cells at 24 and 48 h postinduction (55) and with slight increasing perinuclear cytoplasmic staining at 72 h (Fig. 2A). It thus indicates that the nuclear targeting of BGLF4 might be regulated by some other virus protein(s) during EBV reactivation. In this study, a transient transfection system was used to investigate the functional domains of BGLF4 contributing to its nuclear import. Although a putative NLS was suggested in the region from aa 386 to 393 of BGLF4 (14), the N-terminal deletion mutant d(1-70) was mostly found in the cytoplasm in our previous study (55). To investigate whether both the N- and C-terminal regions of BGLF4 contribute to its nuclear localization, plasmids expressing deletion mutants d(1-26), d(1-70), d(27-70), d(294-429), and d(327-429) of BGLF4 were generated and transfected into HeLa cells (Fig. 2B). For immunofluorescence staining, rabbit antiserum against BGLF4 was used together with β-actin monoclonal antibody for cell morphology and Hoechst 33258 was used for DNA staining. As we previously reported, expression of BGLF4 caused premature chromosome condensation in a kinase activity-dependent manner (33). In immunofluorescence staining and fractionation analysis, WT BGLF4 and K102I (kinase-dead mutant), as well as d(1-26), were distributed predominantly in the nuclei of transfected cells, whereas the cytoplasmic staining of d(1-70) and d(27-70) was increased (Fig. 1B and 2B). However, because the fractionation protocol distributed the nuclear envelope and the nuclear envelope-associated apparatus to the nuclear fraction, d(1-70) appeared to be enriched in the nuclear fraction (Fig. 2C, middle). Both C-terminal deletion mutants, d(294-429) and d(327-429), which had deletions of the putative NLS (aa 386 to 393), NES (aa 342 to 359), and the second SIM (aa 344 to 350), were distributed exclusively in the cytoplasm of transfected cells, as detected with BGLF4 rabbit antiserum in immunofluorescence and fractionation assays. We noticed that the truncation of the N terminus or C terminus seemed to affect BGLF4 protein integrity and cause the presence of some minor protein species on the gel. Nevertheless, data presented here suggest that the C-terminal region of BGLF4 potentially determines its nuclear localization, while the N terminus may also modulate nuclear transportation, possibly through the SIM motif at aa 37 to 40 (37).
Fig 1.
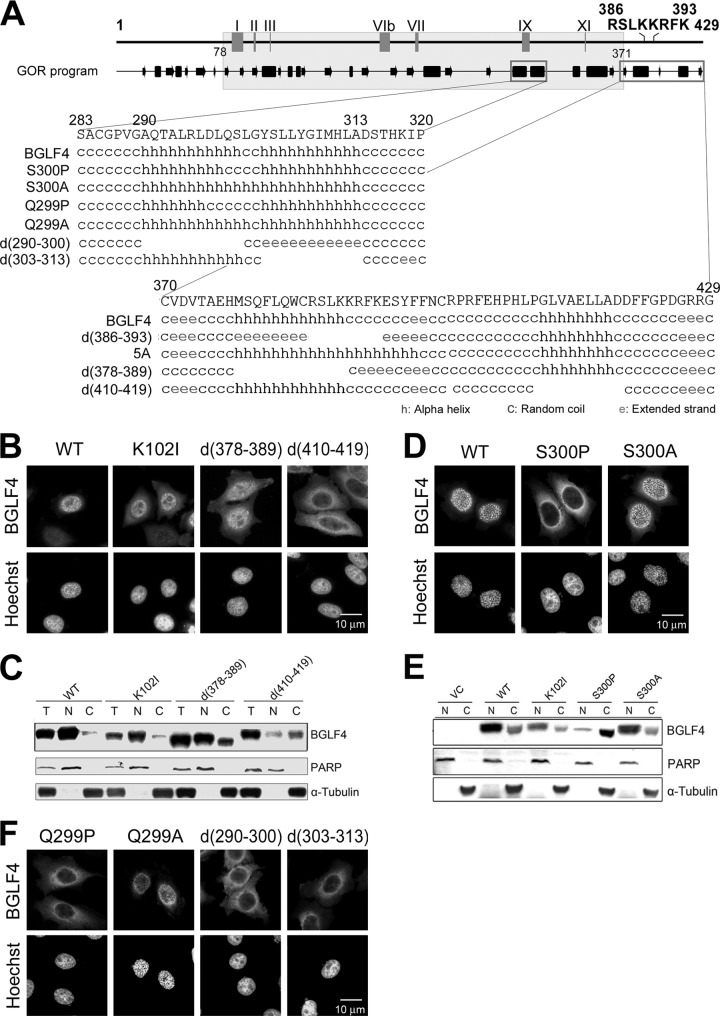
Schematic summary of the conserved kinase motifs of BGLF4 and subcellular distribution of BGLF4 mutants used in this study. (A) Conserved kinase motifs of BGLF4 were identified by alignment with eukaryotic protein kinases. There are 11 conserved motifs defined in eukaryotic protein kinases (19). Only motifs I, II, VIb, VII, IX, and XI can be identified in the kinase domain (gray region) of herpesviral kinases (32). I, ATP binding; II and VIb, catalysis; VII, Mg2+ chelating; IX, catalytic loop maintaining. SIMs at aa 37 to 40 and 344 to 350 and the NES at aa 342 to 359 were identified in a recent study (37). Amino acids 386 to 393 comprise a putative NLS (14).The putative secondary structure of BGLF4 was analyzed by the GOR prediction program (http://www.expasy.ch/tools/). Random coils, alpha helices, and extended strands are indicated as thin lines, boxes, and arrows, respectively. The hatched boxes underneath the predicted secondary structure indicate the critical regions (aa 290 to 313, 386 to 393, and 410 to 419) for BGLF4 nuclear targeting. (B) The subcellular distributions of WT and mutant BGLF4 proteins examined by immunofluorescence assay (IFA) and subcellular fractionation analysis in this study are summarized in the right-hand column. N, nucleus; C, cytoplasm; ND, none detected.
Fig 2.
