Abstract
Human endogenous retroviruses (HERVs) make up 8% of the human genome. The expression of HERV-K (HML-2), the family of HERVs that most recently entered the genome, is tightly regulated but becomes markedly increased after infection with HIV-1. To better understand the mechanisms involved in this activation, we explored the role of the HIV-1 Tat protein in inducing the expression of these endogenous retroviral genes. Administration of recombinant HIV-1 Tat protein caused a 13-fold increase in HERV-K (HML-2) gag RNA transcripts in Jurkat T cells and a 10-fold increase in primary lymphocytes, and the expression of the HERV-K (HML-2) rec and np9 oncogenes was also markedly increased. This activation was seen especially in lymphocytes and monocytic cells, the natural hosts for HIV-1 infection. Luciferase reporter gene assays demonstrated that the effect of Tat on HERV-K (HML-2) expression occurred at the level of the transcriptional promoter. The transcription factors NF-κB and NF-AT contribute to the Tat-induced activation of the promoter, as shown by chromatin immunoprecipitation assays, mutational analysis of the HERV-K (HML-2) long terminal repeat, and treatments with agents that inhibit NF-κB or NF-AT activation. These studies demonstrate that HIV-1 Tat plays an important role in activating expression of HERV-K (HML-2) in the setting of HIV-1 infection.
INTRODUCTION
Human endogenous retroviruses (HERVs) are transposable elements that make up 8% of the total human cellular DNA (58, 62, 90, 117). After a series of germ line infections over millions of years (62, 117), HERVs now exist in the genome in proviral forms (131) consisting of the basic retroviral genes (gag, pro, pol, and env) flanked by two long terminal repeats (LTRs) (8) formed during reverse transcription, with the 5′ LTR serving as the viral transcriptional promoter. Most of these proviral sequences have been rendered nonfunctional through the passage of time due to the acquisition of multiple inactivating mutations and deletions (62). However, many individual HERV genes remain intact, leading to their expression in human cells (41, 62). The endogenous retrovirus HERV-K (HML-2), for example, has been shown to be transcriptionally active (10, 62, 63, 103, 111, 117, 139). It is the most recent entrant into the human genome (9, 128), having last integrated into the human genome between 200,000 and 5 million years ago (8), and it is the only subfamily of endogenous retroviruses with conserved, and therefore potentially functional, open reading frames (ORFs) for all viral proteins (8, 83). There are approximately 91 full-length copies of HERV-K (HML-2) (117) per haploid genome and thousands of solitary LTRs. Copies of HERV-K (HML-2) are found on multiple different chromosomes (8, 13). HERV-K (HML-2) also encodes the accessory oncogenes np9 and rec (nuclear protein of 9 kDa and regulator of expression encoded by cORF) (4, 13, 14, 78), which are expressed, respectively, by the two types of HERV-K (HML-2), type 1 and type 2. Type 1 and type 2 differ only by a 292-bp deletion at the beginning of the envelope gene in type 1 (8, 117).
Endogenous retroviral elements can be involved in physiological processes, such as those regulating the transcription of genes such as INSL4, β1,3-GT, endothelin B receptor, and tissue-specific salivary amylase (11, 35, 62, 84, 124). In addition, expression of certain HERV proteins has important physiological functions, such as in placental development (2, 34, 44, 80), and may also provide mechanisms for protecting against exogenous virus infection (50, 62). However, in general, how or why HERV genes are expressed, and the mechanisms responsible for expression, is not clearly understood. It is known that exogenous viral infections, viral transactivators, processes such as inflammation, chemical agents, cytokines, hormones, and stress conditions can contribute to the activation and transcription of transposable genetic elements, HERV-K (HML-2) being an example (21, 26, 39, 55, 57, 60, 66, 68, 71, 94, 101, 114, 116, 121, 122, 125, 127). A possible role for HERV-K (HML-2) in pathogenesis has been considered in disorders such as systemic lupus erythematosus, rheumatoid arthritis, and neuroinflammation (3, 36, 43, 53, 77, 90, 112, 113). Certain malignancies, most commonly germ cell tumors, melanoma, breast tumors, and prostate cancer, also show high levels of HERV-K (HML-2) antigen expression (18, 59, 62, 104, 110, 132), sometimes accompanied by the production of viral particles (12, 89), and yet the actual contribution of HERVs to disease remains to be characterized.
The HERV-K (HML-2) proteins Rec and Np9 provide a potential link between HERV-K (HML-2) and oncogenesis (4, 6, 18, 31, 41, 52, 67, 101). Both proteins have been shown to stimulate c-Myc expression by binding and inhibiting the c-myc gene repressor promyelocytic leukemia zinc-finger protein (PLZF [31]), and Rec has also recently been shown to interact with the testicular zinc-finger protein, another transcriptional repressor (67). In addition, Rec overexpression leads to testicular carcinoma in situ in transgenic mice (48, 106, 107). Np9 transcripts are detected with high frequency in tumor samples and, although no direct evidence exists that links it to oncogenesis, Np9 has been shown to interact with a member of the cancer-associated Notch signaling pathway (6). Thus, increased expression of the HERV-K (HML-2) proteins Rec and Np9 has the potential to contribute to oncogenesis.
Antibodies against HERV-K have been found in the blood of patients with a number of different clinical conditions (8, 18, 32, 54, 72, 77). One of the highest percentages of antibodies against these retroviruses is seen in HIV-1-infected patients, where ca. 70% show a response against HERV-K (HML-2) antigens (18, 32, 77, 116). We and others have demonstrated that HERV-K (HML-2) RNA levels are significantly increased in the plasma of HIV-1-infected patients (ca. 107 to 108 copies/ml) compared to healthy HIV-1-negative controls (0 to 102 copies/ml) (23–25, 27, 50, 130), and we have detected HERV-K (HML-2) proteins and viral particles in the blood of human patients with HIV-1-associated lymphoma (24, 27). HIV-1 infection of peripheral blood mononuclear cells (PBMCs) isolated from healthy donor blood leads to increased expression of both HERV-K (HML-2) RNA and protein (26). In addition, it has been observed that HERV-K elements are overexpressed in brain tissue of AIDS patients who develop neurological complications due to increased immune activation (63). How HERV-K might be activated by HIV has remained an open question. It is possible that the increased expression of HERV-K in HIV-1 infection is due to immunosuppression, but it could also be a consequence of direct interaction with infectious HIV-1 particles or viral proteins (130). Interestingly, work from the Cullen and Löwer laboratories has provided evidence that HIV-1 Rev recognizes the cis-acting Rec response region in the HERV-K (HML-2) RNA, which is similar to the Rev response element of HIV-1, and can actively export HERV-K (HML-2) RNA from the nucleus to the cytoplasm (79, 135).
The HIV-1 regulatory protein Tat is a potent transactivator of the HIV promoter and is essential for viral replication (42). Tat is produced in the early phase of HIV infection as a 72- or 101-amino-acid protein; depending on the viral isolate, a truncated 86-amino-acid form can also be produced (19, 38, 42, 61, 64). In addition to activating HIV transcription in the cell where it is made, Tat is actively secreted by infected cells into the extracellular surroundings, where it can be taken up by neighboring cells and exert effects on gene expression (38, 42, 60). In addition, HIV-1 Tat has been known to act not only on the HIV-1 and HIV-2 promoters but also on other viral and cellular promoters (60, 102, 108, 120, 137). Interestingly, the HIV-1 Tat protein has been shown to upregulate the transcription of Alu repeat sequences, an effect mediated by interactions of the transcription factor TFIIIC with the Alu promoter (60).
In view of the above, we hypothesized that the HIV-1 Tat protein provides a functional link between HIV-1 infection and the induction of HERV-K (HML-2) gene expression by causing activation of HERV-K LTR-directed transcription. In the studies described below, we show that Tat stimulates HERV-K expression in cell lines and primary lymphocytes. We further demonstrate that the HERV-K (HML-2) transcriptional promoter is responsive to Tat and that this effect is mediated by NF-κB and NF-AT. This may explain, at least in part, why HIV-1 infection is associated with such high levels of HERV-K (HML-2) expression in patients.
MATERIALS AND METHODS
Plasmid constructs.
The HIV-1 molecular clone pNL4-3 has been previously described (Malcolm Martin, NIH AIDS Research and Reference Reagent Program [1]). The HIV-1 Tat coding plasmids pcDNA3.1-Tat72 and pcDNA3.1-Tat86 were made from the parent vector pcDNA3.1+/Tat101-flag (PEV280; Eric Verdin, NIH AIDS Research and Reference Reagent Program [95]) by PCR amplification. The Tat mutants Tat C22G and Tat T23N were made through site-directed mutagenesis of the pcDNA3.1+/Tat101-flag parent vector. The pcDNA3.1(+) empty vector control was made by releasing the Tat insert with BamHI, followed by religation of the backbone. The HIV-1 Rev coding plasmid was made by releasing the rev insert from the pRev-1 plasmid construct (Marie-Louise Hammarskjöld and David Rekosh, NIH AIDS Research and Reference Reagent Program [75]) with BamHI, and ligation into pcDNA3.1(+). The HIV-1 Nef protein is also expressed from pcDNA3.1(+), containing the coding region of HIV-1SF2 Nef, and has been previously described (J. Victor Garcia and John Foster, NIH AIDS Research and Reference Reagent Program [98]). The HIV-1 Vif and Vpu proteins were expressed from pcDNA3.1(−), with both being codon optimized for expression in human cells [called HVif and Vp(h)u] and are further described elsewhere (Stephan Bour and Klaus Strebel, NIH AIDS Research and Reference Reagent Program [91]). Full-length Vpr was cloned into the peGFP-C3 expression vector (Promega, Madison, WI) and is fused to green fluorescent protein (GFP; Warner Greene, NIH AIDS Research and Reference Reagent Program [109]). A vector that expresses HIV-1 Gag derived from NL4-3 in an HIV-1 Rev-dependent manner was kindly provided by Akira Ono at the University of Michigan and has been previously described (pCMVNLGagPolRRE [93]). pCMV (pCMV-PL; Bryan Cullen) was obtained from Addgene (Cambridge, MA). Full-length Env from HIV-1HXB2 was cloned into pSV7D, termed pHXB2Env for simplicity, and has been described previously (Kathleen Page and Dan Littman, NIH AIDS Research and Reference Reagent Program [97]). The molecular clone pHXB2 (pHXB2gpt) was kindly provided by F. Wong-Staal (99, 100). The molecular clone pMtat(−) contains a termination codon (TGA) in place of the ATG (methionine) initiator codon in the Tat coding region, resulting in a mutant unable to synthesize Tat (Reza Sadaie, NIH AIDS Research and Reference Reagent Program [105]). The HERV-K (HML-2) LTR reporter construct (HERV-K LTR-luc) contains a partial version of the HERV-K (HML-2) LTR (GenBank accession no. AF394944) cloned in front of the HSV-1 tk minimal promoter in the pT81 luciferase vector (71) and was kindly provided by Kyung Lib Jang (Pusan National University, Pusan, South Korea). The Renilla luciferase plasmid, pRL-CMV, was obtained from Promega (Madison, WI). The luciferase construct “HERV-K LTR,” used in mutational analyses, was made by addition through PCR amplification of the consensus sequence TGTGGGGAAAAGCAAGAGA to the 5′ end of the partial promoter sequence of the HERV-K (HML-2) LTR from pT81 and subcloning it into the XhoI and HindIII sites of pGL4.10[luc2] (Promega, Madison, WI), which does not contain a tk minimal promoter. Site-directed mutagenesis was performed on the LTR region using the QuikChange Lightning site-directed mutagenesis kit (Stratagene, La Jolla, CA). NF-κB binding site mutations were introduced into the general consensus sequence GGGRNYYYCC (where R = purine, Y = pyrimidine, and N = any nucleotide) as GCTCTAYYCC. NF-AT binding site mutants were obtained by mutating the GGAAA region of the consensus sequence (A/T)GGAAA(A/N)(A/T/C)N to CTCTA. In cases where NF-AT sites were embedded in NF-κB sequences, mutation to CTCTA was used to eliminate both sites simultaneously. For analyses involving the potential Sp1 binding site, two binding site mutants were generated and tested, taking the consensus sequence GGGCGG(G/A)(G/A)(C/T) and changing it to either GAGATCTGC or TTGAGGTGC. PCR primers for introduction of mutations were designed using the QuikChange Primer Design Program (Stratagene). Sequences of the plasmids were confirmed by DNA sequencing.
