Abstract
Herpes simplex virus type 1 (HSV-1) enters cells by fusion of its envelope with a host cell membrane, which requires four viral glycoproteins and a cellular receptor. Viral fusion glycoprotein B (gB) mediates membrane fusion through the action of its ectodomain, while its cytoplasmic domain (cytodomain) regulates fusion from the opposite face of the membrane by an unknown mechanism. The gB cytodomain appears to restrict fusion, because point or truncation mutations within it increase the extent of fusion (syn mutations). Previously, we showed that the hyperfusion phenotype correlated with reduced membrane binding in gB syn truncation mutants and proposed that membrane binding was important in regulating fusion. Here, we extended our analysis to three syn point mutants: A855V, R858H, and A874P. These mutations produce local conformational changes, with some affecting membrane interaction, which suggests that while syn mutants may deregulate fusion by somewhat different mechanisms, maintaining the wild-type (WT) conformation is critical for fusion regulation. We further show that the presence of a membrane is necessary for the cytodomain to achieve its fully folded conformation and propose that the membrane-bound form of the cytodomain represents its native conformation. Taken together, our data suggest that the cytodomain of gB regulates fusion by a novel mechanism in which membrane interaction plays a key role.
INTRODUCTION
Herpes simplex virus type 1 (HSV-1) causes diseases ranging from mild skin lesions to fatal encephalitis and neonatal herpes (28). Like all herpesviruses, HSV-1 enters host cells by fusion of the viral envelope with the plasma membrane or an endocytic vesicle. While the specific viral glycoproteins required for entry differ among herpesviruses, glycoprotein B (gB) and the heterodimer gH/gL are conserved. Entry of HSV-1 requires gD, gB, and gH/gL, in addition to a host cell receptor (Fig. 1A). These five proteins are also sufficient to mediate fusion of transiently transfected cells in the absence of any other viral proteins (47).
Fig 1.
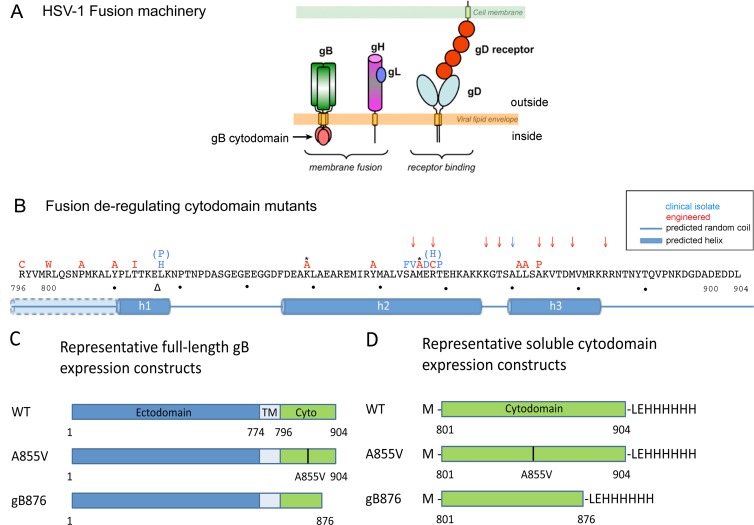
The HSV-1 gB cytodomain is important for fusion regulation. (A) Schematic representation of the five proteins necessary for membrane fusion: viral glycoproteins gB, gD, gH, and gL and the cellular gD receptor. (B) Known syn point mutations (letters), truncations (arrows), and insertions (triangles) in HSV-1 and HSV-2 gB are superimposed on the primary amino acid sequence of the HSV-1 gB cytodomain. Asterisks indicate mutations in HSV-2 where the WT amino acid (not shown) differs from the HSV-1 sequence. Predicted secondary-structure elements (JPred) are shown below the sequence. Helix h1 is predicted to be contiguous with the transmembrane helix when the latter is included in secondary-structure predictions, as indicated by the dotted line and partial shading of helix h1 for residues 796 to 810. (C) Representative constructs used for expression of full-length gB proteins in CHO cells. (D) Representative constructs used to express His6-tagged gB cytodomains in E. coli.
gD initiates entry by binding one of its three cellular receptors (44). The exact role of gH/gL in entry remains unclear; however, recent evidence suggests that binding of a receptor by gD activates gH/gL, which in turn activates gB for fusion (3). gB is a class III viral fusion protein that enables membrane fusion, presumably by undergoing dramatic conformational changes that provide the energy to drive fusion (25, 29). However, unlike fusion proteins of other enveloped viruses, e.g., influenza virus hemagglutinin (HA) or HIV gp160, which accomplish fusion independently (15), HSV-1 gB mediates fusion only in the presence of gD, a gD receptor, and gH/gL (39, 47). gB is a trimeric, type I transmembrane protein with an N-terminal ectodomain displayed on the virion surface, a single-pass transmembrane helix, and a C-terminal cytoplasmic domain (cytodomain) situated on the opposite face of the viral envelope (29) (Fig. 1A). The ectodomain is presumably directly involved in the membrane fusion process: based on the model developed from studies of other viral fusion proteins, its hydrophobic fusion loops are thought to insert into the host cell membrane, bringing the virus and host cell into proximity for fusion (24). In contrast, the cytodomain is thought to play a regulatory role in fusion (41).
Certain mutations within the gB cytodomain enhance membrane fusion and are referred to as syncytial (syn) mutations. These mutations, naturally occurring or engineered (Fig. 1B), can be separated into three classes: point mutations, insertions, and C-terminal truncations (4, 6, 7, 14, 17, 19, 20, 33). While syn mutations are found throughout the cytodomain, naturally occurring mutations cluster in two small regions, or “hot spots” (Fig. 1B). Infection with syn mutant viruses causes formation of multinucleated cells, or syncytia, not generally observed in infections with wild-type (WT) virus (16). Furthermore, in a virus-free cell fusion system, syn gB proteins generate larger syncytia than wild-type gB, a phenotype referred to as hyperfusion. Given that point or truncation syn mutations in the cytodomain increase the extent of fusion, the wild-type cytodomain is thought to restrict fusion whereas syn mutations release this inhibitory effect. Not all cytodomain mutations are hyperfusogenic; a fusion-abrogating mutation caused by truncation at residue 851 has also been previously described (7). This mutant fails to complement an HSV-1 gB-null virus (7), and a comparable truncation in HSV-2 gB fails to induce cell fusion (18). The phenotype of this truncation suggests that the cytodomain plays an essential role in fusion beyond simply restricting it.
Regulation of fusion by the gB cytodomain is an important part of the multilevel regulatory mechanism that controls the complex fusion machinery of herpesviruses. The entry machinery of herpesviruses has been extensively studied, and the crystal structures of the ectodomains of all four proteins required for HSV-1 entry, gD, gB, and gH/gL, are now known (8, 9, 29, 31). And yet, how the cytodomain functions in fusion remains unclear. Previously, we showed that the individually expressed, trimeric cytodomain associates with membranes, with a concomitant increase in helicity (10). Hyperfusogenic C-terminal truncations of the cytodomain reduced membrane association while inducing smaller conformational changes relative to the WT. This led us to hypothesize that membrane association may play an inhibitory role in fusion (10). However, whether hyperfusogenic point mutations deregulate fusion by the same mechanism was not addressed; and yet, point mutations represent the majority of spontaneously occurring syn mutations.
Here, we extended our mutant analyses to include three syn point mutations: A855V, R858H, and A874P. We found that syn truncation and point mutants generated comparable hyperfusion phenotypes that were not due to the overexpression of mutant gB proteins on the cell surface or to a different, potentially more fusogenic, ectodomain conformation. While some mutants mediated low-level fusion in the absence of gH/gL or gD, all mutants required the presence of gD and gH/gL to achieve the hyperfusion phenotype, suggesting that, like WT gB, syn mutants appear to be under the control of the other entry glycoproteins. Unlike truncations, none of the point mutations reduced the extent of membrane association; in fact, two point mutants bound membranes better than the WT. At the same time, we found evidence of local conformational changes in each of the syn mutant cytodomains. Together, these data suggest that while syn mutants may deregulate fusion in different ways, some of which possibly involve changed extent of membrane association, the WT conformation of the cytodomain must be maintained for proper fusion regulation. Importantly, we found that membrane was required for the cytodomain to adopt its fully folded conformation, characterized by a proteolytically resistant core encompassing residues 807 to 872. We propose that the membrane-bound form of the cytodomain is its fully folded, native conformation and present a structural model for the native cytodomain. The large extent of the conformational changes in the gB cytodomain induced upon membrane binding has not been previously seen with cytodomains of other viral fusion proteins. Taken together, our data hint at a unique mechanism of fusion regulation by the gB cytodomain in which membrane interaction plays a key role.
MATERIALS AND METHODS
Reagents.
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphate monosodium salt (PA) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) were purchased from Avanti Polar Lipids, Inc. Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK)-treated chymotrypsin and (l-tosylamido-2-phenyl)ethyl chloromethyl ketone (TPCK)-treated trypsin were purchased from Worthington. Staphylococcus aureus protease V8 was a gift from C. A. Kumamoto.
Plasmids.
Plasmids pPEP98, pPEP99, pPEP100, and pPEP101 carry the full-length HSV-1 (strain KOS) gB, gD, gH, and gL genes, respectively, in a pCAGGS vector and were gifts from P. G. Spear (39). Plasmids pCAGT7 (carrying the T7 polymerase gene) and pT7EMCLuc (carrying the firefly luciferase gene) (38) were also gifts from P. G. Spear. Plasmid pSC386 carrying the HVEM gene (13) and pCAGGS vector were gifts from G. H. Cohen and R. J. Eisenberg. Plasmid pmCherry-C1, a Clontech pAcGFP1-C1 vector carrying mCherry in place of enhanced green fluorescent protein (GFP), was a gift from R. R. Isberg.
Cells.
CHO cells, a gift from J. M. Coffin, were grown in Ham's F-12 media supplemented with 10% fetal bovine serum (FBS) at 37°C in the presence of 5% CO2.
Abs.
IgGs for the HSV-1 gB mouse monoclonal antibodies (MAbs) C226, SS10, SS55, SS144, and SS145 and the rabbit polyclonal antibody (pAb) R68 were gifts from G. H. Cohen and R. J. Eisenberg. A hybridoma cell line expressing the anti-HSV-1 gB MAb DL16 was also a gift from G. H. Cohen and R. J. Eisenberg. DL16 IgG was purified from the hybridoma supernatant by the use of protein A resin at the Tufts Medical Center GRASP facility. Goat anti-rabbit horseradish peroxidase (HRP)-conjugate and goat anti-mouse HRP-conjugate antibodies were purchased from Bio-Rad. Anti-His5 HRP-conjugate Ab was purchased from Qiagen, and anti-actin mouse MAb was a gift from J. Mecsas. Goat anti-rabbit fluorescein isothiocyanate (FITC)-conjugated pAb was purchased from MP Biomedicals, and Alexa-Fluor 488-conjugated goat anti-mouse IgG antibody was purchased from Invitrogen.