Both N- and C-terminal regions of BGLF4 contribute to its nuclear localization. (A) NA cells were treated with TPA (40 ng/ml)-sodium butyrate (3 mM) and harvested at different time points (hours) postinduction (hpi). Cells were fixed and stained for BGLF4 and DNA with rabbit BGLF4 antiserum and Hoechst 33258, respectively. (B) HeLa cells were transfected with vector pSG5, a plasmid expressing WT BGLF4, a kinase-dead mutant (K102I), N-terminal deletion mutants d(1-26), d(1-70), and d(27-70), or C-terminal deletion mutants d(294-429) and d(327-429). At 24 h posttransfection, cells were harvested, fixed, and stained for BGLF4, β-actin, and DNA with rabbit BGLF4 antiserum, β-actin antibody, and Hoechst 33258, respectively. VC, vector control. (C) Transfected HeLa cells were also subjected to subcellular fractionation to separate nuclear (N) and cytosolic (C) fractions. An aliquot of the total lysate (T) was loaded to check the amount of protein expression in 12% SDS-PAGE. WT, K102I, d(1-26), d(1-70), and d(27-70) were detected with BGLF4 2616 antibody; d(294-429) and d(327-429) were detected with rabbit BGLF4 antiserum. PARP and α-tubulin were detected as nuclear and cytoplasmic markers, respectively. *, protein of the expected molecular weight; lane M, molecular weight markers.
The C-terminal amino acids 386 to 393 of BGLF4 are essential for its nuclear distribution but do not contain a classical NLS.
On the basis of the observation described above, we were curious whether the putative NLS motif at aa 386 to 393, RSLKKRFK, can function through the canonical nuclear import mechanism. Therefore, a deletion mutant plasmid encoding d(386-393) of BGLF4 was generated, and its distribution was analyzed. In contrast to WT BGLF4, a predominantly cytoplasmic distribution was observed in cells expressing the d(386-393) protein (Fig. 3A and B), suggesting that the region from aa 386 to 393 contributes to the nuclear import of BGLF4. To determine whether the basic residues within the region from aa 386 to 393 do serve as an NLS for BGLF4 nuclear targeting, alanine substitutions were introduced. Five mutants with point mutations, R386A, K393A, 2A (R386A/K393A), 3A (K389, 390A/R391A), and 5A (R386, 391A/K389, 390, 393A), were generated and expressed in HeLa cells. Surprisingly, all these mutants showed a predominantly nuclear distribution, as revealed by immunofluorescence and fractionation assays (Fig. 3A and B). Taken together, the region from aa 386 to 393 of BGLF4 plays an important role for the nuclear localization of BGLF4, but it does not serve as a conventional NLS.
Fig 3.

Amino acids 386 to 393 of BGLF4 are important for its nuclear localization but do not compose a canonical NLS. (A) HeLa cells were transfected with plasmids expressing wild-type BGLF4 or d(386-393), R386A, K393A, ([R386A/K393A], 2A), ([K389, 390A/R391A], 3A), or ([K386, 391A/K389, 390, 393A], 5A) mutants. At 24 h posttransfection, cells were fixed and stained for BGLF4 and DNA with rabbit BGLF4 antiserum and Hoechst 33258. (B) Transfected HeLa cells were also subjected to subcellular fractionation to separate nuclear (N) and cytosolic (C) fractions. All BGLF4 mutants were detected with BGLF4 monoclonal antibody 2616. PARP and α-tubulin were detected as nuclear and cytoplasmic markers, respectively.
The putative helical regions at the C terminus of BGLF4 are important for its nuclear localization.
Because deletion of aa 386 to 393 affects BGLF4 localization but substitution of the positively charged residues within the putative NLS does not, we propose that the protein structure surrounding aa 386 to 393 may regulate the nuclear import of BGLF4. In searching through the literature for noncanonical nuclear import mechanisms, we found that the amino terminus of the human immunodeficiency virus (HIV) Vpr protein, which contains a predicted alpha-helical structure, is important for nuclear localization of Vpr (49). To determine whether BGLF4 contains important structural domains for regulating nuclear localization, the putative secondary structure of BGLF4 was analyzed using the GOR structure prediction program (http://www.expasy.ch/tools/; Fig. 1A). As illustrated in Fig. 4A, there are two putative helical structures at aa 378 to 389 and aa 411 to 418 of BGLF4. The prediction also suggests that deletion of aa 386 to 393 may cause a structural change of the helix into an extended strand. We propose that the secondary structure at the C terminus of BGLF4 may govern its nuclear localization. To determine whether the helical regions around aa 378 to 389 and 411 to 418 of BGLF4 are important for its nuclear localization, two deletion mutants, d(378-389) and d(410-419), were generated (Fig. 4A). In immunofluorescence and fractionation assays, d(378-389) was expressed predominantly in the nucleus, whereas d(410-419) was expressed predominantly in the cytoplasm (Fig. 4B and C). These results suggest that in addition to the helical regions surrounding aa 386 to 393, the region from aa 410 to 419 of BGLF4 is also crucial for its nuclear localization.
Fig 4.
The C-terminal helical regions of BGLF4 contribute to its nuclear localization. (A) Predicted secondary structures of the BGLF4 C-terminal region (i) aa 283 to 320 and (ii) 370 to 429 of BGLF4. (B) HeLa cells were transfected with plasmids expressing WT, K102I, d(378-389), or d(410-419). At 24 h posttransfection, cells were fixed and stained for BGLF4 and DNA. (C) Subcellular fractionation analysis of WT, K102I, d(378-389), or d(410-419) was performed, and results were analyzed by immunoblotting. (D) Slide-cultured HeLa cells were transfected with plasmids expressing WT, S300P, or S300A. An immunofluorescence assay was used to detect the distribution of BGLF4. (E) Subcellular fractionation of WT, S300P, or S300A was performed and analyzed by immunoblotting. (F) Immunofluorescence detection of the distribution of Q299P, Q299A, d(290-300), or d(303-313) of BGLF4 in transfected HeLa cells.