Cell culture and transient transfections.
Except for 293FT (a fast-growing cell line derived from HEK-293 that contains the simian virus 40 large T antigen [Invitrogen, Carlsbad, CA]), H9 (Robert Gallo, NIH AIDS Research and Reference Reagent Program [82]), and Jurkat-Tat T cells (stably expressing HIV-1 Tat; Antonella Caputo, William Haseltine, and Joseph Sodroski, NIH AIDS Research and Reference Reagent Program [20]), all cell lines used were obtained from the American Type Culture Collection (ATCC; Manassas, VA). All media were obtained from Gibco (Invitrogen). 293FT were maintained in Dulbecco modified Eagle medium, supplemented with 10% fetal bovine serum (FBS; Gibco/Invitrogen, Carlsbad, CA) and 100 U of penicillin and streptomycin (Gibco/Invitrogen)/ml, at 37°C in a 5% CO2 incubator. Jurkat T, U-937, HUT-78, H9 (a derivative of HUT-78), and NCCIT cells were maintained in complete RPMI 1640 medium supplemented as described above. Jurkat-Tat T cells were maintained in complete RPMI 1640 supplemented with 10% FBS, 800 μg of G418 (Invitrogen)/ml, and 100 U of penicillin and streptomycin/ml. When needed, the cells were cultured under stimulatory conditions with PMA (phorbol 12-myristate 13-acetate) and ionomycin, with DMSO (dimethyl sulfoxide) as their vehicle buffer (all from Sigma-Aldrich, St. Louis, MO). Cells with >90% viability were transfected with endotoxin-free plasmids [2 to 5 μg of HIV-1-Tat, Vif, Nef, Rev, Vp(h)u, Vpr, and GagPol plasmids, 2 to 5 μg of empty/control vectors, or 5 μg of pHXB2 or pMtat(−) proviral molecular clones] using the transfection reagents Lipofectamine 2000 (Invitrogen), SuperFect (Qiagen, Valencia, CA), or FuGENE HD (Roche, Indianapolis, IN) according to the manufacturers' protocols. Control experiments included mock transfections.
HIV-1 production and infection.
Infectious HIV-1 was produced by calcium phosphate-mediated transfection of 293FT cells (136) using pNL4-3 (1). Tissue culture medium was harvested at 24, 36, or 48 h posttransfection, pooled, and filtered (0.2-μm pore size) to remove cells and large cell debris. Medium containing HIV-1NL4-3 was frozen and stored at −80°C until used for infections. The relative concentrations of virus in the stocks were determined from the reverse transcription (RT) activity (7) and the ability to induce luciferase activity upon infection of TZM-Bl cells (7). HIV infection of cells (2 × 106) was conducted using 2 ml of virus in a standard spin infection technique (1,048.6 × g) for 2 h at room temperature. Infected cell cultures were diluted in T-25 flasks and maintained at between 0.5 × 106 and 1 × 106 cells per ml for 7 days; then, fresh uninfected cells were added, and the cells were harvested after an additional week. Cellular RNAs were extracted using TRIzol (Invitrogen). HIV-1 env amplification was performed to confirm the infection status with the following primer sequences, in a one-step reverse transcription-PCR (RT-PCR): HIV-1 Env F 1493-1516 (5′-AGGCAAAGAGAAGAGTGGTGCAGA-3′) and HIV-1 Env R 1643-1666 (5′-CCCTCAGCAAATTGTTCTGCTGCT-3′).
Luciferase assays.
All transfections included Renilla luciferase as an internal control (100 ng of pRL-CMV) to assess for variation in transfection efficiency. Transfected cells were assayed for luciferase activity using a dual-luciferase assay kit (Promega, Madison, WI), 4, 6, 8, 12, 24, and 48 h after transfection in a Tecan GENios luminometer plate reader (Phenix Research Products, Candler, NC). The data were normalized to the Renilla luciferase signal and are expressed as standardized luciferase units. The amounts of plasmids used for transfections were as follows: 2.5 μg of luciferase reporter plasmid (HERV-K [HML-2] LTR or HIV-1 LTR), with 2 to 5 μg of Tat expression plasmid or 2 to 5 μg of empty/control vector. Control experiments included mock transfections (Lipofectamine 2000 reagent alone) and no transfection (for background subtraction).
Isolation and culture of primary cells.
PBMCs were obtained by venipuncture from healthy donors and monocytes were separated from peripheral blood lymphocytes (PBLs) by differential adhesion to plates as previously described (119). PBLs were washed three times with phosphate-buffered saline (PBS) and stimulated with 5 μg of phytohemagglutinin (PHA-P; Sigma-Aldrich)/ml for 3 days in RPMI 1640 complete medium containing 10% heat-inactivated FBS and 10 U of interleukin-2 (IL-2; Sigma-Aldrich)/ml.
Addition of exogenous Tat protein.
The purified recombinant 86-amino-acid form of the HIV-1 Tat protein was obtained from the NIH AIDS Research and Reference Reagent Program (the late John Brady and DAIDS, NIAID, [15]) or from ProSpec Protein Specialists (catalog no. HIV-129; ProSpec-Tany TechnoGene, Ltd., East Brunswick, NJ). The protein was resuspended in sterile PBS (Gibco/Invitrogen) containing 1 mg of bovine serum albumin (BSA)/ml and 0.1 mM dithiothreitol (DTT) (both from Sigma-Aldrich), de-aerated, and protected from light. Tat protein was added in a range of concentrations to cells for the specified time points, as indicated in the text and figure legends.
RNA extraction and real-time RT-PCR.
Total cellular RNA was isolated from cells using the RNeasy minikit (Qiagen) and subjected to RNase-free DNase treatment (Qiagen) for 15 min at room temperature. The RNA concentration and purity were measured using a spectrophotometer, calculating the 260/280 ratio. RNA integrity (as well as the absence of DNA contamination) was confirmed by one-step RT-PCR using GAPDH (glyceraldehyde-3-phosphate dehydrogenase) amplification with primers that can bind both genomic and cDNA, under the PCR conditions described below, as well as “no RT” controls. If DNA contamination was detected, another round of DNase treatment was performed using a DNA-free DNase removal kit (Ambion, Austin, TX) following the manufacturer's protocol. To assess the difference in HERV-K (HML-2) RNA transcript expression levels, we performed quantitative real-time RT-PCR using a QuantiTect SYBR green RT-PCR kit (Qiagen) or a Bio-Rad iScript one-step RT-PCR kit with SYBR green (Bio-Rad, Hercules, CA). Briefly, 100 to 500 ng of total cellular RNA and 0.5 μM concentrations (each) of HERV-K (HML-2) gag primers (forward primer, 5′-AGCAGGTCAGGTGCCTGTAACATT-3′; reverse primer, 5′-TGGTGCCGTAGGATTAAGTCTCCT-3′), HERV-K (HML-2) rec primers (forward primer, 5′-ATCGAGCACCGTTGACTCACAAGA-3′; reverse primer, 5′-GGTACACCTGCAGACACCATTGAT-3′), or HERV-K (HML-2) np9 primers (forward primer, 5′-AGATGTCTGCAGGTGTACCCA-3′; reverse primer, 5′-CTCTTGCTTTTCCCCACATTTC-3′) were used in a 20-μl reaction. RNA was reverse transcribed for 30 min at 50°C. PCR consisted of an initial denaturing step of 15 min at 95°C, followed by 35 to 40 cycles with optimal conditions as follows: 94°C for 15 s, 60°C for 30 s, and 72°C for 10 s, as well as optimized data collection steps at 81°C for 10 s (for Gag and Rec amplification) or 78°C for 10 s (for Np9 amplification). Fluorescence captured at 78 or 81°C was determined to be absent of signal generated by primer dimers by a melting-curve analysis. The data were collected and recorded by iCycler iQ software (Bio-Rad). GAPDH amplification was used to normalize samples to an endogenous reference gene, as stated in the figure legends.
Western blot and protein band analysis.