Construction of gB cytodomain mutants.
All gB sequences in this work were derived from the HSV-1 KOS strain. The cytoplasmic domain of gB spans amino acid residues 796 to 904; however, soluble cytodomain protein is proteolytically cleaved after residue 800 during expression in Escherichia coli. Therefore, all soluble cytodomain constructs in this work began with residue 801, preceded by a methionine. Three separate point mutants of the cytodomain were constructed: A855V, R858H, and A874P. Additionally, three previously constructed truncations of the gB cytodomain, 801–876 (gB876), 801–868 (gB868), and 801–851 (gB851) (10) were used.
For expression of soluble cytodomain protein in E. coli, the mutants were generated in a pET24b background. The WT gB cytodomain nucleotide sequence (residues 801 to 904) was cloned into pET24b, resulting in plasmid pKH52, for expression of soluble, C-terminal His6-tagged fusion protein (10). Point mutations in the cytodomain were generated by site-directed mutagenesis of pKH52 plasmid, resulting in plasmids pJLS7 (A855V), pJLS6 (R858H), and pKH63 (A874P). Construction of cytodomain truncations in the pET24b background has been described previously (10).
For expression of full-length gB in CHO cells, all mutants were generated in a pCAGGS background. Point mutations in the cytodomain of the full-length gB gene were generated from pPEP98 by “splicing by overlap extension” PCR (SOE PCR) (27). Forward primer 5′-CTTTGACGAGGCCAAGCTAGCCG-3′ (the NheI site is underlined) was used to generate the 5′ SOE PCR fragment in cloning of all constructs. The reverse primers were 5′-GCGCTCCATGACCGACACCAGG-3′ (A855V mutation), 5′-GTGTTCCGTGTGCTCCATGGCC-3′ (R858H mutation), and 5′-ATGTCGGTGACCTTGGGGCTGAGCA-3′(A874P mutation). Reverse primer 5′-CAGAGGGAAAAAGATCTGCTAGAC-3′ (the BglII site is underlined) was used to generate the 3′ SOE PCR fragment in cloning of all constructs. The forward primers were 5′-CCTGGTGTCGGTCATGGAGCGC-3′ (A855V mutation), 5′-GTGTTCCGTGTGCTCCATGGCC-3′ (R858H mutation), and 5′-TGCTCAGCCCCAAGGTCACCGACAT-3′(A874P mutation). The resulting PCR products were subcloned into NheI and BglII restriction sites of pCAGGS to yield plasmids pJLS9 (A855V), pJLS8 (R858H), and pJLS3 (A874P). Truncations of the cytodomain were amplified by PCR from the full-length gB gene in pPEP98. The same forward primer (5′-AGGAGGGCGGCGACTTTGACGA-3′) was used for all constructs. The reverse primers (a BglII site is underlined) were 5′-AAGATCTTCAGACCTTGGCGCTG-3′ (gB876), 5′-GGAAAAAGATCTTCAGGTGCCCTTCT-3′ (gB868), and 5′-GGAAAAAGATCTTCAGGCCATGTACCG-3′ (gB851). The resulting PCR products were subcloned into NheI and BglII restriction sites of pCAGGS to yield plasmids pJLS12 (gB876), pJLS11 (gB868), and pJLS10 (gB851).
Cell fusion assay.
Cell fusion was assessed using a luciferase reporter gene activation assay (12, 38, 39). CHO cells growing in 24-well plates were transfected with 200 ng each of gB (pPEP98, pJLS3, pJLS8, pJLS9, pJLS10, pJLS11, or pJLS12), gD (pPEP99), gH (pPEP100), gL (pPEP101), and T7 polymerase (pCAGT7) in 200 μl of Opti-MEM with 5 μl of Lipofectamine 2000 (Invitrogen) (“effector cells”). CHO cells seeded in 6-well plates were transfected with 800 ng each of pT7EMCLuc and pSC386 in 1 ml of Opti-MEM with 10 μl of Lipofectamine 2000 (“target cells”). Five hours posttransfection, media were replaced with Ham's F12 medium. After 1 h, target cells were trypsinized and cocultivated with effector cells growing in 24-well plates for 18 h. Cells were washed with phosphate-buffered saline (PBS) and lysed in 200 μl of 1× lysis reagent (Promega). Lysate (190 μl) was combined with 100 μl of luciferase substrate (Promega), and luciferase production was assessed by luminometry. As a control for background levels of fusion, all plasmids except pCAGGS and pCAGT7 were omitted (“vector control”). In each experiment, light output from the vector control was subtracted from each sample (performed in triplicate); values are expressed as a percentage of WT. Values reported here represent averages of the results of at least three experiments. To test whether hyperfusogenic point or truncation mutants could fuse in the absence of other glycoproteins, plasmid pPEP99 or both pPEP100 and pPEP101 or all three plasmids were omitted.
CELISA.
Surface expression of full-length gB bearing hyperfusogenic point mutations was assessed using a competitive enzyme-linked immunosorbent assay (CELISA) (21, 24). CHO cells were seeded in 96-well plates and grown for 24 h in Ham's F12 media. Cells were then transfected with 50 ng each of gB (pPEP98, pJLS3, pJLS8, or pJLS9), gD (pPEP99), gH (pPEP100), gL (pPEP101), and T7 polymerase (pCAGT7) in 50 μl of Opti-MEM media with 0.5 μl of Lipofectamine 2000. All plasmids except pCAGGS were omitted as a vector control. After 5 h, media were replaced with 100 μl of Ham's F12 media, and cells were grown for 48 h before being fixed with 3% paraformaldehyde–PBS overnight at 4°C. Fixed cells were washed twice in PBS and incubated in primary antibody (R68), diluted 1:1,000 in PBS supplemented with 10% FBS (CELISA buffer), for 1 h at room temperature. Cells were washed three times with PBS and incubated in secondary antibody (goat anti-rabbit HRP conjugate; 1:1,000 in CELISA buffer) for 1 h at 4°C. After three washes with PBS, cells were washed with 20 mM sodium citrate (pH 4.5). 2,2′-Azino-di(3-ethylbenzthiazoline) sulfonic acid (ABTS) peroxidase substrate was added to cells, the plate was incubated in aluminum foil at room temperature for 2 h, and the absorbance at 405 nm was recorded. In each experiment, absorbance from the vector control was subtracted from each sample (present in triplicate). The results of triplicate samples were averaged, and the absorbance is expressed as a percentage of WT. Averages of the results of at least three experiments are reported here.
FACS.
Surface expression and ectodomain conformation of gB expressed in CHO cells were assessed by fluorescence-activated cell sorting (FACS) (30). CHO cells were seeded at a density of 0.6 × 106 cells/well in 6-well dishes 24 h prior to transfection. Cells were transfected with 2 μg of gB (pPEP98, pJLS3, pJLS8, pJLS9, pJLS10, pJLS11, or pJLS12) and 667 ng of pmCherry-C1 in 0.5 ml of Opti-MEM with 2 μl of Lipofectamine 2000 for 5 h, followed by incubation in 1 ml of growth media for 20 h. Cells were trypsinized and resuspended in 1 ml of PBS–3% FBS (FACS media). Cells (1 × 106) were transferred to a 5-ml FACS tube, washed with 1 ml of FACS media, and incubated with 50 μl of primary antibody (1:1,000 in FACS media) for 1 h at 4°C. The primary antibody was either the rabbit pAb R68 or a mouse MAb (DL16, C226, SS10, SS55, SS144, or SS145). Cells were washed and incubated in 50 μl of secondary antibody (1:500 in FACS media) covered with aluminum foil for 1 h at 4°C. The secondary antibody was either goat anti-rabbit FITC-conjugated Fc antibody or Alexa-Fluor 488-conjugated goat anti-mouse IgG antibody. Cells were washed, resuspended in 300 μl of FACS media, and analyzed using a Becton, Dickinson LSRII machine. In analyzing our FACS data, we made the assumption that the majority of cells expressing mCherry also received gB plasmid because mCherry was transfected at a 1:3 ratio with gB. FACS data were analyzed as follows. First, a transfected cell population was identified by mCherry fluorescence. Next, a subpopulation of cells with bound secondary antibody, indicating gB expression, was isolated based on FITC fluorescence. To correct for nonspecific binding of both the primary and secondary antibodies, the number of FITC-positive cells in vector-transfected samples was subtracted from the number of FITC-positive cells obtained from samples transfected with gB. For gB expression (R68), data are expressed relative to WT expression. For conformation data (MAbs), the percentage of mCherry- and FITC-positive cells, corrected for vector expression, is reported. All data represent averages of the results of at least three experiments.
Overall gB expression assessed by Western blot analysis.
Expression of gB in CHO cells was assessed by Western blot analysis (43). CHO cells in 1 well of a 6-well dish were transfected with 2 μg of either pPEP98 or pJLS10 as described for FACS experiments. At 22 h posttransfection, cells were trypsinized, washed with 1 ml of PBS, and lysed in 100 μl of reducing buffer by boiling at 95°C for 10 min. Aliquots (20 μl) of lysate were analyzed by gel electrophoresis on a 10% polyacrylamide gel, and the presence of gB was assessed by Western blot analysis using the R68 pAb at a 1:5,000 dilution. The lower portion of the blot was probed with anti-actin mouse MAb (1:10,000) as a loading control. Following incubation in the primary antibodies for 1 h at room temperature, the blots were incubated in goat anti-mouse IgG horseradish peroxidase-conjugated secondary pAb (1:10,000 dilution) for 40 min and developed using a Pierce Western Pico chemiluminescence kit.
Cytodomain protein expression and purification.
Soluble cytodomain proteins were expressed in E. coli Rosetta pLysS cells (Novagen) (10). Freshly transformed Rosetta cells were grown at 37°C in LB supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol at an optical density at 600 nm (OD600) of approximately 0.6. Protein production was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 3 h, cells were harvested and lysed using a French press cell with 20 mM Tris-HCl (pH 8.0), 250 mM NaCl, 3 mM β-mercaptoethanol (BME), and 0.1 M phenylmethylsulfonyl fluoride (PMSF) (lysis buffer). Protein was purified from cell lysates by the use of a 2-ml Ni-Sepharose 6B fast-flow column (GE Healthcare). The column was washed sequentially with 10 column volumes of lysis buffer containing 0, 6, and 20 mM imidazole. Protein was eluted with lysis buffer containing 300 mM imidazole, and 1 mM EDTA was immediately added to the eluate. Eluate was concentrated and imidazole removed by buffer exchange to 20 mM Tris-HCl (pH 8.0), 250 mM NaCl, 1 mM EDTA, and 0.1 mM PMSF in an Ultra-4 10-kDa-cutoff concentrator (Millipore). Concentrated eluate was further purified by size exclusion chromatography (SEC) using a Superdex 200 column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM EDTA (GF buffer). The Superdex 200 column was calibrated with thyroglobulin (670 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), and ovalbumin (44 kDa), and the void volume was determined using blue dextran (GE Healthcare). Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue G250 staining.