The helix regions of aa 290 to 300 and 303 to 313 are important for BGLF4 nuclear targeting.
While we were wondering whether other structural domains may interfere with BGLF4 nuclear targeting, a mutant with a mutation at aa 300 of BGLF4 (S300P) was identified during our PCR cloning. In transiently transfected cells, S300P was exclusively expressed in the cytoplasm (Fig. 4D and E). Because the region from aa 297 to 302 is the catalytic loop-maintaining domain, the orientation of this region may play an important role in the intact structure and biological function of BGLF4 (Fig. 4A). A mutant with the S300A mutation was generated and found to be expressed predominantly in the nuclei of transfected cells, suggesting that substitution of proline at aa 300 disrupts the structure between aa 290 and 313. To determine whether the helix structure is important, Q299P, Q299A, d(290-300), and d(303-313) mutants of BGLF4 were generated and their distributions were monitored. Immunofluorescence staining indicates that Q299P, but not Q299A, displayed a cytoplasmic staining pattern (Fig. 4F). Additionally, both d(290-300) and d(303-313) were mainly expressed in the cytoplasm. This result indicated that the putative helix-coil-helix structure around aa 289 to 314 also plays an essential role in BGLF4 nuclear targeting. Notably, Q299P, S300P, d(290-300), and d(303-313) all displayed a nuclear rim-staining pattern, suggesting that these mutant proteins may associate with the nuclear envelope (Fig. 4D and F).
Because serine 300 is located in the catalytic loop-maintaining domain of BGLF4 and the S300P substitution led to a loss of kinase activity in an immunoprecipitation (IP) kinase experiment (data not shown), we were curious whether the kinase activity or the integrity of the catalytic domains affects nuclear localization. To this end, several mutants, including mutants with D195A, N200K, D219E, D195A/D219E, D195A/N200K, and N200K/D219E mutations, were generated with amino acid substitutions in the catalysis or Mg2+-chelating motifs and examined for their subcellular localization and kinase activities. To evaluate the kinase activity, an immunoprecipitation kinase assay was carried out on these mutants using histone H1 as a substrate (35). All these mutants lost the ability to phosphorylate substrate histone H1 in vitro (data not shown). All mutants were predominantly expressed in the nuclei of transfected cells, except for N200K/D219E, which was distributed in the cytoplasm (Fig. 1 and Fig. 5). Overall, data presented here indicate that the kinase activity is not crucial for nuclear localization of BGLF4 and the cytoplasmic retention of N200K/D219E may be attributable to a conformation change induced by the double mutations.
Fig 5.
The nuclear localization of BGLF4 is not regulated by its kinase activity. HeLa cells were transfected with wild-type BGLF4 and mutant D195A, D219E, N200K, D195A/D219E, D195A/N200K, or N200K/D219E. At 24 h posttransfection, cells were fixed and stained for BGLF4 and DNA.
The C-terminal helical regions of BGLF4 associate with the nuclear envelope apparatus, but the C terminus alone is not sufficient for correct nuclear localization.
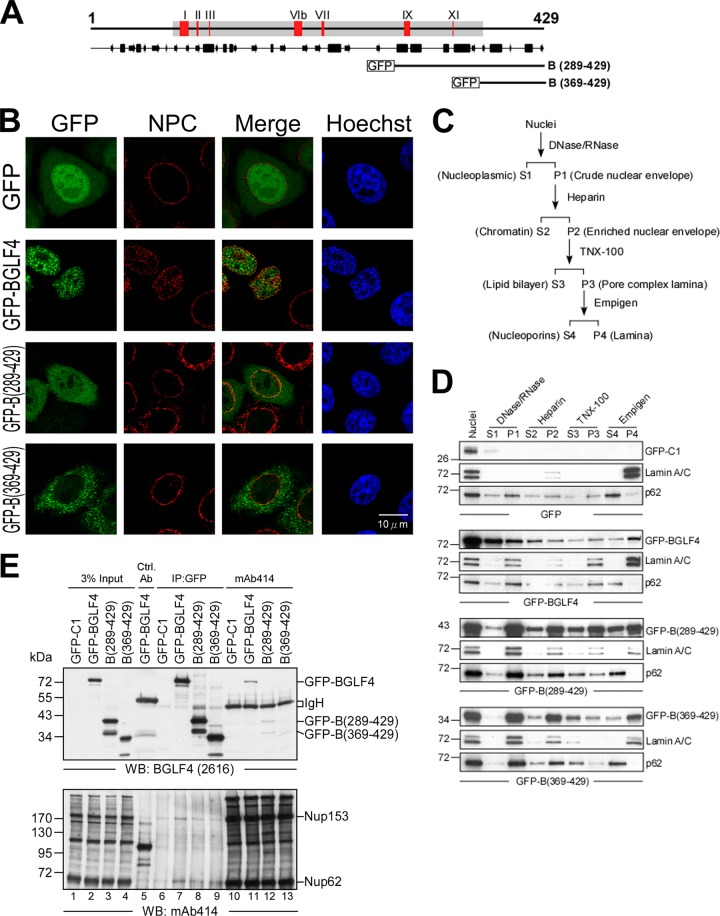
To determine whether the C terminus of BGLF4 containing multiple helices is sufficient for nuclear transport, GFP-tagged wild-type and truncated BGLF4 fusion proteins were generated. GFP-B(289-429) includes six predicted helices within the C-terminal domain, whereas GFP-B(369-429) contains the last two putative helices of BGLF4 (Fig. 6A). In transiently transfected HeLa cells, GFP alone was distributed in both the cytoplasm and the nucleus, coupled with a nuclear envelope-associated staining pattern of NPC. GFP-BGLF4 had a predominantly nuclear distribution, coupled with condensed chromosomes in the nucleus; a loose nuclear envelope association; and some intranuclear staining patterns of FG-Nups (Fig. 6B). Of note, GFP-B(289-429) showed both cytoplasmic and nuclear staining, while GFP-B(369-429) was excluded from the nucleus and displayed GFP signals at the perinuclear region. Taken together, the region from aa 289 to 429 of BGLF4 can facilitate its nuclear localization but is inefficient at mediating nuclear import independently. The intact structure of BGLF4 is required for its complete nuclear targeting.