The antibodies used in the present study, and their respective dilutions, are as follows: mouse anti-HERV-K Gag (HERM-1841-5, 1:1,000 dilution) and mouse anti-HERV-K capsid (HERM-1831-5, 1:200 dilution), both from Austral Biologicals; anti-β-actin-HRP (conjugated to horseradish peroxidase [HRP], 1:25,000 dilution; Abcam, Cambridge, MA); and mouse anti-HIV-1 Tat (1D9; NIH AIDS Research and Reference Reagent Program, Bethesda, MD). After transfection, or recombinant protein addition, Jurkat T cells were washed twice with PBS and then lysed with hot 2% sodium dodecyl sulfate (SDS) buffer (Fisher Scientific, Pittsburgh, PA). The cell lysates were boiled and centrifuged to eliminate DNA-associated viscosity, and the protein concentration was measured using a Pierce BCA protein assay reagent kit (Pierce/Thermo Scientific, Rockford, IL). Equal protein concentrations were loaded and separated on SDS–15% polyacrylamide gels and blotted onto polyvinylidene difluoride membranes. The membranes were blocked with 5% nonfat dry milk in PBS with 0.1% Tween 20 (PBST; Fischer Scientific, Pittsburgh, PA) for 2 h at room temperature. All of the antibodies used were incubated in blocking solution with the blotted membranes overnight at 4°C, with the exception of anti-β-actin-HRP (2 h at room temperature). The membranes were washed three times in PBST and blocked with 5% goat serum for 30 min at room temperature. When necessary, the bound primary antibody was incubated with an HRP-conjugated goat anti-mouse secondary antibody for 1 h. Signal was detected using the Super Signal West Pico system (Pierce/Thermo Scientific). Semiquantification of protein levels was performed by digitization of X-ray films using the Typhoon FLA 7000 scanner (GE Healthcare Life Sciences, Pittsburgh, PA) and subsequent analysis of the gray values of the bands in the resulting images. The ImageQuant TL software was used for analysis of the digitized Western blot images. This software allows the measurement of band volume (average optical density of the band times its area) of the band of interest. Background subtraction was performed using the “image rectangle” method as a user-defined area within the image (size as well as position) and, once defined, it was applied to all lanes of interest. Bands were fitted as tightly as possible, and band volumes are plotted as arbitrary units.
Identification of potential transcription factor binding sites in the LTR of HERV-K (HML-2).
Analyses of the HERV-K (HML-2) LTR for potential transcription factor binding sites were performed using the online prediction software tools ALGGEN PROMO and version 8.3 of TRANSFAC (BioBase Co., Beverley, MA) (40, 85).
NF-κB inhibition assays.
The expression vector pIκBαM (kindly provided by Paul J. Chiao, MD Anderson Cancer Center, Houston, TX), referred to here as IκBα DN (dominant negative), was cotransfected into Jurkat T cells with the HERV-K (HML-2) LTR-luciferase construct with or without Tat (using Renilla luciferase as an internal transfection control). The luciferase activity was measured 48 h after transfection. IκBα DN encodes a phosphorylation site and a degradation site mutant IκBα chain, inhibits translocation of NF-κB from the cytosol to the nucleus upon activation, and has a dominant-negative effect on NF-κB function (33, 46, 47).
NF-κB inhibition was also accomplished through a 1 h pretreatment of experimental samples with 10 μM wedelolactone (7-methoxy-5,11,12-trihydroxycoumestan; Sigma-Aldrich) in DMSO, followed by treatment with recombinant Tat protein or its buffer control. Wedelolactone specifically inhibits NF-κB-mediated gene transcription in cells by blocking the phosphorylation and degradation of IκBα (70).
NF-AT inhibition assays.
Jurkat T cells cotransfected with the HERV-K (HML-2) LTR reporter construct and the Tat expression vector were treated with various doses of the immunosuppressant drug cyclosporine (Sigma-Aldrich) 24 h after transfection, and the luciferase activity was measured 48 h after transfection, as noted above and in the figure legends.
Specific NF-AT inhibition was accomplished through a 1-h pretreatment of experimental samples with 1 μM 11R-VIVIT in DMSO (Calbiochem, La Jolla, CA), followed by treatment with recombinant Tat protein or its buffer control. 11R-VIVIT is a cell-permeable version of the specific NF-AT inhibitor (VIVIT) that is modified at the C terminus with an 11-arginine transduction domain and a 3-glycine linker sequence (92).
ChIP assays.
To examine interactions of NF-κB and NF-AT with the HERV-K (HML-2) LTR, we performed chromatin immunoprecipitation (ChIP) assays using a ChIP-IT Express enzymatic kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. Briefly, experimental samples were lysed and subjected to enzymatic shearing of the DNA (random cleaving). Digestion was performed for 4 h at 37°C, with intermittent vortexing. A 2-μg portion of specific antibody for NF-κB (H-119X; Santa Cruz Biotechnology, Santa Cruz, CA), NF-AT (7A6; Santa Cruz Biotechnology), or vimentin (V9; Santa Cruz Biotechnology) or IgG isotype control antibodies (rabbit IgG or mouse IgG; Sigma-Aldrich) were used for each immunoprecipitation, with overnight incubation. Protein G-beads were added to the overnight incubation as well. Beads were washed three times in ChIP buffer, after which elution of the digested chromatin, reversion of cross-linking, and proteinase K treatment was performed. Immunoprecipitated DNA was detected by PCR, using 5 μl of eluate as a template, to verify success of the precipitation. For amplification of the HERV-K (HML-2) LTR transcription factor binding site areas of interest, the following primers were used (their binding areas are depicted in Fig. 6): KLTR ChiP primer set 1 fwd (5′-TGTGGGGAAAAGCAAGAGA-3′), KLTR ChiP primer set 1 rev (5′-GGTCACAGAATCTCAAGGCAG-3′), KLTR ChiP primer set 2 fwd (5′-GTGACCTTACCCCCAACCCCG-3′), KLTR ChiP primer set 2 rev (5′-TGTTTAACAAAGCACATCCTGC-3′), KLTR ChiP primer set 3 fwd (5′-CTGCCTAGGAAAGCCAGGTA-3′), KLTR ChiP primer set 3 rev (5′-CGGGTATCGGGCTGGGGGACG-3′), KLTR ChiP primer set 4 fwd (5′-CCCTGGGCAATGGAATGTCTCG-3′), KLTR ChiP primer set 4 rev (5′-GCTGCCCGCAGGTCCCACCTC-3′), KLTR ChiP primer set 5 fwd (5′-TGGTTCCCCGGGTCCCCTTAT-3′), and KLTR ChiP primer set 5 rev (5′-CCTACACACCTGTGGGTGTTT-3′).
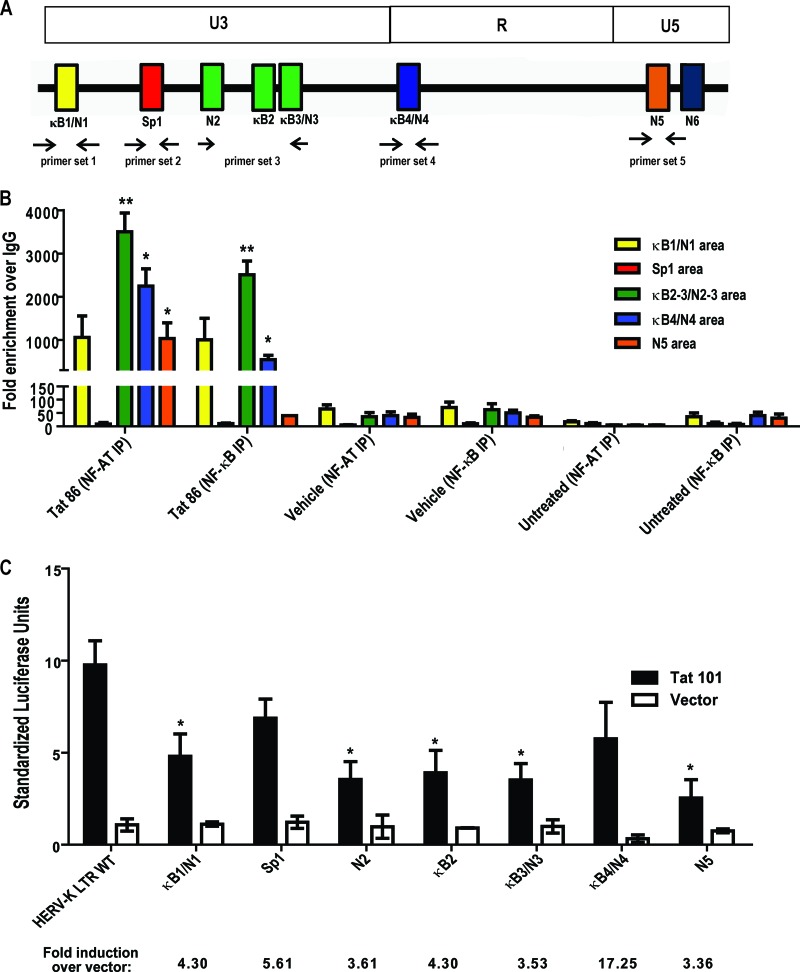
Fig 6.
Tat responsive elements in the HERV-K (HML-2) promoter. ChIP was performed on the cellular HERV-K (HML-2) LTR promoter in Jurkat T cells using antibodies specific for NF-κB or NF-AT (or nonspecific IgG isotype controls) 1 h after treatment with either recombinant HIV-1 Tat protein or PMA and ionomycin. qPCR was performed using primer sets designed to target areas of the promoter where the potential transcription factor binding sites are present. (A) Schematic of primer binding areas in the HERV-K (HML-2) promoter. (B) ChIP analysis of potential NF-κB and NF-AT binding sites with data expressed as the fold enrichment over IgG. Bars are color coded to match primer binding areas. Error bars indicate the SD from three independent immunoprecipitation experiments. (C) Potential transcription factor binding site mutations in the HERV-K (HML-2) LTR and their response to HIV-1 Tat in Jurkat T cells. WT, wild type; κB, NF-κB; N, NF-AT. Luciferase activation was measured 48 h after transfection. Control experiments included a mock transfection and transfection of an empty vector (pcDNA3.1). Error bars indicate the standard errors of the mean from three independent transfections. Significance was calculated using a Student t test comparing Tat treatments to vehicle controls or full-length LTR activity to mutant-LTR activity. Significant results are indicated (*, P < 0.05; **, P < 0.005).