Preparation of liposomes.
Small unilamellar vesicles (SUVs) containing a 1:1 molar ratio of PC:PA were prepared as previously described (10), with some modifications. Chloroform lipid stocks (10 mg/ml) were combined to reach a final concentration of 3 mM and dried into a layered film in a glass vial under vacuum conditions. Lipid layers were further dried by desiccation for 30 min and then hydrated in GF buffer at room temperature overnight. Hydrated lipids were sonicated in an Eppendorf tube using a Branson Sonifier sonicator (duty cycle, 40%; output 4), pulsing in 30-s intervals until the suspension became clear. SUVs were separated from large unilamellar vesicles (LUVs) and sonicator tip titanium debris in a tabletop centrifuge at 18,000 × g for 20 min at room temperature. SUVs were stored at 4°C and used within 1 week. SUVs generated by this method typically have a diameter of 20 nm to 50 nm (32). LUVs containing a 1:1 molar ratio of PC:PA were prepared as described above for SUVs through the hydration step. Hydrated lipids were then downsized to a diameter of 0.1 μm by the extrusion method using an Avanti Polar Mini-Extruder, per the manufacturer's protocol. LUVs were separated from debris in a tabletop centrifuge at 18,000 × g for 5 min at room temperature. LUVs were stored at 4°C and used within 3 weeks.
Gel filtration with liposomes.
The ability of soluble gB cytodomain protein to bind membrane mimetics was assessed by liposome coelution on SEC with SUVs (10). Soluble gB cytodomain protein (2 mg/ml) and a 30-molar excess of PC:PA SUVs were incubated at room temperature for 1 h. The protein/liposome mixture was then applied to a Superose 12 10/30 HR column (GE Healthcare) at 4°C. The elution profiles of gB cytodomain protein alone and liposomes alone were separately determined on the Superose 12 column as controls. Eluate was collected in 0.5-ml fractions, and equal volumes from the void and protein peak fractions were analyzed by SDS-PAGE. The amount of protein in each fraction was quantified by densitometry analysis of scanned gel images. The percentage of protein bound to liposomes was calculated as the ratio of protein in the void peak to the total amount of protein present in both peaks. Experiments were performed in triplicate, with the mean value reported here.
Liposome coflotation.
Association of gB cytodomain protein with membranes was also assessed by liposome coflotation with LUVs (24, 43). Cytodomain proteins (3 μg) were incubated at 37°C for 1 h in 50 μl of PBS with or without 25 μg of LUVs. KCl was added to 1 M to reduce any nonspecific electrostatic interactions, and samples were incubated for an additional 15 min at 37°C. Samples were adjusted to 40% sucrose solution by adding 45% sucrose solution–PBS to reach a final volume of 500 μl. These samples were placed at the bottom of a 5-ml centrifuge tube and overlaid with 4 ml of 25% sucrose solution–PBS and 500 μl of 5% sucrose solution–PBS. The samples were next centrifuged in a Beckman SW-55 Ti rotor at 246,000 × g for 3 h at 4°C, and 700-μl fractions were removed from each gradient, beginning at the top. Fractions were pooled into three groups: top (fractions 1 and 2), middle (fractions 3 to 5), and bottom (fractions 6 to 7). To determine the localization of gB cytodomain protein, protein was precipitated from each pool using 20% trichloroacetic acid (TCA) and analyzed by SDS-PAGE on 15% polyacrylamide gels stained with Coomassie blue. TCA precipitation of the truncation mutant gB851 was inefficient; therefore, this gel was transferred onto a nitrocellulose membrane for Western blot analysis. The membrane was blocked with 0.1% milk–20 mM Tris·HCl (pH 7.5)–500 mM NaCl–0.5% Tween 20 (TBST) for 40 min at room temperature and then incubated with anti-His5 HRP-conjugated MAb at a 1:3,000 dilution for 1 h. The blot was developed using a Pierce Western Pico chemiluminescence kit. To determine the distribution of LUVs throughout the gradient, we used a 1,6-diphenyl-1,3,5-hexatriene (DPH) fluorescence assay. LUVs (25 μg) were spun in a gradient and fractionated as described above. Then, 300 μl of each fraction was placed in a white 96-well plate along with 300 μl of 25% sucrose solution and 300 μl of 25% sucrose solution containing 25 μg of LUVs (negative and positive controls, respectively). Finally, DPH was added to each well to reach a concentration of 10 μM and the fluorescence recorded.
CD.
The effect of liposome binding on the secondary structure of soluble gB cytodomain protein was assessed using circular dichroism (CD) spectroscopy (10). gB cytodomain protein (0.2 mg/ml)–20 mM sodium phosphate buffer (pH 8.0)–100 mM NaF (CD buffer) was incubated either alone or in combination with a 30 M excess of PC:PA SUVs at room temperature for 30 min. The far-UV CD spectrum of the protein was then measured at room temperature on a JASCO model 810 spectropolarimeter using a 50-nm/s scan speed, 1-nm band pass value, and 1-s response time. For each sample, three scans were collected and data averaged, a buffer blank spectrum was subtracted, and the sample was processed for noise elimination. Measurements are presented in units of mean residual ellipticity (ϴ). Protein helical content was estimated from the value of ϴ at 222 nm (ϴ222) using the following formula: percent helix = [(ϴ222 − ϴ2220)/(ϴ222H − ϴ2220)] × 100% (34). The ϴ222 value used for 100% helix was ϴ222H = −38,000° cm2 dmol−1, and the value for 100% random coil was ϴ2220 = 4,000° cm2 dmol−1 (34).
For urea denaturation, 40 μg of cytodomain protein (WT, A855V, or R858H) in CD buffer was incubated with a 30 M excess of PC:PA SUVs at room temperature for 30 min. Urea buffer (8 M urea in 20 mM sodium phosphate buffer [pH 8.0] and 100 mM NaF) was then added to each sample to reach the final urea concentration of between 0 and 7 M in 0.5 M increments, and the final sample volume was 200 μl. Samples were allowed to equilibrate at room temperature for 3 h and then put on ice and the CD spectra at 4°C recorded as described above. The fraction of protein folded (α) was determined using the following equation: α = [ϴi − ϴU]/[ϴF − ϴU], where ϴi is the value of ϴ at 222 nm for a given sample, ϴU is the ϴ222 value when the protein is unfolded (7 M urea), and ϴF is the ϴ222 value when the protein is folded (0 M urea) (22). The folding constant of a trimeric protein (KF) was calculated from the following equation: KF = α/[3Pt(1 − α)]3, where Pt is the molar concentration of the trimer. Free energy of folding was determined from the following formula: ΔGFolding = −RTlnKF, where R is the gas constant in units of cal K−1 mol−1 and T is the temperature in Kelvin (22). To estimate the free energy of folding in the absence of urea, the fraction of protein unfolded verses the urea concentration was plotted, and ΔGFolding values for samples in the transition region of this curve were plotted against the urea concentration. Extrapolation of the best fit line for the latter plot yielded the estimated ΔGFolding value at 0 M urea (26).
Limited proteolysis.
Cytodomain protein (3 μg of WT, A855V, R858H, A874P, or R882S/R884S) in 8 μl of GF buffer was incubated for 1 h at room temperature with 1.5, 3, 6, 15, 30, 60, or 120 ng of TLCK-treated chymotrypsin (Worthington), TPCK-treated trypsin (Worthington), or V8 protease (Sigma). These amounts correspond to protease/protein ratios of 1:2,000, 1:1,000, 1:500, 1:200, 1:100, 1:50, and 1:25, respectively. Samples were spun briefly at 18,000 × g in a tabletop centrifuge prior to incubation. For samples containing LUVs, cytodomain protein was incubated with a 30 M excess of LUVs for 1 h at room temperature prior to the addition of protease. Digestion reactions were stopped by adding 8 μl of 2× reducing tricine sample buffer and boiling at 95°C for 5 min. Denatured, reduced proteolytic products were resolved by gel electrophoresis on 10% Tris-tricine gels, followed by staining with Coomassie blue G250. For mass spectrometry analysis, digests with a protease/protein ratio of 1:1,000 (without LUVs) or 1:500 (with a 30 M excess of LUVs) were scaled up to 75 μg of protein. Digestion reactions were quenched by precipitation with acetone (protein alone) or a mixture of 90% acetone and 10% methanol (protein with LUVs) to dissolve liposomes and dried at room temperature overnight before mass spectrometry analysis. Digested samples were then dissolved in an acetonitrile-water mixture acidified with 0.1% TFA and analyzed by direct infusion electrospray ionization quadrupole Fourier transform ion cyclotron resonance (ESI-Q-FT-ICR) mass spectrometry (Bruker) (9.4-T magnet).
RESULTS
Choice of hyperfusogenic mutants.
Biochemical properties of the WT cytodomain, syn truncation mutant cytodomains gB876 and gB868, and the fusion-null mutant cytodomain gB851 (the listed amino acid corresponds to the last residue in the truncated protein) were previously characterized by our laboratory (10). To determine whether the same biochemical properties underlie the hyperfusion phenotype of gB syn point and truncation mutants, we chose to characterize the hyperfusogenic point mutants A855V, R858H, and A874P (Fig. 1B and C). The first two point mutations were found in clinical isolates (6, 17), while the hyperfusogenic A874P mutation was previously engineered to break the predicted helix h3, believed to play a role in fusion regulation (Fig. 1B) (19).
Quantification of cell fusion induced by hyperfusogenic point and truncation mutants.
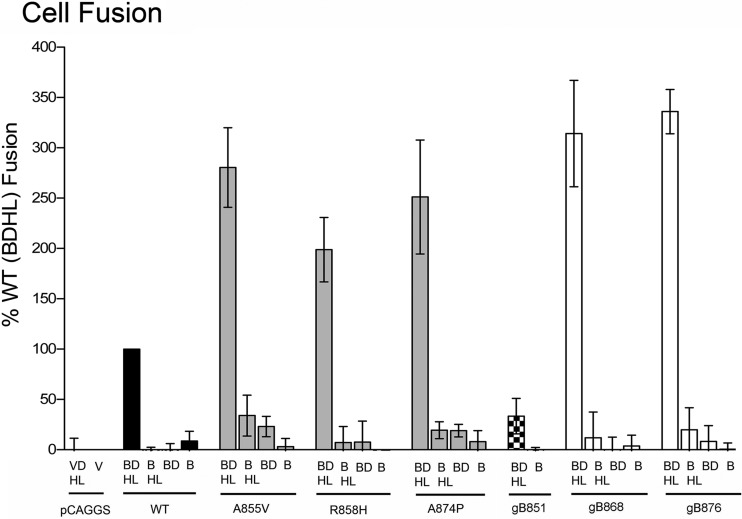
The fusion phenotypes of three point mutants studied in this work (A855V, R858H, and A874P) and the three truncation mutants (gB851, gB868, and gB876) were previously determined by counting the average number of nuclei per syncytium in transiently transfected mammalian cells or by observing plaque formation in cells infected with mutant viruses (4, 6, 7, 17, 19). To confirm the fusion phenotypes of all six mutants (Fig. 1C), we tested them in a quantitative luciferase cell fusion assay (12, 38, 39). All previously characterized syn mutants (gB868, gB876, A855V, R858H, and A874P) were hyperfusogenic, exhibiting a 2-to-3-fold increase in fusion relative to the WT protein (Fig. 2). In contrast, the truncation mutant gB851 mediated only ∼30% of WT fusion (Fig. 2).