Fig 6.
The C terminus of BGLF4 associates with the nuclear envelope. (A) Schematic illustration of full-length and truncated GFP-BGLF4 constructs. (B) Slide-cultured HeLa cells were transfected with plasmid expressing GFP-BGLF4, GFP-B(289-429), or GFP-B(369-429) or vector control. At 24 h posttransfection, cells were fixed and stained for NPC with FG repeat-containing nucleoporin-specific mAb414 antibody, and DNA was stained with Hoechst 33258. The distribution of GFP-BGLF4 fusions and NPC was observed with a confocal microscope. (C) Scheme of fractionation of the various compartments of the nuclear envelope. The proteins associated with the nuclear envelope were finally fractionated into soluble nucleoporins (S4) or insoluble nuclear lamina (P4) fractions. (D) Nuclear envelope fractionation of HeLa cells expressing GFP, GFP-BGLF4, GFP-B(289-429), or GFP-B(369-429). Lamin A/C and nucleoporin p62 serve as the markers of nuclear lamina and nuclear pore complex, respectively. S, supernatant; P, pellet. (E) Cell lysates harvested from HeLa cells transfected with the GFP-BGLF4, GFP-B(289-429), or GFP-B(369-429) plasmid or GFP vector control were subjected to coimmunoprecipitation assay. Protein complexes were immunoprecipitated with rabbit anti-GFP antiserum or mAb414 antibody, and the immunocomplexes were detected in immunoblots using BGLF4-specific MAb 2616 or mAb414. The control antibody (Ctrl. Ab.) is a monoclonal antibody against GST. WB, Western blot.
We noticed that some cellular macromolecules, such as unphosphorylated Stat1 and Vpr, are translocated to the nucleus through direct interaction with FG repeat-containing nucleoporins (13, 36, 43). Because partial colocalization of GFP-BGLF4 and FG-Nups was observed (Fig. 6B), we suspected that BGLF4 may associate directly with the nuclear envelope apparatus. Thus, the nuclear envelope fractionation protocol established by Matunis (45) was adapted to analyze the distribution of full-length and truncated GFP-BGLF4 fusions (Fig. 6C and D). The final supernatant fraction (S4) contained solubilized nucleoporins and NPC-associated proteins. In our previous study, we found that a portion of BGLF4 associates with and can cause disassembly of nuclear lamina through phosphorylation of lamin A/C (35). Here, similar to the result with GFP-BGLF4, both GFP-B(289-429) and GFP-B(369-429) were cofractionated with FG-nucleoporin p62 in the Empigen-soluble fractions (S4; Fig. 6D), in addition to associating with nuclear lamina (P4). To further determine whether GFP-BGLF4 interacts with FG-Nups, GFP-BGLF4 or truncated mutants GFP-B(289-429) and GFP-B(369-429) were coimmunoprecipitated with FG-Nups in reciprocal directions. We found that GFP-BGLF4 interacted with both Nup153 and Nup62, while the interaction ability was attenuated in GFP-B(289-429) and was much weaker in GFP-B(369-429) (Fig. 6E, lanes 6 to 9). Similarly, GFP-BGLF4 was more efficiently coimmunoprecipitated with FG-Nups by mAb414 antibody than GFP-B(289-429) and GFP-B(369-469) (Fig. 6E, lanes 10 to 13). Data presented here suggest that the C terminus of BGLF4 potentially mediates its association with FG repeat-containing nucleoporins and contributes to its nuclear translocation.
BGLF4 interacts directly with FG repeat-containing nucleoporins.
Because FG repeat-containing nucleoporins are important mediators for nuclear import, we wondered whether the nuclear targeting of the various BGLF4 mutants correlates with their abilities to interact with FG-Nups in vivo. To this end, mAb414 and BGLF4-specific antibody 2224 were used reciprocally to immunoprecipitate the cell lysates containing wild-type or mutant BGLF4 proteins. We found that nucleus-distributed wild-type BGLF4, BGLF4(5A), and d(378-389) and cytoplasmically distributed d(386-393) and d(410-419) can be coimmunoprecipitated with Nup153 and Nup62 to similar extents (Fig. 7A and B). We postulate that there might be more than one FG repeat-interacting region on BGLF4 and the coordination between different interacting regions and various FG-Nups may be required for proper nuclear targeting of BGLF4. This may also explain why the single replacement of serine 300 or aspartic acid 299 by proline may disrupt the coordination and lead to the complete abolition of the nuclear translocation of BGLF4. Indeed, we found that Nup153 was coimmunoprecipitated with BGLF4 S300P less efficiently than BGLF4 WT and S300A (data not shown).
Fig 7.
BGLF4 interacts with FG repeat-containing nucleoporins. (A and B) HeLa cells were transfected with plasmids expressing BGLF4, d(386-393), d(378-389), d(410-419), or BGLF4 (5A). At 24 h posttransfection, cells were harvested and lysed in RIPA buffer. Protein complexes were immunoprecipitated with BGLF4-specific antibody 2224 (A) or mAb414 (B) and the immunocomplexes were detected in immunoblots using BGLF4-specific antibody 2224 and mAb414 antibody. (C) Purified GST-BGLF4, GST-K102I, GST–importin-β, or GST control was incubated with [35S]Met-labeled Nup62 and Nup153. Proteins that interacted with various GST fusion proteins were analyzed by SDS-PAGE and detected by autoradiography. (D) HeLa cells were permeabilized with digitonin and incubated with purified yeast recombinant GST-LacZ-NLS, GST-BGLF4, or GST-K102I in the absence (left) or presence (middle) of cytosolic factors; (right) permeabilized cells were incubated with 250 μg/ml WGA for 15 min before adding the nuclear import substrates. After 1 h of incubation, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% TNX-100. GST-BGLF4 and GST-K102I were detected using BGLF4-specific antibody 2224, and GST-LacZ-NLS was detected with anti-GST antibodies.