Real-time PCR was performed with the Bio-Rad iQ SYBR green Supermix (Bio-Rad) with a 0.3 μM final primer concentration in a 20-μl final reaction volume and the following cycle conditions: 1 cycle of 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 57°C for 30 s, and 72°C for 30 s, followed in turn by 1 cycle of 72°C for 10 min. A melting-curve analysis was also performed to verify specific product amplification. Primers targeting the HERV-K env gene and the IL-6 promoter were used as controls, with PCR conditions as stated above.
Statistical analysis.
The mean number of HERV-K (HML-2) mRNA and standardized luciferase units between Tat treatments and controls were compared using an independent Student t test for samples exhibiting normally distributed values. Two-tailed P values were considered significant at P < 0.05.
RESULTS
HERV-K (HML-2) RNA expression is increased in HIV-1-infected cell lines.
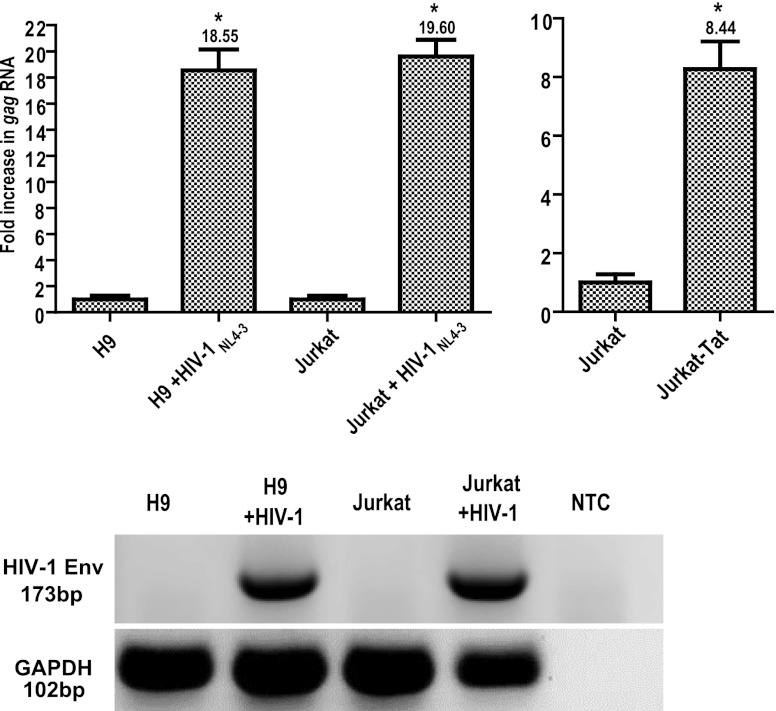
Recent studies by our group and others have shown that HIV-1 infection increases HERV-K (HML-2) gene expression, both in cell culture and in patients (22–27, 50). However, the underlying mechanism for this increased expression has remained unknown. To begin to address this issue, we first ascertained what the levels of HERV-K (HML-2) RNA expression were after HIV-1 infection of 2 different T cell lines. Quantification of HERV-K (HML-2) gag RNA by quantitative RT-PCR (qRT-PCR) in cells not infected with HIV-1 showed RNA levels of ∼103 copies per 500 ng of total RNA (data not shown). These levels increased up to ∼20-fold with HIV-1 infection (Fig. 1). Higher expression of HERV-K (HML-2) gag RNA was consistently seen in all HIV-1-infected cells compared to their uninfected counterparts (P < 0.001, Fig. 1). Interestingly, uninfected Jurkat T cells that stably express the Tat protein from HIV-1 (Jurkat-Tat) showed 8-fold more HERV-K (HML-2) gag RNA expression than did Jurkat T cells lacking Tat (Fig. 1). Since Tat has been shown to activate both viral and cellular genes (15, 52, 86, 90, 110), and since we observed that Jurkat-Tat cells have higher HERV-K (HML-2) gag RNA expression compared to the parental Jurkat counterpart, we hypothesized that the HIV-1 Tat protein might play a role in activating HERV-K (HML-2) expression during HIV-1 infection.
Fig 1.
HIV-1 infection and Tat stimulate HERV-K (HML-2) gene expression. Total cellular RNA was isolated from cells that were infected with HIV-1NL4-3 (for 1 week) or left uninfected. RNA was amplified using primers specific for HERV-K (HML-2) gag through one-step qRT-PCR and quantified using a standard curve generated by amplification of HERV-K (HML-2) gag RNA standards. The data are expressed as the fold increase over uninfected cells, with uninfected cells shown as normalized to 1 for simplicity of comparison. The rightmost panel shows Jurkat-Tat HERV-K (HML-2) gag RNA levels compared to levels in the parental Jurkat T cell line, with Jurkat T cell gag RNA levels normalized to 1. HIV-1 env and GAPDH one- step RT-PCR amplifications were also performed on the RNA to verify infection status and integrity of the material (NTC, nontemplate control). Error bars indicate the standard deviations (SD) for the results of three independent experiments. Significance was calculated by comparing infected samples with their uninfected counterparts using a Student t test and significant results are indicated (*, P < 0.005).
HIV-1 Tat and Vif independently cause an increase in HERV-K (HML-2) gag RNA expression.
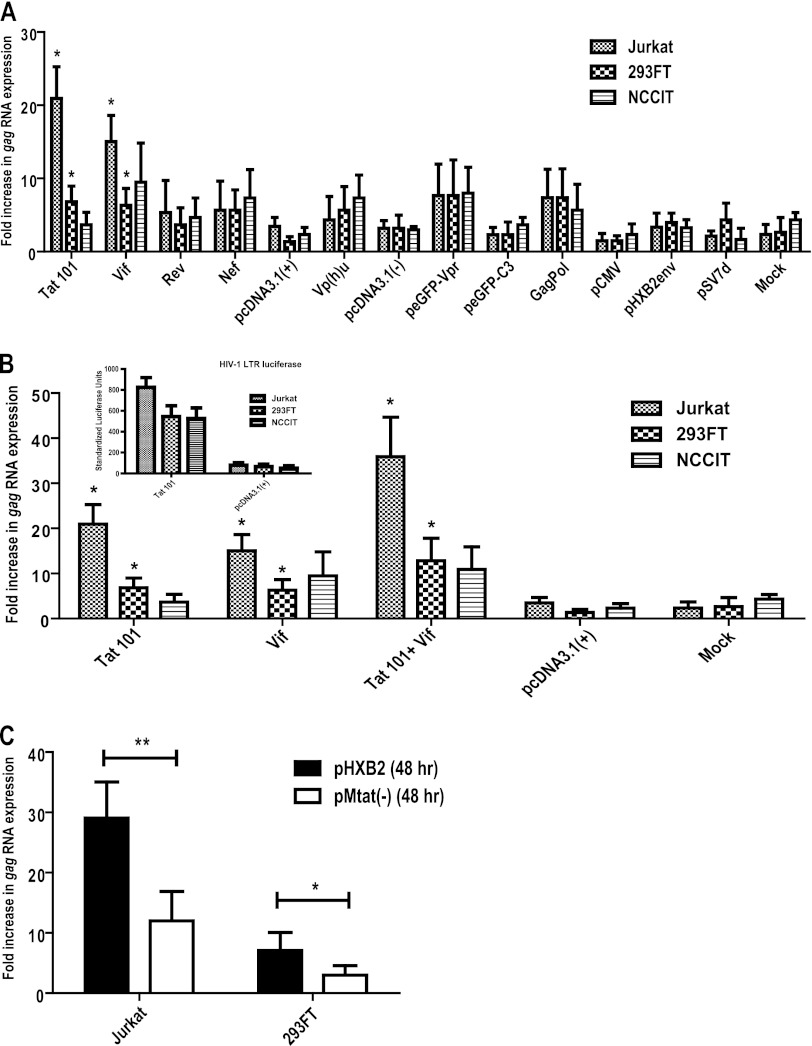
To ascertain whether our hypothesis that HIV-1 Tat activates the synthesis of HERV-K (HML-2) gag RNA is correct, we first transfected different cell types with plasmids encoding each of the regulatory, accessory, and structural proteins from HIV-1 and measured the levels of cellular HERV-K (HML-2) gag RNA after 24 and 48 h. We used a T cell line permissive for HIV-1 infection (Jurkat T cells), an easily transfectable line (293FT), and a cell line known for its high expression of HERV-K (HML-2) transcripts and proteins (NCCIT teratocarcinoma cells). Expression of Tat (from a plasmid encoding the full-length, 101-amino-acid form) or Vif yielded a significant increase in HERV-K (HML-2) RNA expression (Fig. 2A). The presence of Tat or Vif increased HERV-K (HML-2) gag RNA by about 21- or 15-fold, respectively, compared to untreated Jurkat cells (P < 0.01). Similar responses to Tat and Vif were seen in 293FT cells, whereas the teratocarcinoma NCCIT showed a response only to Vif (Fig. 2A). Similar increases in HERV-K (HML-2) gag RNA expression were observed 48 h after transfection (data not shown). The expression of any of the other HIV-1 proteins resulted in no significant increase in HERV-K (HML-2) RNA.
Fig 2.
HIV-1 Tat and Vif proteins activate HERV-K (HML-2) RNA expression. (A) Total cellular RNA was isolated from cells 24 h after transfection with plasmids encoding the individual regulatory and accessory proteins from HIV-1, and subjected to one-step Sybr green qRT-PCR with primers specific for HERV-K (HML-2) gag. HIV-1 full-length Tat (Tat 101), Rev, and Nef were cloned into the pcDNA3.1(+) expression vector. HIV-1 full-length Vif and Vpu were cloned into the pcDNA3.1(−) expression construct. Full-length Vpr was cloned into the peGFP-C3 expression vector and is fused to GFP. Full-length HIV-1 Gag and Pol were expressed from the HIV-1 gag-pol RNA sequence cloned into pCMV and expressed with cotransfection of HIV-1 Rev. Full-length Env from HIV-1HXB2 was cloned into pSV7D and termed pHXB2Env for simplicity. Respective empty vectors are shown, as well as mock transfections, with data expressed as the fold increase in RNA over untreated cells. (B) Cells were transfected with full-length Tat and/or Vif encoding plasmids, and total RNA was extracted 24 h later and subjected to one-step Sybr green qRT-PCR using primers specific for HERV-K gag. The inset shows the Tat activity further verified by cotransfection with an HIV-1 LTR-luciferase reporter vector. The relative luciferase units were normalized to an internal transfection control vector encoding Renilla luciferase and expressed as standardized luciferase units. (C) HERV-K (HML-2) gag RNA expression from cell lines transfected with an infectious HIV-1 molecular clone (pHXB2) or a mutant version lacking the Tat protein [pMtat(−)]. At 48 h after transfection, total RNA was isolated and subjected to amplification by one-step Sybr green qRT-PCR. The data are expressed as the fold increase in RNA over untreated cells. All qRT-PCR results were normalized to the GAPDH reference gene after analysis using the 2−ΔΔCT method, and the relative expression is plotted. Error bars indicate the SD for the results of three independent experiments. Significance was calculated by comparing protein treatments to the empty vector controls (i.e., pcDNA3.1, peGFP-C3, pCMV, and pSV7D) or by comparing the full-length molecular clone to the Tat mutant using a Student t test. Significant results are indicated (*, P < 0.05; **, P < 0.005).