Fig 2.
Syn point and truncation mutants mediate comparable levels of cell fusion. Luciferase production was used to quantify fusion of CHO cells mediated by WT gB, truncated gB, or gB bearing a syn point mutation, in the presence or absence of gD and gH/gL. Transfected glycoproteins (abbreviated as B, D, H, and L) are indicated below each bar, and the gB construct used is indicated below each set of bars. In the “vector” control (V), all plasmids except pCAGGS and pCAGT7 were omitted. In each experiment, the vector control was subtracted from each sample (performed in triplicate), and the values are expressed as a percentage of WT. Reported values represent averages of the results of at least three experiments.
gB cytodomain mutants require gD, gH, and gL for hyperfusogenicity.
A hyperfusogenic truncation mutant of gB from Epstein-Barr virus (EBV) (23, 36), corresponding to HSV-1 gB864, has been previously found to mediate ∼30% of WT fusion in the absence of EBV gH/gL, which is normally required for fusion (36). We tested whether any HSV-1 gB syn point or truncation mutants studied here could mediate fusion in the absence of HSV-1 gD or gH/gL or both.
As expected, in the absence of either gD or gH/gL, WT gB had background fusion levels (Fig. 2). In contrast, several hyperfusogenic mutants exhibited low-level fusion even in the absence of gD or gH/gL (Fig. 2). Both the A855V and A874P mutants mediated ∼30% of WT fusion in the absence of gD and ∼20% of WT fusion in the absence of gH/gL, while gB876 mediated ∼20% of WT fusion in the absence of gD. In all cases, however, the standard deviation was high relative to the magnitude of observed fusion, which has also been seen with the hyperfusogenic EBV gB truncation mutant discussed above (36). While the low-level fusion accomplished by some HSV-1 gB syn mutants in the absence of gD or gH/gL may suggest a certain degree of independence from these proteins, all mutants required both gD and gH/gL to achieve high levels of fusion, indicating that they are likely positively regulated by these proteins.
The hyperfusogenic mutations do not increase cell-surface expression of gB.
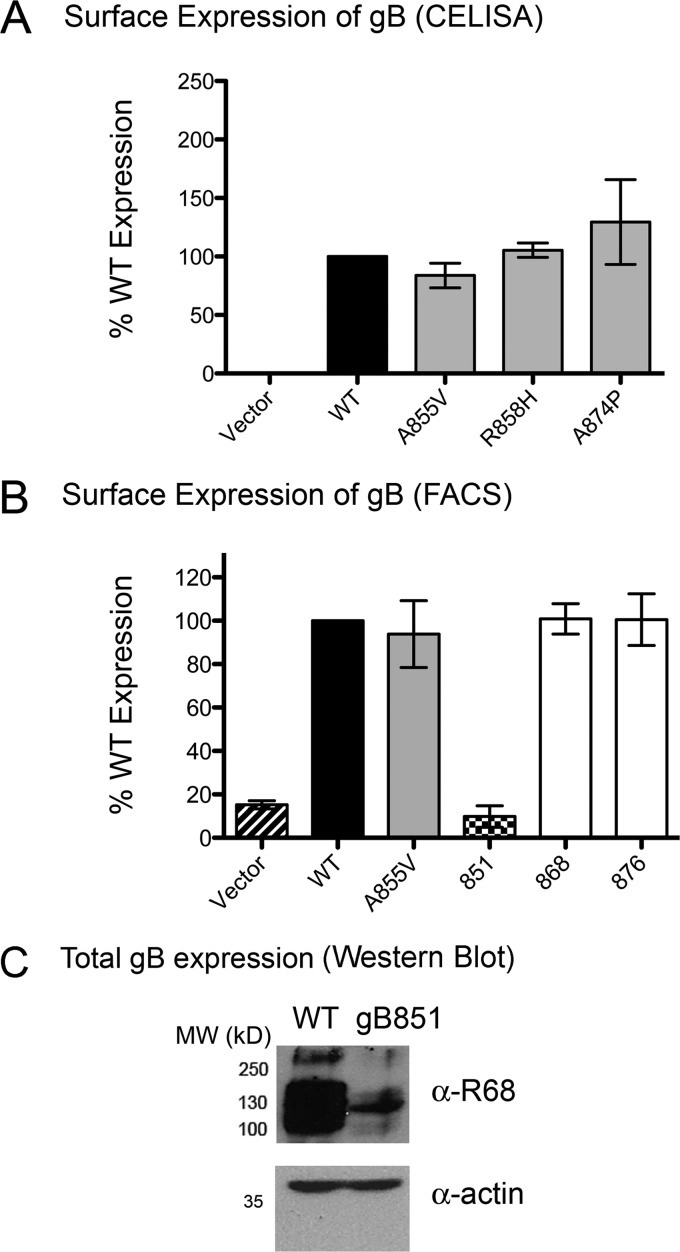
The hyperfusion phenotype of the syn mutants could be due to their overexpression on the cell surface. To test this, we measured the expression of WT and mutant HSV-1 gB proteins on the surface of transiently transfected CHO cells by the use of CELISA (Fig. 3A and Table 1) or FACS (Fig. 3B, Table 1, and Table 2). Point mutants A855V and R858H and both truncation mutants were expressed at WT levels, while point mutant A874P showed 1.3-fold-higher surface expression than the WT (Fig. 3A and B and Table 1). Thus, the hyperfusion phenotype was not accounted for by higher surface expression, in agreement with what was previously seen with other syn mutations in HSV-2 (18, 37, 41).
Fig 3.
Syn mutants are not overexpressed on the cell surface. (A) Surface expression in CHO cells of WT gB and the three syn point mutants was assessed by CELISA with the pAb R68. Nonspecific binding of R68 to cells was measured with cells transfected with pCAGGS vector instead of gB. In each experiment, the vector control was subtracted from each sample (present in triplicate). The results determined with triplicate samples were averaged, and the absorbance is expressed as a percentage of WT (100%). An average of the results of at least three experiments is reported. (B) Surface expression of WT gB, the syn point mutant A855V, and the three truncation mutants was assessed by FACS with the pAb R68. The percentage of transfected cells positive for FITC signal, indicating bound secondary antibody, was measured, and data are presented relative to WT gB (100%). (C) Overall levels of expression of WT gB and gB851 in CHO cell lysates were compared by Western blot analysis using the pAb R68. Actin was used as a loading control. MW, molecular weight (in thousands).
Table 1.
Summary of biochemical and functional findings for the WT, syn mutant, and fusion-null gB cytodomainsa
| Parameter | Value(s) for indicated cytodomain |
||||||
|---|---|---|---|---|---|---|---|
| WT | Truncation mutant |
Point mutant |
|||||
| 876 | 868 | 851 | A855V | R858H | A874P | ||
| Fusion (%) | 100 | 336 ± 22.0 | 314.2 ± 52.9 | 33.4 ± 17.6 | 280.5 ± 39.5 | 198.8 ± 32.0 | 251.2 ± 56.6 |
| CELISA (% WT) | 100 | ND | ND | ND | 83.8 ± 10.5 | 105.4 ± 6.2 | 129.6 ± 36.3 |
| FACS (% WT) | 100 | 100.5 ± 11.9 | 100.9 ± 7.0 | 9.8 ± 5.0 | 93.8 ± 15.4 | ND | ND |
| Membrane binding (% total protein) | 43.7 ± 7.7 | 34.2b | 21.7 ± 5.3 | 12b | 88.2 ± 20.9 | 47.1 ± 6.5 | 60.2 ± 9.6 |
| Increase in helicity upon membrane binding (%) | 19.4 ± 3.9 | 7.2 | 14.6 ± 1.8 | 0.4 ± 1.1 | 19.0 ± 5.7 | 17.0 ± 4.7 | 12.0 ± 6.2 |
| ΔGFolding (kcal/mol) | −21.3 ± 1.0 | ND | ND | ND | −20.7 | −20.9 | ND |
| Trypsin cleavage sites with membrane | K875, R885 | ND | ND | ND | K875, R885 | K807, R844, K875, R885 | K875, R885 |
WT and WT-like results (determined to be within 50% of the WT value for fusion and surface expression or within 1 standard deviation of the mean for membrane binding, increased helicity, and free energy of folding) are shown in regular font. Lower-than-WT results are shown in italics; higher-than-WT results are shown in bold. ND, not determined.
Data are from reference 10.
Table 2.
Expression and ectodomain conformation of gB on the surface of CHO cells by FACS
| Sample with full-length gB | % cells transfecteda | % transfected cells with bound secondary antibodyb |
||||||
|---|---|---|---|---|---|---|---|---|
| R68 | DL16 | C226 | SS10 | SS55 | SS144 | SS145 | ||
| Vector | 84.1 ± 2.3 | 4.6 ± 1.9 | 3.2 ± 1.9 | 3.4 ± 2.2 | 3.8 ± 1.1 | 2.8 ± 1.3 | 2.3 ± 2.2 | 3.1 ± 2.1 |
| WT | 85.1 ± 4.8 | 28.2 ± 3.8 | 34.9 ± 7.7 | 56.7 ± 6.5 | 57.1 ± 6.3 | 44.6 ± 7.2 | 41.7 ± 2.5 | 50.6 ± 8.1 |
| gB876 | 87.7 ± 1.9 | 27.2 ± 2.1 | ND | ND | ND | ND | ND | ND |
| gB868 | 84.5 ± 4.7 | 29.6 ± 3.3 | 37.0 ± 9.6 | 55.5 ± 5.9 | 58.4 ± 10.1 | 49.2 ± 10.0 | 49.6 ± 6.4 | 53.5 ± 5.4 |
| gB851 | 87.3 ± 0.9 | 2.7 ± 1.4 | ND | ND | ND | ND | ND | ND |
| A855V | 85.3 ± 1.1 | 27.2 ± 1.7 | 33.1 ± 5.2 | 51.0 ± 5.4 | 55.2 ± 1.5 | 40.0 ± 5.4 | 41.7 ± 8.7 | 50.1 ± 6.4 |
Calculated as the percentage of total cells positive for mCherry, which indicates transfection.