To further demonstrate direct interactions between BGLF4 and FG-Nups, purified bacterially recombinant GST, GST-BGLF4, or GST-K102I or positive-control GST–importin-β was used to pull down the in vitro-translated [35S]Met-labeled Nup62 or Nup153. The result showed that Nup62 was pulled down by both GST-BGLF4 and GST-K102I at levels similar to those of GST–importin-β, whereas the abilities of GST-BGLF4 and GST-K102I to pull down Nup153 were slightly weaker than the ability of GST–importin-β (Fig. 7C).
BGLF4 translocates into the nuclei of permeabilized cells in the absence of cytosolic factors.
To demonstrate that BGLF4 may target the nucleus through direct interaction with FG repeat-containing nucleoporins, the in vitro nuclear import assay was performed in the presence or absence of cytoplasmic factors according to a previous protocol (1). To this end, HeLa cells were permeabilized with digitonin to wash out most of the soluble cytosolic components and incubated with purified yeast GST-BGLF4 and GST-K102I. As a control, GST-LacZ-NLS, which expresses the LacZ protein fused with the NLS sequence of SV40 large T antigen (PKKKRKV), could be translocated into the nucleus only in the presence of cytosolic factors after 60 min incubation (Fig. 7D). Notably, after the incubation, GST-BGLF4 and GST-K102I were detected in the nucleus even in the absence of other exogenous factors (Fig. 7D), suggesting that BGLF4 translocates into the nucleus in a cytosolic factor-independent manner. To further confirm that the FG repeats on the nucleoporins mediate BGLF4 translocation, WGA, which binds to N-acetyl-d-glucosamine-modified nucleoporins, including Nup153 and Nup62, was used as the classical inhibitor of FG-Nups-dependent nuclear translocation (1). As expected, WGA inhibited the process of BGLF4 nuclear import (Fig. 7D), suggesting that the binding of BGLF4 to FG repeat-containing nucleoporins is required for its nuclear localization.
Overall, we propose that BGLF4 may translocate into the nucleus through interaction with various FG-Nups, including Nup62 and Nup153. A sequential displacement for interactions between BGLF4 and various nucleoporins may be required for proper nuclear targeting of BGLF4.
DISCUSSION
BGLF4 appears to be a very competent kinase that can phosphorylate and regulate the function of multiple viral and cellular factors. In virus-replicating cells, BGLF4 displays a predominant nuclear distribution, while cytoplasmic partition is increased at the late stage of virus replication. With a CDK1-like activity, BGLF4 translocates into the nucleus through a pathway very distinct from that of CDKs. In this study, data from mutants with deletion and point mutations revealed that both the N- and C-terminal regions of BGLF4 play important regulatory roles for nuclear targeting (Fig. 1). Although d(386-383) of BGLF4 lost its nuclear localization, replacement of basic amino acids within the putative NLS (aa 386 to 393) by alanine did not affect the nuclear targeting of BGLF4. Because d(386-393) also lost its kinase activity in an IP kinase assay (data not shown), we speculate that deletion of aa 386 to 393 distorted the protein structure of BGLF4. This suggests that the native protein structure is important for nuclear targeting of BGLF4. It was supported by the cytoplasmic distribution of S300P, Q299P, d(290-300), d(303-313), d(386-393), and d(410-419) and the nuclear distribution of S300A and Q229A (Fig. 4). It is very likely that structural distortion caused by proline protrusion or small deletions of the putative helices between aa 290 and 313, 386 and 393, and 410 and 419 affect the nuclear transport of BGLF4. In our mechanistic analysis, we found that the nuclear translocation of BGLF4 is independent of its kinase activity, importin, or other cytoplasmic factors. BGLF4 appeared to use an unconventional mechanism that may ensure its nuclear targeting without the limitation of cellular factors.
In the canonical nucleus-targeting process, proteins first need to bind to FG-Nups through the interaction with importins/Ran-GTP. The final step is mediated by the release of importin-β from FG-Nups to distribute the substrate protein into nucleoplasm. Nevertheless, there are alternative mechanisms for nuclear import control, in addition to the Ran- and importin-dependent nuclear import (for a review, see reference 52). For example, the nuclear targeting of the small HIV protein Vpr, which contains two discrete nucleus-targeting signals in its C and N termini, appears to involve access of the NPC directly without a requirement for soluble factors for the nuclear import (13, 23, 36). Cellular β-catenin and unphosphorylated Stat1 translocate into the nucleus through direct interaction with FG-Nups (12, 43, 62). As demonstrated by an in vitro import assay, the transport of β-catenin is energy dependent, whereas unphosphorylated Stat1 and extracellular signal-regulated kinase (ERK) appeared to translocate into nucleus through direct interaction with Nup153 and Nup214 in an energy-independent manner (43, 44, 59). On the basis of the nuclear import assay, both WT and kinase-dead BGLF4 appeared to translocate into the nucleus independently of cytoplasmic factors, while the energy dependence of their translocation is not clear (Fig. 7D).
The in vitro pull-down assay demonstrated that BGLF4 interacts with Nup153 and Nup62 (Fig. 7C). In the GFP fusion analysis, although the region from aa 289 to 429 of BGLF4 is not sufficient for appropriate nuclear targeting of a GFP fusion protein (Fig. 6), the C terminus of BGLF4 was associated with the NPC in coimmunoprecipitation analysis (Fig. 6E). The abilities of GFP fusions to interact with Nup153 showed a decline of GFP-BGLF4 > GFP-B(289-429) > GFP-B(369-429) when both directions of antibodies were used for IP (Fig. 6E). Because the interaction between nuclear targeting of WT BGLF4 or BGLF4(5A) to Nup153 or Nup62 appeared to be similar to that of cytoplasmic targeting of d(386-393) or d(410-419) in the coimmunoprecipitation experiments (Fig. 7A and B), we propose that multiple regions within BGLF4 mediate the interactions to FG-Nups. However, successful nuclear transport may depend on coordinated sequential binding between different helices and various Nups.