To determine whether Tat and Vif synergistically increase HERV-K (HML-2) transcript levels, we cotransfected plasmids encoding these proteins and observed the effect on transcription after 24 h. For both Jurkat T cells and 293FT, Tat and Vif had an additive, not synergistic, effect with regard to HERV-K (HML-2) transcription, whereas in NCCIT no significant difference was seen compared to Vif-induced expression alone (Fig. 2B). Tat functionality was verified in NCCIT cells in parallel cotransfections using an HIV-1 LTR-luciferase reporter assay (Fig. 2B, inset). These data show that both Tat and Vif can activate HERV-K (HML-2) transcription and suggest that Tat alone is sufficient for activation in HIV-1-relevant targets of infection (i.e., Jurkat T cells).
To further test the importance of Tat in HIV-1-mediated activation of HERV-K (HML-2), we transfected an HIV-1 infectious molecular clone (pHXB2) or a mutant version of it lacking the Tat protein [pMtat(−)] into Jurkat T cells and 293FT cells and measured HERV-K (HML-2) gag RNA 48 h later. As can be seen in Fig. 2C, Tat expression in Jurkat T cells is important for higher HERV-K (HML-2) RNA expression, since its absence greatly diminishes levels of transcripts (by more than half, P < 0.05). Of course, this experiment must be interpreted with caution since the lack of Tat also leads to a decrease in the expression of other HIV-1 proteins. However, these data are consistent with the observation that HIV-1 Tat expression is sufficient to increase HERV-K (HML-2) transcript expression.
Recombinant HIV-1 Tat increases HERV-K (HML-2) expression.
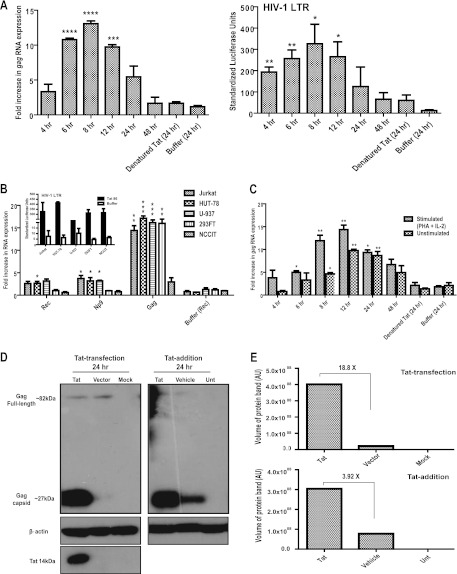
We next analyzed whether addition of physiologically relevant levels of recombinant HIV-1 Tat to cells could activate HERV-K (HML-2) transcription. We took advantage of the fact that, unlike most transcription factors, HIV-1 Tat is able to cross intact cellular membranes when it is present in the extracellular milieu (37, 38, 42, 138). Time course experiments in Jurkat T cells to which we added a recombinant 86-amino-acid form of Tat and measured HERV-K (HML-2) gag RNA expression showed that Tat addition caused a 10.8-fold increase in HERV-K (HML-2) gag RNA production after 6 h (Fig. 3A, left panel). The effect peaked at 8 h (13-fold increase) and then gradually declined (Fig. 3A, left panel). Recombinant Tat activity was further verified in parallel experiments by measuring reporter activity from Jurkat T cells transfected with a vector containing the HIV-1 LTR fused to the luciferase reporter gene, which showed the protein to be fully active (Fig. 3A, right panel). To verify that endotoxin or other extraneous material in the Tat protein samples were not responsible for HERV-K (HML-2) gag activation, we heat denatured Tat and added it to cells (endotoxin is highly heat stable, whereas Tat is not). No significant increase in HERV-K (HML-2) gag RNA was detected under these conditions, suggesting that activation of HERV-K (HML-2) gag gene expression was due to the Tat protein and not to endotoxin (Fig. 3A).
Fig 3.
Recombinant HIV-1 Tat activates HERV-K (HML-2) gene expression. Total cellular RNA was isolated from cells that were subjected to different Tat treatments and amplified by one-step Sybr green real-time qRT-PCR using primers specific for HERV-K gag, rec, np9, envelope type 1, and envelope type 2. (A) The left panel shows the HERV-K (HML-2) gag RNA expression from Jurkat T cells treated with 100 ng of purified Tat protein/ml for the specified periods of time. The right panel shows the results for Jurkat T cells transfected with an HIV-1 LTR-luciferase reporter construct, which 24 h later were treated with 100 ng of purified Tat protein/ml for the specified periods of time. The relative luciferase units were normalized to an internal transfection control vector encoding Renilla luciferase and are expressed as standardized luciferase units. Denatured Tat protein and the Tat vehicle buffer were used as negative controls, and only the 24-h time point is shown for those as representative results. (B) RNA expression of the HERV-K (HML-2) genes rec, np9, gag, envelope type 1, and envelope type 2 in different cell lines after 8 h of treatment with 100 ng of Tat protein/ml. The inset shows the Tat activity verified by transfection with an HIV-1 LTR-luciferase reporter vector. The relative luciferase units were normalized to an internal transfection control vector encoding Renilla luciferase and expressed as standardized luciferase units. (C) HERV-K (HML-2) gag RNA expression from PBLs. PBLs were split in half and either treated with 100 ng of Tat protein/ml for 8 h or prestimulated with PHA and IL-2 for 3 days, followed by an 8-h Tat treatment. All qRT-PCR results were normalized to the GAPDH reference gene after analysis using the 2−ΔΔCT method, and the relative expression is plotted as the fold increase over untreated cells. Error bars indicate the SD for the results of three independent experiments. Significance was calculated by comparing Tat treatments to buffer controls, at the same time points, using a Student t test, and significant results are indicated (****, P < 0.001; ***, P < 0.005; **, P < 0.01; *, P < 0.05). (D) Western blot showing HERV-K (HML-2) Gag protein expression in Jurkat T cells after treatment with HIV-1 Tat. For the detection of HERV-K (HML-2) Gag protein, two monoclonal antibodies were used simultaneously: anti-HERV-K Gag and anti-HERV-K capsid. The left panel shows the HERV-K (HML-2) Gag protein expression in cell lysates 24 h after transfection with Tat 101 (“Tat”) or a control vector (“Vector”, which is pcDNA3.1) or after mock transfection. The right panel shows the HERV-K (HML-2) Gag protein expression in cell lysates 48 h after the addition of recombinant Tat protein. Control lanes include the vehicle buffer (“Vehicle”, PBS with BSA and DTT), and untreated cell lysate (“Unt”). Respective Gag proteins and sizes are shown. Tat protein expression in transfected cells is shown, and β-actin protein expression is shown as a loading control. (E) Densitometry analysis of the HERV-K (HML-2) Capsid protein bands seen in the Western blots in panel D.
Activation of HERV-K (HML-2) RNA synthesis by Tat was not limited to gag transcripts in Jurkat T cells, nor was it limited to that cell type alone, since we also detected significantly increased HERV-K (HML-2) transcripts for gag, rec, and np9 in HUT-78 lymphoblasts, U-937 monocytes, and 293FT fibroblasts after Tat was added for 8 h (Fig. 3B). This effect was not seen with all cell types tested, since the teratocarcinoma cell line NCCIT did not show any significant increase in HERV-K (HML-2) gene expression with Tat treatment, a finding consistent with the results of the transfection experiments (Fig. 2A). Further evidence that this lack of effect was not due to the cells being nonpermissive or nonresponsive to Tat comes from parallel experiments involving transfection of the HIV-1 LTR-luciferase reporter vector, and subsequent Tat exposure, which showed that Tat could be internalized and activates the HIV-1 LTR (Fig. 3B, inset).
Since the experiments described above were all performed in cell lines, we sought to verify that the increase in HERV-K (HML-2) transcripts in response to Tat also occurs in primary cells, especially those most relevant to HIV infection. PBLs from healthy individuals were either exposed to stimulatory conditions (PHA and IL-2) or not and then treated with recombinant Tat protein. RNA expression analyses from unstimulated PBLs showed that Tat treatment leads to expression of HERV-K (HML-2) gag RNA at as early as 6 h, which continues to rise and peaks by 12 h, with an ∼10-fold increase over cells not exposed to Tat (Fig. 3C). We observed that stimulation of PBLs with PHA and IL-2 alone for 3 days activates transcription of HERV-K (HML-2) (data not shown), but this effect was sustained for only about a total of 74 h in culture (3 days of prestimulation and then experimental treatments), with RNA expression returning to basal levels by 96 h (Fig. 3C, buffer [24 h]).
Since our data show that Tat activated HERV-K (HML-2) transcription, we next assessed whether the increases in HERV-K (HML-2) transcripts would result in increased protein expression. Using untreated Jurkat T cell lysate with a mix of commercially available antibodies against the full-length HERV-K (HML-2) Gag and its capsid form, little protein expression was detected in untreated cells, as has previously been reported (16). When Tat was transfected into Jurkat T cells, full-length HERV-K (HML-2) Gag protein was still minimally expressed but the cleaved capsid form of Gag (∼30 kDa) showed an ∼18.8-fold increase in expression over the empty vector control (Fig. 3D and E). A similar increase in capsid expression was seen when recombinant Tat was added to the cells, although treatment with the vehicle buffer (which contains the reducing agent DTT) in this case also increases the expression to some degree (Fig. 3D). Protein increases with Tat treatment in this setting were ∼3.92-fold over the vehicle control (Fig. 3E). Although not as impressive as the RNA increases seen with Tat, these results show that Tat can substantially increase the expression of HERV-K (HML-2) Gag, and this protein is detected in its capsid form after it is further processed by a protease. Indeed, it has previously been established that cell lysates can contain both the precursor form of HERV-K (HML-2) Gag, as well as the cleaved capsid (12, 16).