Calculated as the percentage of mCherry-positive cells also positive for FITC signal, which indicates both transfection and binding of secondary antibody. ND, not determined.
Interestingly, gB851, which was impaired for fusion, was expressed on the surface of CHO cells at only ∼10% of WT levels (Fig. 3B and Table 1). Low expression at the cell surface correlated with low overall protein expression (Fig. 3C). Therefore, the low level of fusion observed for the gB851 mutant was due to poor expression. Low expression of gB851 would probably lead to poor incorporation of gB851 into virions, which may explain the inability of gB851 to complement gB-null virus (7).
Hyperfusogenic mutations do not alter the conformation of the ectodomain.
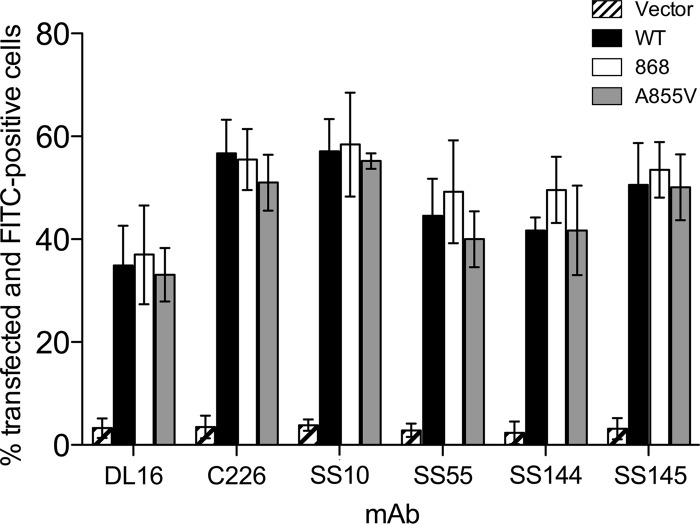
In some viral fusion proteins, such as paramyxovirus F protein and Moloney murine leukemia virus (MoMLV) Env protein, removal of the inhibitory C-terminal sequences in the cytoplasmic domain is thought to activate the ectodomain for fusion by a process termed “inside-out signaling” (1, 35, 48). We hypothesized that syn mutations in the cytoplasmic domain could alter the conformation of the ectodomain, possibly rendering it more fusogenic. Previously, it was shown using CELISA that insertion of five amino acids after either residue T868 or N886 in the cytodomain altered reactivity of the gB ectodomain with some monoclonal antibodies (MAbs) (33). To test this, we expressed WT or syn mutant gB proteins gB868 and A855V in CHO cells and tested their reactivity with a panel of MAbs in a FACS assay. These MAbs recognize conformational epitopes within four of the five domains of the ectodomain (5). Moreover, one antibody, DL16, is trimer-specific and requires intact protomer contacts (5). Thus, if the conformation of the ectodomain in any mutant were different from that of the WT, we would have expected to see a difference in reactivity with at least one conformational MAb.
Each MAb showed slightly different reactivity with the WT gB ectodomain, ranging from 30% to 60% FITC-positive transfected cells (Fig. 4 and Table 2). Overall, there were no significant differences in monoclonal antibody reactivity between the mutants gB868 and A855V and the WT (Fig. 4 and Table 2). Similar results were obtained for mutant R858H (data not shown). These data suggest that there are no significant conformational differences between the ectodomains of WT gB, the truncation mutant gB868, and the point mutants A855V and R858H. Therefore, these three representative mutations in the cytoplasmic domain do not change the conformation of the gB ectodomain, making it more fusogenic. We further hypothesize that most syn mutations do not render the ectodomain more fusogenic.
Fig 4.
Syn gB cytodomain mutations do not alter the ectodomain conformation. MAb reactivity of the ectodomain of gB expressed on the surface of CHO cells was determined by FACS analysis. Cells were transfected with WT gB, the syn truncation mutant gB868, the syn point mutant A855V, or vector. The percentage of transfected cells positive for FITC signal, indicating bound secondary antibody, was measured, and data are presented relative to the total population of transfected cells.
Hyperfusogenic point mutants stably interact with membranes.
Having found no differences in surface expression or the ectodomain conformation between syn mutant and WT gB proteins, we tested whether these mutations instead influenced the conformation or other biochemical properties of the cytodomain itself. To do so, we expressed and purified the cytodomains of all five mutants and the WT gB as soluble proteins with C-terminal His6 tags (Fig. 1D). On the size exclusion column, all point mutant cytodomains eluted at the expected molecular mass of the trimeric species, 38 kDa (data not shown), indicating that, like the WT cytodomain (10), these point mutant cytodomains were trimers in solution.
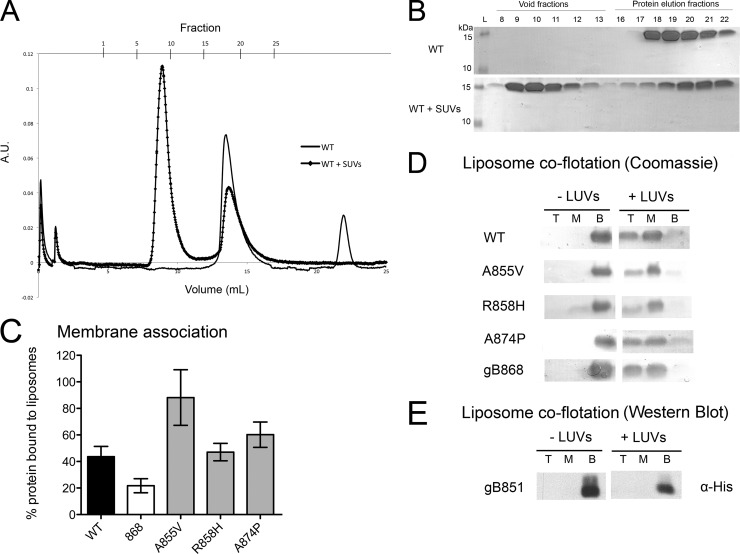
We next tested membrane association of cytodomains by coelution with small unilamellar vesicles (SUVs) by the use of size exclusion chromatography (SEC) (Fig. 5A and B) (10). Consistent with our previous report (10), approximately 44% of WT protein and only 22% of the gB868 truncation mutant coeluted with SUVs (Fig. 5C and Table 1). Point mutant R858H bound SUVs as well as the WT, whereas point mutants A855V and A874P bound SUVs better than the WT, with 88% and 60% of total protein bound, respectively (Fig. 5C and Table 1). Better binding of the A855V mutant could be potentially explained by its increased hydrophobicity.
Fig 5.
Membrane binding by the WT and mutant gB cytodomains. (A to C) Association of soluble WT or mutant cytodomain proteins with anionic SUVs as assessed using liposome coelution in size exclusion chromatography. Soluble WT protein was passed over a Superose 12 column alone (solid line) or following incubation with anionic SUVs (squares). A.U., absorbance units. (B) Representative Coomassie-stained gels of fractions from the chromatogram shown in panel A. The presence of protein in the void fractions indicates binding of the protein to the SUVs. (C) Extent of binding was determined as bound/(bound + unbound) × 100%. (D and E) Stable association of soluble cytodomain proteins with (+) or without (−) anionic LUVs as assessed using liposome coflotation in a sucrose gradient. Total protein was precipitated with TCA out of the top (T), middle (M), and bottom (B) fractions of the gradient, analyzed by SDS-PAGE, and detected by Coomassie staining (D) or by Western blotting performed with anti-His antibody (E).
Using a liposome coflotation assay, we further verified that protein coeluting with SUVs was stably bound to the liposomes rather than simply aggregating in their presence. SUVs are incompatible with the liposome coflotation assay because they do not partition properly in the sucrose gradient, likely due to compression and fusion during centrifugation. Instead, we used large unilamellar vesicles (LUVs) of the same lipid composition, which are more commonly used in coflotation experiments. As expected, in the presence of LUVs, WT cytodomain protein was found in both the middle and the top fractions of the gradient, indicating stable association with the LUVs, and was largely absent from the bottom fraction (Fig. 5D). Localization of the WT protein to the middle fraction was likely due to incomplete partitioning of the liposomes into the top fraction, as seen using 1,6-diphenyl-1,3,5-hexatriene (DPH) fluorescence (data not shown). Our negative control, the fusion-impaired truncation mutant gB851, did not float with LUVs (Fig. 5E), consistent with the lack of binding to SUVs observed by SEC (10). Somewhat surprisingly, all of the cytodomain point mutants as well as the truncation mutant gB868 cofloated with the LUVs, with little, if any, protein remaining at the bottom (Fig. 5D). This discrepancy between the coflotation and coelution assays could have been due to differences in the cytodomain binding to LUVs (coflotation assay) and SUVs (SEC coelution assay). Nevertheless, while it appears that there are no pronounced differences in binding to LUVs, unlike the coelution assay, the coflotation assay does not provide quantitative results. Thus, there could still be differences, even if modest, in binding of mutant and WT cytodomains to LUVs.
In summary, none of the point mutants showed a decreased association with SUVs relative to the WT (Fig. 5C), which contrasts with the result obtained for the truncation mutants (10). Moreover, the point mutants A855V and A874P bound SUVs better than the WT. These results suggest that truncation mutants and point mutants may deregulate fusion by different mechanisms, in which altered membrane association plays an important role. Nevertheless, we cannot rule out the alternative scenario that emphasizes LUV binding results rather than SUV binding results, in which differences in membrane binding are too small to account for the syn phenotype.
Hyperfusogenic point mutants undergo WT-like conformational changes in the presence of membrane.
Secondary-structure algorithms (JPred [11]) predicted three helices in the cytoplasmic domain (Fig. 1B), with approximately 50% overall α-helical content. Previously, we showed that the helical content of the WT cytodomain approached the predicted value only in the presence of anionic membrane mimetics (10), which suggested that in the presence of a membrane, certain regions of the cytodomain underwent conformational changes from random coil to helix. Syn truncation mutants showed a reduced increase in helicity in the presence of membranes, which correlated with lower membrane binding (10).
In the presence of liposomes, two point mutants (A855V and R858H) showed a WT increase in helicity, 17% to 19%, while mutant A874P showed a smaller increase in helicity, 12% (Table 1 and Table 3). The lower α-helical content in the third point mutant, A874P, was probably due to the introduction of a proline within predicted helix h3 (19) that could kink this helix, thereby decreasing its helical CD signal.
Table 3.