While there is no sequence homology between BGLF4 and importin-β, BGLF4 nuclear transport may use a strategy similar to that of importin-β. Importin-β is imported into the nucleus by direct interaction with FG repeat-containing nucleoporins through its HEAT repeats. Crystal structure analysis showed that importin-β is composed of 19 tandem HEAT repeats, each consisting of one turn of a superhelix by 2 α helices (2). The HEAT repeats 5 and 6, 6 and 7, 14 and 15, and 15 and 16 of importin-β form four pockets for binding to FG repeats on FG-Nups (5, 50). Individual pockets have different affinities for FG-Nups and different sensitivities to Ran-GTP-induced conformational changes to release the importin-β–FG–Nup complex (50). Using three-dimensional (3D) structure prediction software (PS)2 (version 2; http://ps2v2.life.nctu.edu.tw/), casein kinase 2 alpha 1 polypeptide (Protein Data Bank [PDB] accession number 3bqcA) was identified as a template for the structural alignment of BGLF4. The predicted model shows that the conserved catalytic domains of protein kinase locate between aa 85 and 315 of BGLF4. Beyond the catalytic domain, some helix-turn-helix structures were predicted within the C terminus of BGLF4 (aa 333 to 354 and 380 to 406; Fig. 8). The region from aa 296 to 312 within the catalytic loop was identified as a helix structure in the 3D prediction and as a helix-turn-helix structure in the secondary structure prediction (Fig. 8). Taken together, we propose that the C terminus of BGLF4 containing multiple helix regions, including aa 296 to 312 and 380 to 406, is responsible for binding FG repeat-containing Nups. BGLF4 may target the nucleus through sequential direct interactions with FG repeat-containing nucleoporins such as Nup62 and Nup153. Further experiments are required to map the interacting domains among BGLF4 and FG-Nups and to reveal whether other FG-Nups, non-FG-Nups, or cellular factors are involved in the nuclear transport process.
Fig 8.
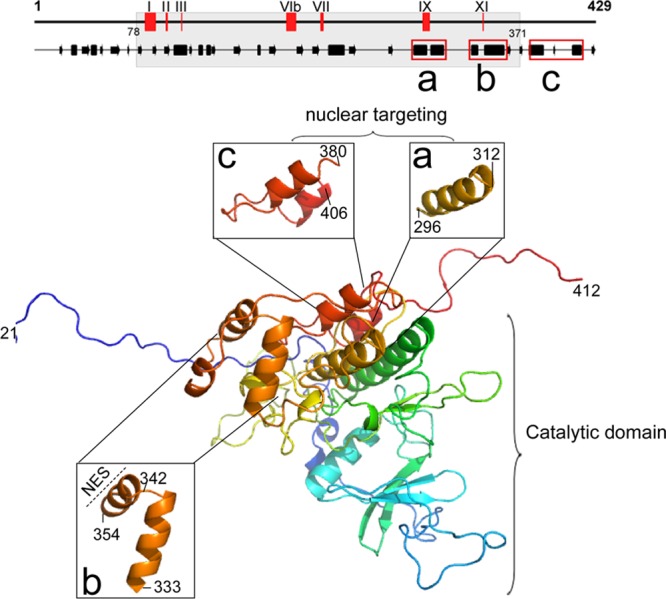
Predicted 3D structure of BGLF4. The 3D structure of BGLF4 was predicted from Protein Structure Prediction Server (PS)2 (version 2; http://ps2v2.life.nctu.edu.tw/) using casein kinase 2 alpha 1 polypeptide (PDB accession number 3bqcA) as the template for aa 21 to 412 of BGLF4. The critical helical regions for BGLF4 nuclear targeting are shown in enlarged 3D images for aa 296 to 312 (a) and aa 380 to 406 (c). (b) Helix-loop-helix region from aa 333 to 354 containing the NES (aa 342 to 359).
BGLF4 is a component of the viral tegument, and a small portion of BGLF4 was detected in the cytoplasm of virus-replicating cells at the late stage of virus replication (24, 55) (Fig. 2A). We suggest that the recently identified SIMs (aa 37 to 40 and 344 to 360) and NES (aa 342 to 359) of BGLF4 potentially regulate the proper localization of BGLF4 at different virus replication stages through either modulation of the BGLF4 structure or interaction with other viral components. According to our 3D modeling, the BGLF4 C-terminal helices (aa 290 to 313, 386 to 393, and 410 to 419) contributing to FG-Nup interaction are separated from the NES and SIMs. The SIMs seem unlikely to regulate the FG-Nup-interacting ability of BGLF4 directly. One possibility would be that the newly synthesized BGLF4 can be transported into the nucleus though binding to FG-Nups and then interact with other intranuclear sumoylated proteins through its SIMs, leading to the masking of NES and a stable nuclear distribution at the early stage of virus replication. Once the SUMO binding ability of BGLF4 provided by the coordination of both SIM motifs is disrupted at the later stage, the NES will distribute BGLF4 into the cytoplasm.