Taken together, these observations demonstrate that HIV-1 Tat stimulates HERV-K (HML-2) gene expression at the RNA and the protein level in what appears to be a cell type-specific manner, with cells that are relevant to HIV biology being the most affected, and with significant transcriptional activation seen in primary lymphocytes, a major target for HIV infection.
Activation of the HERV-K (HML-2) promoter by HIV-1 Tat.
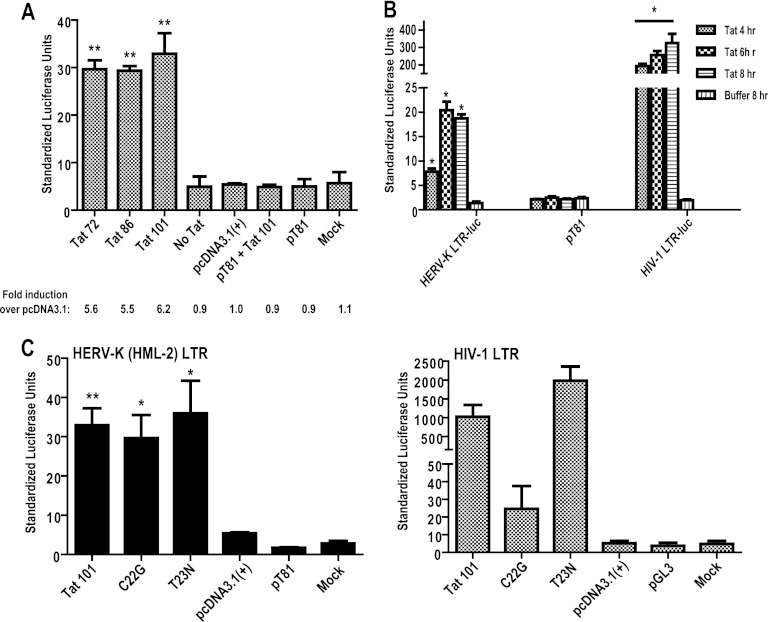
Since HIV-1 Tat is known to act upon both viral and cellular promoters to increase or decrease gene expression (17, 19, 60, 73, 74, 102, 108, 115, 137), we tested whether the effect of Tat on HERV-K (HML-2) occurred at the level of the transcriptional promoter. Using a construct containing a HERV-K (HML-2) LTR promoter driving the expression of the luciferase reporter gene, Jurkat T cells were cotransfected with plasmids encoding one of three isoforms of Tat (72, 86, or 101 amino acids). This was done since it is known that all isoforms do not necessarily behave the same with regard to cellular gene activation (e.g., full-length Tat 101, but not one-exon Tat 72, represses MHC-I expression) (19, 56, 133). The expression of Tat RNA and protein was confirmed by RT-PCR and immunoblot analyses (data not shown), and its functional integrity was confirmed by showing transactivation of the HIV-1 LTR in luciferase reporter gene assays as described above. Consistent with the findings shown in Fig. 2 and 3 above, we found that all HIV-1 Tat isoforms transactivated the HERV-K (HML-2) promoter with almost equal efficiency, approximately 5- to 6-fold over the empty vector control (Fig. 4A). This effect was corroborated by the addition of purified, recombinant Tat protein (Tat 86 isoform) to Jurkat T cells that were transfected with the HERV-K (HML-2) LTR reporter construct, which led to an increase in luciferase activity of approximately 4- to 8-fold over the signal generated by the buffer control alone (Fig. 4B). Tat showed little effect on the HERV-K (HML-2) promoter after transfection into 293FT or NCCIT cells (data not shown), which suggests that the Tat-mediated stimulation of HERV-K (HML-2) is cell type specific. Interestingly, the degree of activation seen from the HERV-K (HML-2) promoter used in the luciferase reporter assays was consistent with the observed Tat-induced increases in gag RNA. These promoter activation data thus support our observations that HIV-1 Tat activates HERV-K (HML-2) at the transcriptional level.
Fig 4.
HIV-1 Tat activates the HERV-K (HML-2) promoter by a different mechanism than it uses to activate HIV-1. (A) Jurkat T cells were transfected with a HERV-K (HML-2) LTR-luciferase construct (except for the luciferase backbone construct pT81 and mock samples) and cotransfected with the construct shown on the x axis (Tat 72, Tat 86, Tat 101, or pcDNA3.1). “Mock” corresponds to treatment with transfection reagent alone, whereas “No Tat” corresponds to untreated cells. Activation of the luciferase construct was measured 24 h after transfection, normalized to Renilla luciferase signal, and shown as standardized luciferase units. The fold induction was calculated over the empty vector, pcDNA3.1. Control experiments included a mock transfection and transfection of the backbone luciferase vector (pT81) alone or with a Tat-expressing vector. (B) Jurkat T cells were transfected with the HERV-K (HML-2) LTR-luciferase construct, or a similar HIV-1 LTR luciferase construct as a positive control, and 24 h after transfection were treated with 500 ng of purified Tat protein/ml for the specified periods of time. The buffer control is an 8-h treatment. (C) Jurkat T cells were transfected with the HERV-K (HML-2) LTR- or HIV-1 LTR-luciferase reporter construct and cotransfected with the Tat mutants C22G or T23N, and the cells were harvested at 24 h. These mutants were expressed from the pcDNA3.1 expression vector and are either unable to activate transcription from the HIV-1 LTR (Tat C22G) or cause an increase in HIV-1 LTR transcriptional activity (Tat T23N). The data are shown as standardized luciferase units. Error bars indicate the SD from three independent experiments. Significance was calculated using a Student t test comparing Tat transfection/treatment to pcDNA3.1 (A and C) or buffer control (B), and significant results are indicated (*, P < 0.05; **, P < 0.01).
In order to understand whether HERV-K (HML-2) promoter activation occurs by a mechanism similar to that which Tat uses to activate transcription from the HIV-1 LTR, we cotransfected the HERV-K (HML-2) LTR-luciferase constructs with two different Tat mutants: Tat C22G and the naturally occurring Tat T23N. Tat C22G contains a mutation in a cysteine at position 22 (to a glycine) in the transactivation domain, which renders it unable to interact with cyclin T1, and thus it is HIV-1 LTR activation deficient (49, 88). Tat T23N, on the other hand, contains a mutation in the threonine at residue 23 (to asparagine) that increases Tat's ability to activate the HIV-1 LTR by increasing binding of Tat to the cellular kinase-positive transcription elongation factor b (P-TEFb [49]). Neither of these mutations in Tat affected its ability to drive transcription from the HERV-K (HML-2) promoter, whereas they had the predicted effect on the HIV-1 LTR (Fig. 4C). These data show that activation of HERV-K (HML-2) transcription by Tat occurs in a different manner than that of HIV-1, and does not appear to involve Tat's interaction with Cyclin T1 or P-TEFb. This is consistent with the data described below.
Activation of the HERV-K (HML-2) promoter by Tat is mediated by NF-κB and NF-AT.
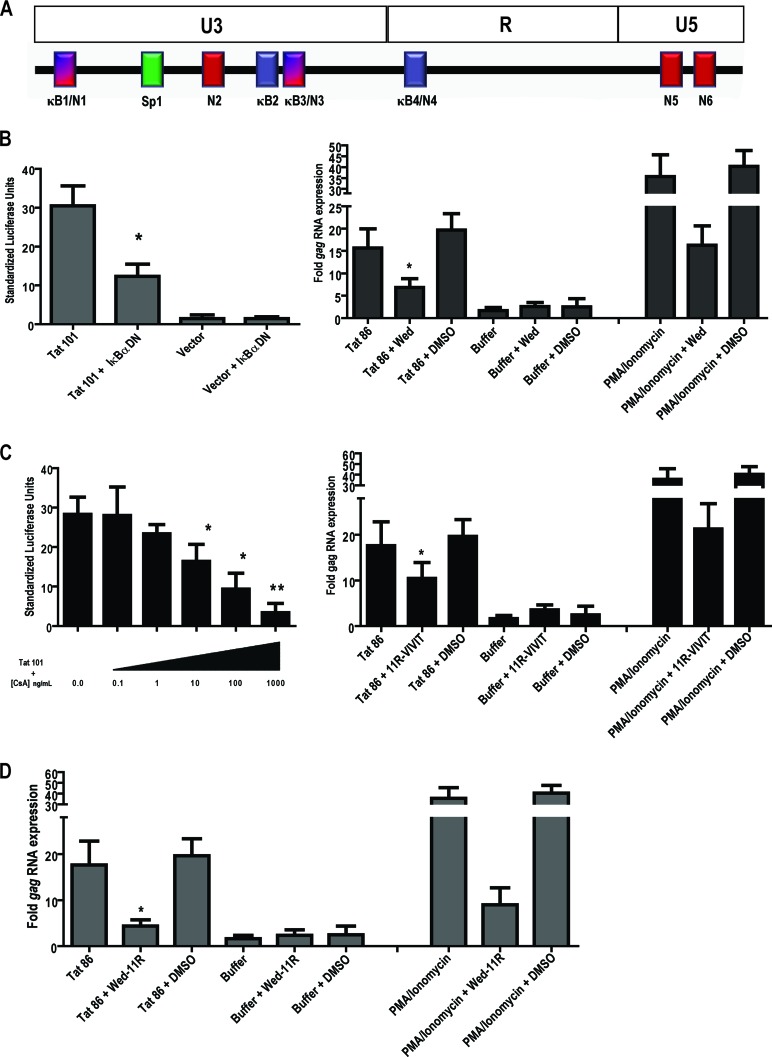
In addition to stimulating HIV transcriptional elongation through its interactions with cyclin T1 and P-TEFb, Tat is known to activate cellular genes through regulation and/or interaction with upstream cellular transcription factors. Furthermore, it has been shown that Tat can, in the absence of a functional TAR, remain an important factor for HIV-1 transcription via Sp1 sequence elements in the U3 promoter region (29). Tat can additionally interact directly with NF-κB, with this interaction not only demonstrating TAR-independent transactivation in HIV-1 but also pointing toward a mechanism of Tat-mediated modulation of cellular genes (28). To understand the mechanism by which Tat activates HERV-K (HML-2) LTR-directed transcription, we analyzed the sequence of the promoter for potential transcription factor binding sites. Utilizing the ALGGEN-PROMO software (40, 85) and the sequence prediction algorithm TRANSFAC database software (BioBase, Beverley, MA), we analyzed the promoter sequence and found that a number of transcription factors might potentially interact with the HERV-K (HML-2) promoter, including AP-1, CREB, CEBP (C/EBPα), c-Rel, NF-AT, CEBPβ, NF-κB(p50:p52), Rel-A, p53, YY1, c-Myc, Sp1, Sp3, and the STATs. Of the potential sites present, we decided to focus on ones previously shown to be particularly associated with HIV-1 Tat activation: Sp1, NF-κB, and NF-AT (30, 51, 69, 86, 87, 118, 129, 134). Two potential NF-κB binding sites, with NF-AT sites embedded in them (here referred to as κB1/N1 and κB3/N3), are found in the most upstream part of the U3 region of the promoter, along with single NF-AT (N2), NF-κB (κB2), and Sp1 sites (Fig. 5A). The R region contains a lone NF-κB/NF-AT site (κB4/N4), and the U5 region has two potential NF-AT binding sites present (N5 and N6, Fig. 5A).