Changes in the secondary structure of soluble cytodomains upon association with liposomes
| Soluble cytodomain | Fusion phenotype | PC:PA-to-protein molar ratio | % helicitya | Absolute increase in helicity (%) |
|---|---|---|---|---|
| gB904 (WT) | WT | No lipids | 26.8 ± 1.7 | |
| 30:1 | 46.3 ± 3.9 | 19.4 ± 3.9 | ||
| gB868 | Hyperfusion | No lipids | 28.0 ± 0.7 | |
| 30:1 | 42.6 ± 1.2 | 14.6 ± 1.8 | ||
| gB851 | Fusion-null | No lipids | 21.7 ± 3.2 | |
| 30:1 | 22.1 ± 4.1 | 0.4 ± 1.1 | ||
| A855V | Hyperfusion | No lipids | 26.9 ± 1.8 | |
| 30:1 | 45.9 ± 7.2 | 19.0 ± 5.7 | ||
| R858H | Hyperfusion | No lipids | 26.1 ± 1.2 | |
| 30:1 | 43.2 ± 5.0 | 17.0 ± 4.7 | ||
| A874P | Hyperfusion | No lipids | 25.6 ± 2.5 | |
| 30:1 | 37.6 ± 8.2 | 12.0 ± 6.2 |
% helicity = [(Θ222 − Θ2220)/(Θ222H − Θ2220)] × 100% (34).
Interestingly, in point mutants, there was no correlation between the extent of membrane binding and the magnitude of the increase in helicity (Table 1). While the A855V point mutant showed a WT increase in helicity in the presence of membrane (Table 1), it bound liposomes better than the WT (Fig. 5C and Table 1). Conversely, the A874P point mutant demonstrated a lower-than-WT increase in helicity in the presence of membrane (Table 1), and yet this mutant also bound liposomes better than the WT (Fig. 5C and Table 1). These observations suggest that regions that undergo random coil-to-helix transitions in the presence of membranes are not the only ones contributing to binding and that additional regions must contribute to binding as well. For example, residue A855 may be located either in the region that directly interacts with the membrane or in the region that indirectly contributes to membrane interaction.
The membrane-bound conformation of the cytodomain is protease resistant.
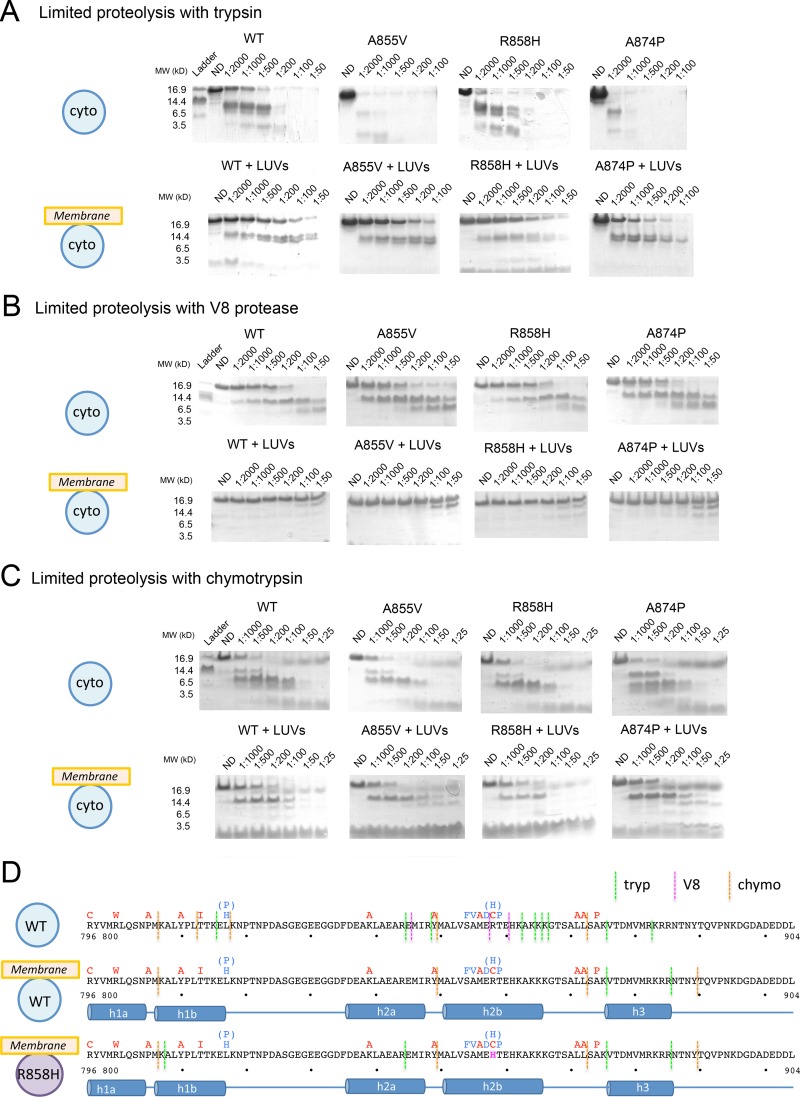
To better map the structural changes that occur in the presence of liposomes, we used limited proteolysis with three proteases with different specificities: trypsin, chymotrypsin, and V8. In the absence of liposomes, the soluble WT cytodomain was highly susceptible to degradation by all three proteases (Fig. 6), which all cleaved throughout the length of the protein. In the presence of liposomes, however, the cytodomain was significantly protected from all three proteases (Fig. 6). Specifically, the entire cytodomain was resistant to V8 cleavage whereas residues 801 to 875 were resistant to trypsin cleavage. Membrane-bound protein was more sensitive to chymotrypsin than to trypsin, with cleavage sites after residues 807, 849, and 872 (Fig. 6A, C, and D). Interestingly, residue Y849, located in the middle of the predicted helix h2, was the site of chymotrypsin cleavage with or without membrane, suggesting that helix h2 likely contains a break near this residue.
Fig 6.
The cytodomain has a proteolytically resistant core in the presence of membrane. (A to C) Digest patterns of soluble WT or syn point mutant gB cytodomains in the absence or presence of anionic LUVs with trypsin (A), V8 protease (B), or chymotrypsin (C). Representative gels are shown. The protease-to-protein ratio is indicated above each lane. ND, nondigested. (D) Sequence of WT HSV-1 gB cytodomain with superimposed syn point mutations and locations of experimentally determined protease cleavage sites in the absence or presence of LUVs. The sequence of the R858H mutant cytodomain with superimposed syn point mutations and locations of experimentally determined protease cleavage sites in the presence of LUVs is also shown. The proposed secondary structure is shown beneath the sequences.
Protection from proteolysis implies that the membrane-bound conformation of the cytodomain includes a proteolytically resistant core that encompasses residues 807 to 872 (Fig. 6D). The alternative explanation, that protection is achieved solely by association of the cytodomain with the membrane, e.g., via liposome shielding or insertion of protected residues into the lipid bilayer, is less likely, considering the extent of protection. Surprisingly, residues 818 to 844 were protected from proteolysis under all conditions (Fig. 6D), despite the presence of five potential V8 protease cleavage sites within this region. This region is likely concealed within the cytodomain trimer even in the absence of membrane.
The C terminus of the cytodomain remained proteolytically susceptible even in the presence of the membrane and is probably not a part of the protein core. Nevertheless, the C terminus also changed its conformation in the presence of the liposomes. Trypsin cleaved after residue R882 in the absence of liposomes but after residue R885 in the presence of liposomes (Fig. 6D). Additionally, in the presence of liposomes a new chymotrypsin cleavage site appeared after residue Y889 (Fig. 6D). Thus, membrane interaction likely induced a conformational change within the C terminus of the cytodomain that shielded residue R882 and exposed residues R885 and Y889.
The R858H mutation altered protease resistance of the membrane-bound cytodomain.
In the absence of membrane, the protease cleavage pattern of each of the point mutants was either the same as or very similar to the WT pattern (Fig. 6A to C). The A855V and A874P mutants were somewhat more susceptible to trypsin cleavage than the WT, but their cleavage products matched those observed for the WT by SDS-PAGE analysis (Fig. 6A). Mutant R858H lost three trypsin cleavage sites after residues K862, K866, and K875 but gained an additional cleavage site after residue K807 (data not shown). Thus, while the mutants possibly had slightly different conformations in solution relative to the WT, in the absence of membrane they all were still significantly susceptible to proteolysis.
In the presence of LUVs, all point mutants, like the WT, were significantly protected from proteolysis (Fig. 6A to C). This suggested that in both the WT cytodomain and the point mutants, membrane binding induced large conformational changes resulting in formation of a protease-resistant core. A855V and A874P mutants had cleavage patterns similar to the WT patterns with all three proteases (Fig. 6A to C), which indicated that their membrane-bound conformations were similar to that of the WT. In contrast, the R858H mutant was more susceptible to trypsin digestion, being cleaved at two additional sites, after residues K807 and R844 (Fig. 6A and D). Increased protease susceptibility of R858H likely stemmed from mutation-induced conformational changes in the membrane-bound form.
Hyperfusogenic point mutations do not affect the stability of the cytodomain.
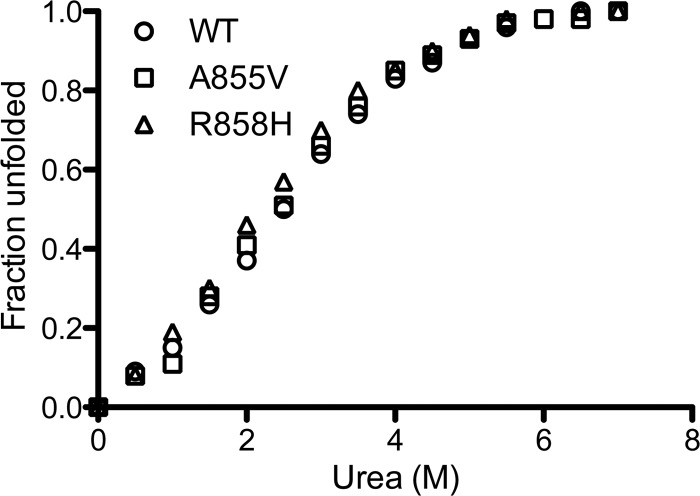
Increased proteolytic sensitivity of the R858H mutant could reflect a more loosely packed protein core. We tested the overall stability of the WT and the A855V and R858H mutant cytodomains by CD using chemical denaturation with urea (Fig. 7). Interestingly, both point mutants were as stable as the WT (Fig. 7), with similar free energies of folding (Table 1). While the WT stability of the A855V mutant is consistent with its WT-like resistance to proteolysis, the WT stability of the R858H mutant suggests that this mutation has a local rather than a global effect on the membrane-bound conformation of the cytodomain.
Fig 7.
Stability of the WT, A855V, and R858H mutant cytodomains was assessed using urea denaturation. The percentage of the protein unfolded at each concentration of urea is plotted against the urea concentration.
Taken together, our results suggest that point mutations R858H and A874P cause local conformational changes in the membrane-bound cytodomain, detected by limited proteolysis and CD, respectively, that could potentially contribute to the hyperfusion phenotype. The A855V mutation may also cause a local conformational change that manifests itself as 2-fold-higher binding to SUVs. We hypothesize that, while altered membrane binding could contribute to the hyperfusion phenotype of the A855V and A874P mutants, the hyperfusion phenotype of all mutants is likely caused by local perturbation of the cytodomain conformation.