In a protein array phosphorylation analysis, some nucleoporins were identified as BGLF4 substrates, including Nup62 and Nup35, but not Nup107 and Nup133 (38). Nup62 is located at the center of the nuclear pore and functions as a major barrier for molecular gating into the nucleus. Nup35 is located at the inner ring of NPC and is important for nuclear pore complex assembly and nuclear envelope formation (20). In this study, BGLF4 appeared to induce the reorganization of NPC (Fig. 6B), and possible effects of BGLF4 in modulating NPC function also are of interest. During herpesvirus replication, the distribution of NPC on the nuclear envelope is changed, but the major composition and functions of NPC were not altered significantly (21). Almost half of the nuclear pores in herpes simplex virus 1-infected cells were larger than 140 nm, with the protrusion of some nuclear material into the cytoplasm being detected. However, it remains unclear whether viral capsids with an average size of 115 to 130 nm can bud through the dilated pores or whether the virus egresses from the nucleus through an envelope-dependent process and the coupled NPC enlargement may facilitate the transport of other materials required for nucleocapsid assembly (60). In a preliminary experiment, the nuclear retention of yellow fluorescent protein (YFP)-LacZ-NLS was not affected by the expression of BGLF4 (data not shown). Thus, the nuclear import or export of other proteins should be analyzed further. As a tegument protein, BGLF4 may also modify NPC function upon initial infection for translocation of the viral genome through the NPC into the nucleus. Indeed, phosphorylation of nucleoporins was observed upon signaling activation or virus infection. ERK is a mammalian conserved mitogen-activated protein kinase (MAPK). Upon phosphorylation and activation by the upstream kinase MEK, ERK is further phosphorylated and translocated into the nucleus through direct binding to importin-7 (9, 63). Many FG-Nups contain multiple S/TP motifs (with serine or threonine before proline) near FG repeats. ERK can bind to Nup153 and Nup214 and phosphorylate Nup50 in vitro and in vivo at Ser-221 and Ser-315 within the FG repeat domain, and this is related to its reduced affinity for importin-β and transportin. Thus, ERK-induced phosphorylation may reduce the hydrophobicity of the FG repeats and disrupt the nuclear transport receptor-FG interaction (29). It was also revealed using various kinase inhibitors that the encephalomyocarditis virus leader protein triggers nucleoporin phosphorylation in a MAPK-mediated pathway (51), suggesting that viruses may modulate NPC function to facilitate their replication. Further investigation of the possible effects of CHPKs in regulating NPC distribution and individual nucleoporin phosphorylation may help provide an understanding of the possible regulation of NPC function by CHPKs.
ACKNOWLEDGMENTS
We are grateful to Tim J. Harrison of University College London for critical reading and editing of the manuscript. We thank Mitsuhiro Kawata at Kyoto Prefectural University of Medicine for the pEYFP-LacZ-NLS plasmid, Yoshihiro Yoneda at Osaka University for the GST–importin-β plasmid, Diane S. Hayward at the Johns Hopkins University School of Medicine for the pEG-BGLF4 plasmid, and J. Lu at Academia Sinica for the Nup62 cDNA clone. We appreciate the technical advice for yeast recombinant protein expression and purification from Zih-Jie Shen and Shu-Chun Teng (National Taiwan University).
This study was supported by the National Science Council (NSC98-2320-B-002-054-MY3), National Health Research Institutes (NHRI-EX98-9609BI, NHRI-EX99-9928BI), and National Taiwan University (98R0302 and 99R71423).
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Adam SA, Marr RS, Gerace L. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309:1–18 [DOI] [PubMed] [Google Scholar]
- 3. Asai R, et al. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 80:5125–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baldwin AS., Jr 1996. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649–683 [DOI] [PubMed] [Google Scholar]
- 5. Bayliss R, Littlewood T, Stewart M. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102:99–108 [DOI] [PubMed] [Google Scholar]
- 6. Chang Y, et al. 1999. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J. Virol. 73:8857–8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chee MS, Lawrence GL, Barrell BG. 1989. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J. Gen. Virol. 70(Pt 5):1151–1160 [DOI] [PubMed] [Google Scholar]
- 8. Chen MR, Chang SJ, Huang H, Chen JY. 2000. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J. Virol. 74:3093–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chuderland D, Konson A, Seger R. 2008. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol. Cell 31:850–861 [DOI] [PubMed] [Google Scholar]
- 10. Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis LI, Blobel G. 1986. Identification and characterization of a nuclear pore complex protein. Cell 45:699–709 [DOI] [PubMed] [Google Scholar]
- 12. Fagotto F, Gluck U, Gumbiner BM. 1998. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr. Biol. 8:181–190 [DOI] [PubMed] [Google Scholar]
- 13. Fouchier RA, et al. 1998. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 72:6004–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gershburg E, Marschall M, Hong K, Pagano JS. 2004. Expression and localization of the Epstein-Barr virus-encoded protein kinase. J. Virol. 78:12140–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gershburg E, Pagano JS. 2008. Conserved herpesvirus protein kinases. Biochim. Biophys. Acta 1784:203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gershburg E, Raffa S, Torrisi MR, Pagano JS. 2007. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 81:5407–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gershburg S, Murphy L, Marschall M, Gershburg E. 2010. Key motifs in EBV (Epstein-Barr virus)-encoded protein kinase for phosphorylation activity and nuclear localization. Biochem. J. 431:227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gorlich D, Henklein P, Laskey RA, Hartmann E. 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810–1817 [PMC free article] [PubMed] [Google Scholar]
- 19. Hanks SK, Quinn AM. 1991. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200:38–62 [DOI] [PubMed] [Google Scholar]
- 20. Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW. 2008. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol. Biol. Cell 19:1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hofemeister H, O'Hare P. 2008. Nuclear pore composition and gating in herpes simplex virus-infected cells. J. Virol. 82:8392–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imamoto N, et al. 1995. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 368:415–419 [DOI] [PubMed] [Google Scholar]
- 23. Jenkins Y, McEntee M, Weis K, Greene WC. 1998. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J. Cell Biol. 143:875–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johannsen E, et al. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaku N, Matsuda K, Tsujimura A, Kawata M. 2008. Characterization of nuclear import of the domain-specific androgen receptor in association with the importin alpha/beta and Ran-guanosine 5′-triphosphate systems. Endocrinology 149:3960–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kato K, et al. 2001. Epstein-Barr virus-encoded protein kinase BGLF4 mediates hyperphosphorylation of cellular elongation factor 1delta (EF-1delta): EF-1delta is universally modified by conserved protein kinases of herpesviruses in mammalian cells. J. Gen. Virol. 82:1457–1463 [DOI] [PubMed] [Google Scholar]