Fig 5.
Inhibition of the NF-κB or NF-AT transcription factors suppresses Tat-mediated activation from the HERV-K (HML-2) LTR. (A) The HERV-K (HML-2) LTR promoter with U3, R, and U5 regions, as well as potential transcription factor binding sites indicated as follows: κB1/N1, first potential NF-κB/NF-AT binding site (one site embedded in the other); Sp1, potential Sp1 binding site; N2, second potential NF-AT binding site; κB2, second potential NF-κB binding site; κB3/N3, third potential NF-κB/NF-AT binding site; κB4/N4, fourth potential NF-κB/NF-AT binding site; and N5 and N6, fifth and sixth potential NF-AT binding sites, respectively. (B) The left panel shows the results for a dominant-negative construct coding for the inhibitor of NF-κB alpha (IκB-α DN) that was cotransfected into Jurkat T cells with the full-length HERV-K (HML-2) LTR-luciferase reporter and a Tat expression vector. The luciferase activity was measured 24 h after transfection, normalized to Renilla luciferase, and expressed as standardized luciferase units. For the right panel, Jurkat T cells were pretreated for 1 h with 10 μM wedelolactone (a specific inhibitor of NF-κB) and then treated with recombinant HIV-1 Tat for 8 h, followed by total RNA isolation and HERV-K (HML-2) gag amplification by qRT-PCR. (C) For the left panel, Jurkat cells cotransfected with the full-length HERV-K (HML-2) LTR-luciferase construct and Tat were subsequently treated with various concentrations of cyclosporine (CsA), and the luciferase activity was measured after 24 h, normalized, and expressed as standardized units. For the right panel, Jurkat T cells were pretreated for 1 h with 4 μM 11R-VIVIT (a specific inhibitor of NF-AT) and then treated with recombinant HIV-1 Tat for 8 h, followed by total RNA isolation and HERV-K (HML-2) gag amplification by qRT-PCR. (D) Jurkat T cells were pretreated for 1 h with both 10 μM Wedelolactone and 4 μM 11R-VIVIT prior to an 8-h recombinant Tat treatment and subsequent RNA amplification. PMA and Ionomycin treatments served as positive controls for transcription factor activation. Error bars indicate the SD from three independent experiments. Significance was calculated using a Student t test comparing Tat transfection/treatment in the absence of inhibitors to Tat transfection/treatment in the presence of inhibitors. Significant results are indicated (*, P < 0.05; **, P < 0.01). MTT assays performed to assess the toxicity of the drugs used in the experiments showed no significant cell death.
In view of the multiple NF-κB sites found in the HERV-K (HML-2) promoter and the known contribution of NF-κB to Tat-mediated activation of the HIV-1 promoter (30, 76), we tested whether NF-κB mediates the Tat effect on the HERV-K (HML-2) LTR. To do so, we first examined whether a dominant-negative inhibitor of NF-κB nuclear translocation would block Tat stimulation of the HERV-K (HML-2) promoter. This dominant-negative construct (referred to in the figure as IκBα DN) codes for the inhibitor of NF-κB alpha (IκBα) and sequesters NF-κB in the cytosol, preventing its translocation into the nucleus upon activation. At 48 h after cotransfection, the activation of the HERV-K (HML-2) promoter construct in response to Tat was significantly decreased, by approximately 65%, in the presence of the NF-κB inhibitor (Fig. 5B, left panel, P < 0.05). This dependence on NF-κB was corroborated in experiments in which Jurkat T cells were pretreated for 1 h with a specific, irreversible inhibitor of IKKα and β kinase activity, wedelolactone (70) before the addition of recombinant Tat protein. After 6 h, RNA was isolated and quantitated by qRT-PCR. Figure 5B (right panel) shows that inhibition of NF-κB diminishes the increase in HERV-K (HML-2) gag RNA seen in response to Tat by half (P < 0.01) but does not completely abolish it. As a control, we also observed that PMA and Ionomycin-induced transcription is similarly diminished, but not abolished, in the presence of wedelolactone. Thus, NF-κB mediates, in part, activation of HERV-K (HML-2) by Tat.
Since the HERV-K (HML-2) LTR also contains potential NF-AT binding sites, NF-AT activation could additionally contribute to HERV-K (HML-2) Tat-driven expression and might compensate for the absence of NF-κB activity. We therefore examined whether inhibiting NF-AT activation would also result in diminished HERV-K (HML-2) responsiveness to Tat. Treatment of Jurkat T cells that were cotransfected with the HERV-K (HML-2) reporter construct and Tat with the immunosuppressive drug cyclosporine (a calcineurin inhibitor that prevents dephosphorylation of NF-AT and therefore its activation) showed a ose-dependent inhibition of Tat-mediated HERV-K (HML-2) promoter activation (Fig. 5C, CsA, left panel). This suggests that NF-AT is also involved in HERV-K (HML-2) activation. An MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay was performed on cells treated with the described doses of cyclosporine and showed no significant cell death at any of the concentrations tested (data not shown). However, knowing that cyclosporine is not an NF-AT-specific drug, we also pretreated Jurkat T cells with 11R-VIVIT, a cell-permeable peptide inhibitor specific for NF-AT (92) for 1 h, added recombinant Tat protein for 6 h, and then isolated RNA. We observed a reduction in gag RNA to about half of the starting levels in cells treated with the NF-AT inhibitor in the presence of Tat (Fig. 5C, right panel). Treatment with both wedelolactone and 11R-VIVIT simultaneously prior to Tat addition had the strongest effect on diminishing gag RNA levels, since this treatment decreased expression by approximately 75% (Fig. 5D). Overall, these data show that both NF-κB and NF-AT activation in response to HIV-1 Tat drive transcription from the HERV-K (HML-2) promoter.
To verify the physical interaction of NF-κB and NF-AT with the HERV-K (HML-2) promoter, we performed ChIP assays. After Jurkat T cells were treated with HIV-1 Tat or PMA and ionomycin for 1 h, the cells were treated with formaldehyde for chromatin-protein cross-linking and lysed, and the cellular DNA was then purified. DNA-protein complexes were precipitated using anti-NF-κB, anti-NF-AT, or their respective IgG isotype controls, the cross-links were reversed, and the DNA was precipitated and purified. We then performed qPCR on the regions of the HERV-K (HML-2) promoter where these transcription factors should bind, using five different sets of primers that span the sites of interest (Fig. 6A) and provide fragments of optimal size for DNA amplification for ChIP. By measuring the fold enrichment in amplification of the specific antibody immunoprecipitation over the IgG control immunoprecipitation in a specific region of the promoter of interest, we can detect whether the transcription factors under study bind to that particular region and can compare the binding to other regions of the promoter. We detected that, in response to HIV-1 Tat, the order of enrichment (from highest to lowest) for NF-κB was as follows: the κB2-3/N2-3 position, followed by the κB1/N1 and then the κB4/N4 position (Fig. 6B). Thus, NF-κB appears to preferentially bind to the κB2-3/N2-3 position in the HERV-K (HML-2) promoter in response to Tat. Similar results were obtained for PMA- and ionomycin-induced NF-κB activation (data not shown). For NF-AT, enrichment is also highest at the κB2-3/N2-3 position, followed by the κB4/N4 position (Fig. 6B). Enrichment levels were of equal proportions when comparing positions κB1/N1 and N5 (Fig. 6B). No signal was detected when trying to amplify a product around the N6 position, suggesting that this is not an actual binding site for NF-AT. The specificity of the immunoprecipitation was further assessed by amplification of the region containing the potential Sp1 transcription factor binding site. qPCR amplification of the Sp1 area never yielded a product or gave a strong signal above background, thus it served as a control for immunoprecipitation (data not shown, and Fig. 6B). Comparing the NF-AT immunoprecipitation to the NF-κB immunoprecipitation shows more enrichment for NF-AT in all sites except at the first position (which seems equal to NF-κB enrichment). These data show that there is activation of NF-κB and NF-AT in response to HIV-1 Tat and that these two transcription factors interact directly with the HERV-K (HML-2) LTR promoter.
We next introduced site-directed mutations into the NF-κB, NF-AT, and/or Sp1 sites in the HERV-K (HML-2) promoter construct. We then transfected the mutated HERV-K (HML-2) promoter constructs with or without HIV-1 Tat into Jurkat T cells and measured the effects on luciferase reporter activity. It should be noted that some mutations of NF-κB sites also destroy the potential binding sites for NF-AT proteins, since they were embedded in their DNA sequence. Interestingly, site-specific mutation of most of these sites led to a clear decrease in Tat responsiveness compared to the wild-type promoter (Fig. 6C). Although others have reported a role for Sp1 in regulation of the basal expression of HERV-K (HML-2) (45), mutation of the Sp1 site did not significantly decrease promoter activity in response to Tat, a finding consistent with our ChIP data (Fig. 6B). Whereas mutation of the fourth NF-κB/NF-AT (κB4/N4) binding site did not affect the response to Tat, mutation of the first NF-κB/NF-AT (κB1/N1) and the second NF-κB (κB2) sites led to a decrease in activity (approximately 2- to 2.5-fold). The most pronounced decrease occurred when the first NF-AT site of the U5 region (N5) was mutated alone (∼3.8-fold), further suggesting that NF-AT is a crucial modulator of the response of HERV-K (HML-2) to HIV-1 Tat. Mutation of both potential NF-AT sites in the U5 region simultaneously did not affect the response to Tat any more than did mutation of the single N5 site (data not shown), suggesting that the N6 site is not a functional NF-AT binding site, which is consistent with the ChIP data. Taken together, these data show that both NF-κB and NF-AT are important for HIV-1 Tat-mediated activation of the HERV-K (HML-2) promoter, that one transcription factor may compensate for loss of the activity of the other, and that multiple NF-AT and NF-κB binding sites mediate much of the response to Tat.