DISCUSSION
Previous studies of hyperfusogenic gB mutants from several herpesviruses, including HSV-1, HSV-2, and EBV (4, 6, 7, 14, 17–20, 23, 36), suggested that the cytoplasmic domain of gB plays a regulatory role in fusion. To gain insight into the mechanism by which the cytoplasmic domain of HSV-1 gB regulates membrane fusion, we took advantage of a panel of previously identified C-terminal truncations and point mutations in the cytodomain that alter the extent of fusion. Our previous work identified a correlation between certain biochemical properties, namely, membrane association and concomitant changes in regular secondary structure, and the hyperfusion phenotype of several syn truncation mutants of HSV-1 gB (10). Here, we expanded our mutant analysis to syn point mutants, several of which occur spontaneously, to test whether this correlation existed for them.
Syn mutations do not affect gB surface expression or the conformation of the ectodomain.
All tested syn mutants, truncation and point, were hyperfusogenic, exhibiting a 2-to-3-fold increase in fusion relative to the WT. The hyperfusion phenotype of all five tested syn mutants required the presence of gD and gH/gL, even though some mutants mediated low-level fusion in the absence of gH/gL or gD.
The most obvious explanation for the hyperfusion phenotype of gB mutants would be their increased expression on the cell surface. Previously, it was proposed that the hyperfusion phenotype of an EBV gB truncation mutant was due to increased surface expression (36), presumably because the truncation eliminated the putative ER localization signal (RKRR) (23, 36). We did not find this to be the case for any of the tested syn mutants, truncation or point, consistent with previous observations of HSV-2 syn mutants, including mutations in the YXXϕ endocytosis motif (18, 37, 41). While increased surface expression may cause the hyperfusion phenotype in EBV gB mutant, it does not underlie the hyperfusogenicity of the HSV-1 gB syn mutants.
We next hypothesized that syn mutations in the cytodomain of gB may alter the conformation of the ectodomain. This type of communication between domains separated by membrane, often referred to as inside-out signaling, has been observed for other viral fusion proteins such as the F protein from paramyxoviruses PIV5 (48) and Nipah virus (2), Env from several retroviruses, including MoMLV (1, 35), HIV (49), and simian immunodeficiency virus (SIV) (45), and even several HSV-1 gB insertion mutants (33). Inside-out signaling also exists in nonfusion proteins such as integrins (42, 50). In all these cases, mutations in the cytoplasmic domain result in an altered conformation of the ectodomain and, as a consequence, its altered function. For HSV-1 gB, a viral fusion protein, this altered conformation could mean that the ectodomain is more fusogenic because it adopts an intermediate conformation along the path from the prefusion to the postfusion conformation. We found no significant differences in the antigenic properties of the WT and several syn mutant gB ectodomains and conclude that they all adopt similar conformations. Therefore, syn mutations in the gB cytodomain do not appear to convert its ectodomain into a more fusogenic conformation. Nevertheless, we cannot exclude the possibility that inside-out signaling may take place during the fusion process or require other entry glycoproteins. Future research will address these and other possibilities.
The fusion-null phenotype of gB851 is due to poor expression.
The gB truncation mutant gB851 was previously described as null with respect to fusion due to its inability to complement a gB-null virus (7), which led to the idea that residues 851 to 868 were essential for fusion. We found that gB851 mutant was expressed on the surface of CHO cells at 10% of WT levels due to poor overall protein expression. This may have been due to the low-level expression of the gene or to expression of a misfolded protein that is rapidly degraded. Indeed, the corresponding truncation mutant 829T from HSV-2 does not mediate cell fusion and may, perhaps, be misfolded upon expression in mammalian cells (18). We propose that limited protein expression and the resulting poor incorporation of the gB851 mutant into the virions likely accounts for its inability to complement a gB-null virus. Given that the gB851 mutant mediated 30% of WT fusion despite being expressed at only 10% of WT levels, it should not be referred to as fusion null. These results highlight the importance of conducting mechanistic studies with point mutants, as point mutations, unlike truncations, typically have a less severe effect on protein expression.
The cytoplasmic domain adopts its fully folded conformation only in the presence of membrane.
According to secondary-structure predictions, the gB cytodomain is approximately 50% α-helical. However, this secondary-structure content is achieved only in the presence of anionic membrane mimetics (10). We also showed previously (10) and confirmed here that the cytodomain stably associates with membranes. To characterize the membrane-bound conformation of the ectodomain in more detail, we used limited proteolysis. In the absence of membrane, the soluble WT cytodomain was highly susceptible to degradation by multiple proteases. These proteases have different sequence specificities, but all cleave throughout the length of the protein. In the presence of membrane, however, the bulk of the cytodomain was significantly protected from all three proteases, with only its C terminus remaining proteolytically susceptible. While such protection could, in principle, be explained simply by association of the cytodomain with the membrane, e.g., via liposome shielding or insertion of protected residues into the lipid bilayer, it is more likely that the membrane-bound conformation of the cytodomain includes a stable, proteolytically resistant core that encompasses residues 807 to 872. We propose that the cytodomain is fully folded only in the presence of the membrane. Considering that in the viral envelope or in cellular membranes, the cytodomain of gB is always in proximity to the membrane, the fully folded, membrane-bound form of the cytodomain likely represents its native conformation.
Surprisingly, we found that residues 818 to 844 were not susceptible to proteolytic cleavage in either the absence or presence of membrane, despite the presence of five potential V8 protease cleavage sites within this region. A large portion of this region, residues 819 to 835, is predicted not to have any defined secondary structure and is mostly hydrophilic. This region is probably shielded from proteolysis by other regions of the cytodomain. Of note, only one syn point mutant, D839A in HSV-2, falls within this range. In contrast, the majority of the cytodomain sequence contains a number of point mutations that induce hyperfusion. We hypothesize that residues 818 to 844 may be important for the overall stability of the cytodomain such that mutations in this region may be deleterious to viral replication and, therefore, selected against.
A model for the fully folded conformation of the cytodomain.
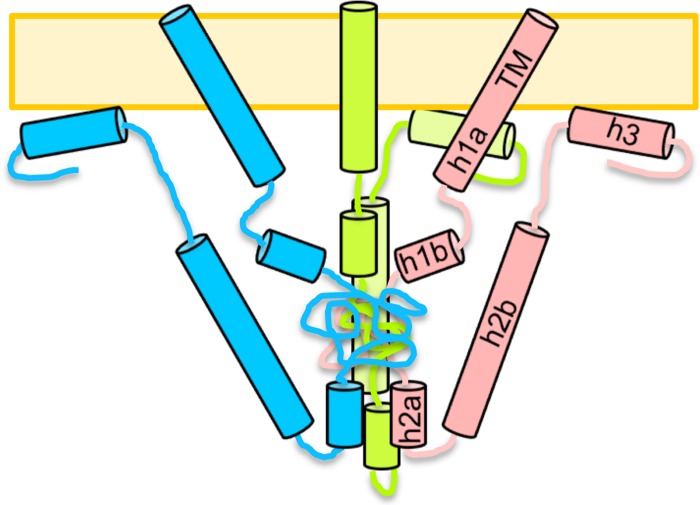
Combining our proteolysis and CD data with secondary-structure predictions, we propose the following model for the native, membrane-bound conformation of the cytodomain (Fig. 8). The N terminus of the cytodomain, residues 796 to 817, is predicted to form a continuous helix with the transmembrane region. However, helix h1 may have a kink or a break near residue M806, which is cleaved by chymotrypsin even in the presence of the membrane (Fig. 6D), and is therefore shown as helices h1a and h1b in the model (Fig. 8). Because we found that attaching a small trimerization domain to the N terminus of the cytodomain resulted in misfolded protein (data not shown), the N termini of the cytodomain are shown as not interacting. This result also implies that the TM helices probably do not interact within the plane of the membrane. Interestingly, the crystal structure of the prefusion form of glycoprotein G of vesicular stomatitis virus, another class III fusion protein, suggests that its transmembrane segments are too far apart to interact (40).
Fig 8.
Model of the native, membrane-bound conformation of the gB cytodomain. Three protomers are shown in different colors. Helices are depicted as cylinders and labeled within one protomer.
Further along the cytodomain polypeptide, residues 818 to 844 are resistant to protease cleavage and appear protected regardless of whether the membrane is present. Since no secondary structure is predicted for this region, our model shows it as a random coil core in the middle of the cytodomain trimer. The predicted helix h2 is shown as two helices, h2a and h2b, in our model (Fig. 6D and 8), because residue Y849 is susceptible to chymotrypsin cleavage even in the presence of membrane. Interestingly, the R858H mutation within putative helix h2b makes residue R844 within putative helix h2a sensitive to trypsin cleavage. It is tempting to speculate that these helices may interact, possibly through salt bridges that could be formed by many positively and negatively charged amino acids within both putative helices. Disruption of a salt bridge by the R858H mutation could potentially explain why R844 is more accessible to trypsin.
The putative helix h3 is predicted to include residues 870 to 883; however, due to the presence of protease cleavage sites after residues 872, 875, and 885, we hypothesize that helix h3 encompasses residues 876 to 885. This helix is likely involved in membrane interaction, because its removal decreases membrane binding (10). It is also possible that helix h3 interacts with other regions of the protein. Finally, the C terminus of the cytodomain likely remains unordered but may associate with the membrane.
Although our model incorporates several important observations, it does not explain the nature of extensive conformational changes within the cytodomain that accompany membrane binding. This model underscores the need to determine the structure of the cytodomain, which would help identify precisely which residues are part of the protein core and which are available to associate with membrane. The structure would reveal which regions of the protein interact, perhaps explaining the mechanism by which mutations R858H and A874P may induce local conformational changes. The structure would also clarify why residues 818 to 844, which are predicted to be unstructured, are proteolytically resistant. Finally, the structure may also provide some insight into why naturally occurring syn mutations are confined to two “hot spots” within the cytodomain sequence and why mutations in other regions of the protein, e.g., residues 818 to 844, are selected against. Our results highlight the importance of determining the structure in the presence of membranes to ensure that the native conformation is being studied.
Altered membrane binding may play a role in the mechanism of fusion deregulation.
We previously observed a correlation between the hyperfusion phenotype, reduced membrane binding, and lower helicity in the presence of membrane in the syn truncation mutants gB876 and gB868 (10). Unlike syn truncation mutants, none of the syn point mutants showed reduced membrane association. On the contrary, two mutants (A855V and A874P) bound membrane better than the WT, while one point mutant (R858H) displayed WT membrane association. Thus, while reduced membrane binding may be a part of the mechanism by which syn truncation mutants deregulate fusion, this is not the case for the syn point mutants. Altered membrane binding by four of five syn mutants suggests that membrane binding, specifically, membrane binding with WT affinity, may play a role in the mechanism of fusion regulation. One can envision an “on/off mechanism” whereby the cytodomain is required to release its association with membrane to some extent during the fusion process.