- 27. Kato K, et al. 2003. Identification of protein kinases responsible for phosphorylation of Epstein-Barr virus nuclear antigen leader protein at serine-35, which regulates its coactivator function. J. Gen. Virol. 84:3381–3392 [DOI] [PubMed] [Google Scholar]
- 28. Kawaguchi Y, et al. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 77:2359–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kosako H, Imamoto N. 2010. Phosphorylation of nucleoporins: signal transduction-mediated regulation of their interaction with nuclear transport receptors. Nucleus 1:309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krajewski S, et al. 1993. Investigation of the subcellular distribution of the bcl-2 oncoprotein: residence in the nuclear envelope, endoplasmic reticulum, and outer mitochondrial membranes. Cancer Res. 53:4701–4714 [PubMed] [Google Scholar]
- 31. Kudoh A, et al. 2006. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J. Virol. 80:10064–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leader DP. 1993. Viral protein kinases and protein phosphatases. Pharmacol. Ther. 59:343–389 [DOI] [PubMed] [Google Scholar]
- 33. Lee CP, et al. 2007. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J. Virol. 81:5166–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee CP, Chen MR. 2010. Escape of herpesviruses from the nucleus. Rev. Med. Virol. 20:214–230 [DOI] [PubMed] [Google Scholar]
- 35. Lee CP, et al. 2008. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 82:11913–11926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Le Rouzic E, et al. 2002. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J. Biol. Chem. 277:45091–45098 [DOI] [PubMed] [Google Scholar]
- 37. Li R, et al. 2012. SUMO binding by the Epstein-Barr virus protein kinase BGLF4 is crucial for BGLF4 function. J. Virol. 86:5412–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li R, et al. 2011. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 10:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu CC, Chen MR. 2006. Lytic replication of Epstein-Barr virus. Future Virol. 1:435–446 [Google Scholar]
- 40. Lu CC, Chen YC, Wang JT, Yang PW, Chen MR. 2007. Xeroderma pigmentosum C is involved in Epstein Barr virus DNA replication. J. Gen. Virol. 88:3234–3243 [DOI] [PubMed] [Google Scholar]
- 41. Lu CC, et al. 2006. Genome-wide transcription program and expression of the Rta responsive gene of Epstein-Barr virus. Virology 345:358–372 [DOI] [PubMed] [Google Scholar]
- 42. Makarova O, Kamberov E, Margolis B. 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29:970–972 [DOI] [PubMed] [Google Scholar]
- 43. Marg A, et al. 2004. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 165:823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsubayashi Y, Fukuda M, Nishida E. 2001. Evidence for existence of a nuclear pore complex-mediated, cytosol-independent pathway of nuclear translocation of ERK MAP kinase in permeabilized cells. J. Biol. Chem. 276:41755–41760 [DOI] [PubMed] [Google Scholar]
- 45. Matunis MJ. 2006. Isolation and fractionation of rat liver nuclear envelopes and nuclear pore complexes. Methods 39:277–283 [DOI] [PubMed] [Google Scholar]
- 46. Michel D, et al. 1998. Functional regions of the human cytomegalovirus protein pUL97 involved in nuclear localization and phosphorylation of ganciclovir and pUL97 itself. J. Gen. Virol. 79(Pt 9):2105–2112 [DOI] [PubMed] [Google Scholar]
- 47. Mitchell DA, Marshall TK, Deschenes RJ. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9:715–722 [DOI] [PubMed] [Google Scholar]
- 48. Moore MS, Schwoebel ED. 2001. Nuclear import in digitonin-permeabilized cells. Curr. Protoc. Cell Biol. Chapter 11:Unit 11.7 [DOI] [PubMed] [Google Scholar]
- 49. Nie Z, et al. 1998. The putative alpha helix 2 of human immunodeficiency virus type 1 Vpr contains a determinant which is responsible for the nuclear translocation of proviral DNA in growth-arrested cells. J. Virol. 72:4104–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Otsuka S, Iwasaka S, Yoneda Y, Takeyasu K, Yoshimura SH. 2008. Individual binding pockets of importin-beta for FG-nucleoporins have different binding properties and different sensitivities to RanGTP. Proc. Natl. Acad. Sci. U. S. A. 105:16101–16106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Porter FW, Brown B, Palmenberg AC. 2010. Nucleoporin phosphorylation triggered by the encephalomyocarditis virus leader protein is mediated by mitogen-activated protein kinases. J. Virol. 84:12538–12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robbins J, Dilworth SM, Laskey RA, Dingwall C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623 [DOI] [PubMed] [Google Scholar]
- 53. Sorokin AV, Kim ER, Ovchinnikov LP. 2007. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc.) 72:1439–1457 [DOI] [PubMed] [Google Scholar]
- 54. Wang JT, et al. 2009. Epstein-Barr virus BGLF4 kinase suppresses the interferon regulatory factor 3 signaling pathway. J. Virol. 83:1856–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang JT, et al. 2005. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 86:3215–3225 [DOI] [PubMed] [Google Scholar]
- 56. Webel R, et al. 2011. Two isoforms of the protein kinase pUL97 of human cytomegalovirus are differentially regulated in their nuclear translocation. J. Gen. Virol. 92:638–649 [DOI] [PubMed] [Google Scholar]
- 57. Weis K, Ryder U, Lamond AI. 1996. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15:1818–1825 [PMC free article] [PubMed] [Google Scholar]
- 58. Wente SR, Rout MP. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harbor Perspect. Biol. 2:a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Whitehurst AW, et al. 2002. ERK2 enters the nucleus by a carrier-independent mechanism. Proc. Natl. Acad. Sci. U. S. A. 99:7496–7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wild P, et al. 2009. Exploring the nuclear envelope of herpes simplex virus 1-infected cells by high-resolution microscopy. J. Virol. 83:408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang PW, Chang SS, Tsai CH, Chao YH, Chen MR. 2008. Effect of phosphorylation on the transactivation activity of Epstein-Barr virus BMRF1, a major target of the viral BGLF4 kinase. J. Gen. Virol. 89:884–895 [DOI] [PubMed] [Google Scholar]
- 62. Yokoya F, Imamoto N, Tachibana T, Yoneda Y. 1999. beta-Catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell 10:1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoon S, Seger R. 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 24:21–44 [DOI] [PubMed] [Google Scholar]
- 64. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]
- 65. Yue W, Gershburg E, Pagano JS. 2005. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J. Virol. 79:5880–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu J, et al. 2009. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J. Virol. 83:5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]