These differences in the extents of membrane binding by four syn mutants were detected by coelution with SUVs but not by cofloatation with LUVs. An alternative explanation, which places more weight on the cofloatation results, is that altered membrane binding is not a part of the mechanism by which syn mutations deregulate fusion. While we do not favor this conclusion due to the qualitative nature of the coflotation results as opposed to the quantitative data obtained using coelution experiments, we concede that altered membrane binding probably plays only a small role in fusion deregulation by the syn mutants.
Preservation of the WT conformation is necessary for fusion regulation.
The three point mutants studied here differed from the WT cytodomain in different ways, which initially suggested that they may deregulate fusion by completely different mechanisms. Specifically, only mutant R858H demonstrated increased proteolytic sensitivity; only mutant A874P had lower α-helical content; and only mutant A855V showed a dramatic increase in extent of binding to SUVs. Importantly, however, all mutations appear to induce conformational changes within the cytodomain.
Point mutant R858H displayed differences in proteolytic sensitivity in predicted helices h1b and h2a, which points to local conformational changes caused by the R858H mutation, located in predicted helix h2b. Therefore, the hyperfusion phenotype of the R858H mutant could potentially be explained by the altered cytodomain conformation.
The lower α-helical content in the point mutant A874P was initially attributed to the helix-kinking effect of the proline introduced in the middle of the predicted helix h3 (19). Our proteolysis data suggest, however, that residue A874 lies outside putative helix h3; if this is, indeed, the case, A874P mutation could reduce helicity by reorienting helix h3 in a way that would render it unable to interact with another region of the cytodomain required for its stabilization.
Better binding of SUVs by the A855V mutant could be explained by the increased hydrophobicity of residue 855. According to our model (Fig. 8), this mutation lies within predicted helix h2b, and although our model does not depict helix h2b interacting directly with membrane, it is possible that altered hydrophobicity of helix h2b could disturb other intramolecular contacts important for membrane binding.
Taken together, these data suggest that while the precise mechanisms by which syn point mutants deregulate fusion may differ somewhat, the underlying theme that unites these mechanisms is a perturbation of interactions within the cytodomain itself. We have found evidence of conformational changes in each of the three syn point mutants studied here: increased proteolytic sensitivity of helices h1b and h2a in the R858H mutant; decreased overall helicity of the A874P mutant due to a possible destabilization of predicted helix h3 as well as other helices; and increased binding of SUVs by the A855V mutant, which may have resulted from increased hydrophobicity within the predicted helix h2b. Each of these mutations likely alters the intramolecular contacts of the cytodomain, and we hypothesize that these perturbations of the fully folded, membrane-bound cytodomain conformation lead to fusion deregulation.
The conformational changes induced by these mutations may affect the cytodomain in other ways that, while not detected by our assays, can influence the fusion phenotype. For example, these mutations may alter contacts with predicted helix h1a, which is predicted to be contiguous with the transmembrane helix but may not be fully folded in its absence. Obtaining the structure of the membrane-bound cytodomain will be necessary to fully understand the effect of syn point mutations on the structure of the cytodomain and, therefore, its function.
Cytoplasmic domains of several viral fusion proteins have been shown to regulate fusion through regulating stability of the cytoplasmic tail. For example, in paramyxovirus fusion protein F, fusion is regulated by the length of its cytoplasmic tail such that viral isolates with longer cytoplasmic tails are less fusogenic than those with shorter tails (48). Fusogenicity is controlled by the stability of the F protein trimer, with a longer cytoplasmic tail repressing fusion by increasing the stability of F protein trimer and a shorter cytoplasmic tail promoting fusion by reducing the trimer stability (48). Similarly, in MoMLV Env, the C-terminal R-peptide represses fusion, presumably by stabilization of the Env trimer (46). Our data show that syn mutations do not affect cytodomain stability, and thus this model does not apply to the HSV-1 gB cytodomain. Furthermore, unlike the other viral cytodomains, which consist of relatively short, somewhat unstructured tails, our data show that the cytodomain of HSV-1 is an independently folded domain.
Our data do not support the idea of the presence of inside-out signaling mechanisms or the trimer stabilization mechanisms employed by some paramyxoviruses and retroviruses. Instead, the intimate association of the gB cytodomain with the membrane and the large conformational changes induced by membrane association hint at a unique mechanism. Direct structural information on the cytoplasmic domain will be indispensable for full elucidation of this mechanism, which, in turn, may aid the development of antiviral therapeutics aimed at preventing herpesvirus entry.
ACKNOWLEDGMENTS
We thank Arti Tewari for assistance with figures and Gary Cohen and Roselyn Eisenberg for critical readings of the manuscript. We also thank Stephen Kwok and Allen Parmelee at the Tufts Laser Cytometry Core facility for their assistance with FACS experiments, Alison Davis for sharing her tricine gel expertise, Jim Baleja for valuable discussions regarding CD, and Tirumala Chowdary for valuable discussions with regard to this project.
This work was funded by NIH grant 1DP20D001996 and by the Pew Scholar Program in Biomedical Sciences (E.E.H.).
J.L.S. and E.E.H. designed the experiments, J.L.S. and N.G.G. carried out the experiments, J.L.S. and E.E.H. contributed new reagents, D.S.K. carried out the mass spectrometry analysis, and J.L.S. and E.E.H. wrote the manuscript.
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Aguilar HC, Anderson WF, Cannon PM. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J. Virol. 77:1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguilar HC, et al. 2007. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J. Virol. 81:4520–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84:12292–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baghian A, Huang L, Newman S, Jayachandra S, Kousoulas KG. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bender FC, et al. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J. Virol. 81:3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bzik DJ, Fox BA, DeLuca NA, Person S. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185–190 [DOI] [PubMed] [Google Scholar]
- 7. Cai WH, Gu B, Person S. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carfí A, et al. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169–179 [DOI] [PubMed] [Google Scholar]
- 9. Chowdary TK, et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chowdary TK, Heldwein EE. 2010. Syncytial phenotype of C-terminally truncated herpes simplex virus type 1 gB is associated with diminished membrane interactions. J. Virol. 84:4923–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Connolly SA, et al. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J. Virol. 77:8127–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Connolly SA, et al. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diakidi-Kosta A, Michailidou G, Kontogounis G, Sivropoulou A, Arsenakis M. 2003. A single amino acid substitution in the cytoplasmic tail of the glycoprotein B of herpes simplex virus 1 affects both syncytium formation and binding to intracellular heparan sulfate. Virus Res. 93:99–108 [DOI] [PubMed] [Google Scholar]
- 15. Earp LJ, Delos SE, Park HE, White JM. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357–364 [DOI] [PubMed] [Google Scholar]
- 17. Engel JP, Boyer EP, Goodman JL. 1993. Two novel single amino acid syncytial mutations in the carboxy terminus of glycoprotein B of herpes simplex virus type 1 confer a unique pathogenic phenotype. Virology 192:112–120 [DOI] [PubMed] [Google Scholar]
- 18. Fan Z, et al. 2002. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J. Virol. 76:9271–9283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foster TP, Melancon JM, Kousoulas KG. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18–29 [DOI] [PubMed] [Google Scholar]
- 20. Gage PJ, Levine M, Glorioso JC. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geraghty RJ, Jogger CR, Spear PG. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147–158 [DOI] [PubMed] [Google Scholar]
- 22. Greenfield NJ. 2006. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protoc. 1:2891–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haan KM, Lee SK, Longnecker R. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106–114 [DOI] [PubMed] [Google Scholar]
- 24. Hannah BP, et al. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrison JS, Koellhoffer JF, Chandran K, Lai JR. 2012. Marburg virus glycoprotein GP2: pH-dependent stability of the ectodomain alpha-helical bundle. Biochemistry 51:2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932 [DOI] [PubMed] [Google Scholar]
- 28. Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heldwein EE, et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 30. Krummenacher C, et al. 2004. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 322:286–299 [DOI] [PubMed] [Google Scholar]
- 31. Krummenacher C, et al. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lichtenberg D, Barenholz Y. 1988. Liposomes: preparation, characterization, and preservation. Methods Biochem. Anal. 33:337–462 [DOI] [PubMed] [Google Scholar]
- 33. Lin E, Spear PG. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 104:13140–13145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindberg M, Jarvet J, Langel U, Graslund A. 2001. Secondary structure and position of the cell-penetrating peptide transportan in SDS micelles as determined by NMR. Biochemistry 40:3141–3149 [DOI] [PubMed] [Google Scholar]
- 35. Löving R, Li K, Wallin M, Sjoberg M, Garoff H. 2008. R-Peptide cleavage potentiates fusion-controlling isomerization of the intersubunit disulfide in Moloney murine leukemia virus Env. J. Virol. 82:2594–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McShane MP, Longnecker R. 2004. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. U. S. A. 101:17474–17479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muggeridge MI, Grantham ML, Johnson FB. 2004. Identification of syncytial mutations in a clinical isolate of herpes simplex virus 2. Virology 328:244–253 [DOI] [PubMed] [Google Scholar]
- 38. Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235–244 [DOI] [PubMed] [Google Scholar]
- 39. Pertel PE, Fridberg A, Parish ML, Spear PG. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324 [DOI] [PubMed] [Google Scholar]
- 40. Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848 [DOI] [PubMed] [Google Scholar]
- 41. Ruel N, Zago A, Spear PG. 2006. Alanine substitution of conserved residues in the cytoplasmic tail of herpes simplex virus gB can enhance or abolish cell fusion activity and viral entry. Virology 346:229–237 [DOI] [PubMed] [Google Scholar]
- 42. Shattil SJ. 2009. The beta3 integrin cytoplasmic tail: protein scaffold and control freak. J. Thromb. Haemost. 7(Suppl. 1):210–213 [DOI] [PubMed] [Google Scholar]
- 43. Silverman JL, Sharma S, Cairns TM, Heldwein EE. 2010. Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation. J. Virol. 84:2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spear PG, Eisenberg RJ, Cohen GH. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1–8 [DOI] [PubMed] [Google Scholar]
- 45. Spies CP, Ritter GD, Jr, Mulligan MJ, Compans RW. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 68:585–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor GM, Sanders DA. 2003. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology 312:295–305 [DOI] [PubMed] [Google Scholar]
- 47. Turner A, Bruun B, Minson T, Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Waning DL, Russell CJ, Jardetzky TS, Lamb RA. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. U. S. A. 101:9217–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wyss S, et al. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 79:12231–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang J, et al. 2009. Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl. Acad. Sci. U. S. A. 106:17729–17734 [DOI] [PMC free article] [PubMed] [Google Scholar]