DISCUSSION
Eight percent of the human genome consists of fixed retroviral elements, termed HERVs, acquired throughout thousands of years of human evolution. Although most HERV genes seem to be nonfunctional, there are some endogenous retroviral genes that are expressed, even becoming important contributors to human physiological processes, as can be seen in trophoblast development (2, 34, 44, 80). However, HERV gene expression appears to be tightly controlled under normal circumstances, and only when cellular fitness or integrity is compromised (e.g., with synthetic chemical agents, radiation, stress, cytokines/chemokines, or biological agents acting upon human cells) is there pronounced or increased expression of endogenous retroviral elements (26, 57, 62, 63, 81, 121, 127). Under conditions where an exogenous virus, such as HIV-1, establishes a productive infection, the repression of HERV expression can be lifted (130). HIV-1-infected individuals have abnormally high levels of the endogenous retrovirus HERV-K (HML-2) expressed in cells, and vastly increased RNA titers are present in their plasma (24, 25, 27, 50, 130). The consequences of this activation in the setting of HIV-1 infection are still a matter of speculation, but since HERV-K (HML-2) encodes two putative oncogenes and has been associated with several autoimmune, inflammatory, and neurological diseases, it could potentially participate in the development of HIV-1-associated disease (3, 36, 43, 53, 77, 90, 113). Understanding the initial steps in the activation of HERV-K (HML-2) expression after HIV-1 infection is the first step toward deciphering how HERV-K (HML-2) elements might play a role in HIV-1 pathogenesis.
Previous research has shown how several proteins from HIV-1 interact with HERV-K products. The HIV-1 protease, for example, can cleave HERV-K Gag (96, 126). In addition, HIV-1 Rev can actively transport HERV-K RNA from the nucleus to the cytoplasm of cells that express it (135). However, these interactions likely only partially explain the massive increase in HERV-K (HML-2) RNA expression seen in HIV-1-infected patients (24, 25, 27). Therefore, we reasoned that another protein from HIV-1 might be involved in stimulating HERV-K (HML-2) expression. Since the HIV-1 Tat protein has been shown to be a potent activator of viral (120) and cellular gene expression (17, 108), we thought it to be a likely candidate to stimulate HERV-K (HML-2) transcription.
We confirm here that HIV-1 infection leads to increased expression of HERV-K (HML-2) and show that the HIV-1 Tat and Vif proteins activate HERV-K (HML-2) expression at the RNA level, with Tat also causing an increase in the expression of at least one HERV-K (HML-2) protein, capsid. In addition, treating primary lymphocytes (which are more biologically relevant than continuously passaged cell lines) with HIV-1 Tat also results in increased HERV-K (HML-2) RNA expression. This effect of Tat on HERV-K (HML-2) activation was pronounced but dependent on the cell-type. For cells that constitutively express high levels of HERV-K (HML-2) transcripts and proteins (such as the teratocarcinoma cell lines), little response to Tat was detected at the RNA level. However, in cell lines normally expressing low levels of HERV-K (HML-2) or in primary cells with undetectable or extremely low levels of transcripts, Tat treatment upregulated transcription of these proviral sequences. Importantly, this was seen in primary lymphocytes, a major target of HIV-1 infection. This activation occurred whether Tat was added exogenously to the cells or if the cells were transfected with constructs encoding it. The effect occurred at the level of the transcriptional promoter, with cooperative involvement between Tat and the cellular transcription factors NF-κB and NF-AT. Since the promoter also contains a number of other potential transcription factor binding sites, we cannot discount the possibility of other factors also contributing to activation in the presence of Tat. We must note that other biological agents, chemicals, exogenous infections, or stress conditions can activate HERV expression in cells (39, 55, 66, 68, 94, 101) and that, while Tat is one important mechanism for HERV-K activation during HIV-1 infection, it is but one contributor among the whole spectrum of factors that cause an increase in HERV expression in a biological system. Interestingly, a recent study has found that the HTLV-1 Tax protein has been shown to potently activate the LTRs of different HERVs, including HERV-K (125), demonstrating that other exogenous retroviral transactivators can modulate endogenous retroviral expression.
Lastly, we should point out that in the future it will be necessary to verify which HERV-K (HML-2) proteins are expressed at a higher level in the presence of Tat, but the current status of the available antibodies precludes quantitatively accurate experiments.
Our research strongly suggests a role for the HIV-1 Tat protein in the activation of HERV-K (HML-2) transcription, which helps to explain the basis for the increased expression of HERV-K (HML-2) RNA seen during HIV-1 infection. Interestingly, though our data show marked increases in RNA expression in response to Tat treatment, the degree of stimulation still does not reflect the very high increase in the number of HERV-K (HML-2) RNA copies seen in the plasma of HIV-1-infected patients compared to uninfected individuals (25, 27). Thus, other HIV-1 proteins could also increase production of HERV-K (HML-2). For example, as we have demonstrated above, Vif could contribute to activation of HERV-K (HML-2) in patients. Further, a potential cooperativity between HERV-K (HML-2) Rec and HIV-1 Rev (both of which can transport HERV-K [HML-2] RNA out of the nucleus) could lead to increased HERV-K (HML-2) protein expression. Thus, additional interactions between HIV-1 and HERV-K likely contribute to the marked increase in HERV-K (HML-2) expression seen in patients with HIV-1 infection.
The consequences of the high levels of HERV-K (HML-2) expression seen in HIV-1-infected individuals are potentially important, since increased expression of HERV-K has been associated with a number of different pathologies (3, 18, 36, 43, 53, 54, 113). HERV-K (HML-2) encodes the Rec protein, a Rev-like protein that could augment the transport of RNAs out of the nucleus (14, 78, 135), thus altering the global processing of cellular RNA. Further, HERV-K (HML-2) encodes two putative oncogenes, rec and np9 (5, 13, 101), either of which might play a role in HIV-related malignancies. However, it has recently been shown that HERV-K antigens promote a T cell response against HIV-1 (50, 123), although Gag- and Env-specific T cell responses are infrequent (65). Thus, increased expression of HERV-K (HML-2) following HIV-1 infection might actually be helpful in controlling the replication of HIV. Further studies need to be conducted in order to fully ascertain the true impact of HERV-K (HML-2) activation in the setting of HIV-1 infection and its contribution to HIV-1-mediated pathogenesis.
ACKNOWLEDGMENTS
We thank Ferdinand Kappes, Nirit Mor-Vaknin, Maureen Legendre, Koh Fujinaga, Derek Dube, Seagal Teitz-Tennenbaum, and Akira Ono for their help in experimental design, reagent supplies, and data interpretation and for their thoughtful comments. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3, pcDNA3.1+/Tat101-flag (PEV280), pRev-1, pcDNA-HVif, pcDNA-Vphu, pEGFP-Vpr, pHXB2-env, pcDNA3.1SF2Nef, pMtat(−), clade B recombinant HIV-1 Tat protein, HIV-1 Tat monoclonal antibody (1D9), the H9 cell line, and the Jurkat-Tat cell line.
M.J.G-H. was supported by a Rackham Merit Fellowship, the Mechanisms of Microbial Pathogenesis Training Grant from the University of Michigan, and by an NIH Ruth L. Kirschstein NRSA Individual Predoctoral Fellowship to Promote Diversity in Health-Related Research grant 1F31CA150523-01. Part of this research was performed while on appointment as a U.S. Department of Homeland Security (DHS) Graduate Student Fellow under the DHS Scholarship and Fellowship Program, administered by ORISE, through an interagency agreement with the U.S. Department of Energy (DOE), contract number DE-AC05-00OR22750. This research was additionally supported by a generous grant from the Concerned Parents for AIDS Research and by grants RO1AI062248 and RO1CA144043 to D.M.M. from the National Institutes of Health. D.M.M. was also the recipient of a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
All opinions expressed here are the authors' and do not necessarily reflect the policies and views of the DHS, DOE, or ORISE.
Footnotes
Published ahead of print 16 May 2012
REFERENCES
- 1. Adachi A, et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson AC, et al. 2002. Developmental expression of HERV-R (ERV3) and HERV-K in human tissue. Virology 297:220–225 [DOI] [PubMed] [Google Scholar]
- 3. Antony JM, Deslauriers AM, Bhat RK, Ellestad KK, Power C. Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants? Biochim. Biophys. Acta. 2010 doi: 10.1016/j.bbadis.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Armbruester V, et al. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800–1807 [PubMed] [Google Scholar]
- 5. Armbruester V, et al. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800–1807 [PubMed] [Google Scholar]
- 6. Armbruester V, et al. 2004. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J. Virol. 78:10310–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arnold BA, Hepler RW, Keller PM. 1998. One-step fluorescent probe product-enhanced reverse transcriptase assay. Biotechniques 25:98–106 [DOI] [PubMed] [Google Scholar]
- 8. Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101(Suppl 2):14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barbulescu M, et al. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861–868 [DOI] [PubMed] [Google Scholar]
- 10. Berkhout B, Jebbink M, Zsiros J. 1999. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J. Virol. 73:2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bieche I, et al. 2003. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol. Reprod. 68:1422–1429 [DOI] [PubMed] [Google Scholar]
- 12. Bieda K, Hoffmann A, Boller K. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82:591–596 [DOI] [PubMed] [Google Scholar]
- 13. Boese A, et al. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328–4336 [DOI] [PubMed] [Google Scholar]
- 14. Boese A, Sauter M, Mueller-Lantzsch N. 2000. A rev-like NES mediates cytoplasmic localization of HERV-K cORF. FEBS Lett. 468:65–67 [DOI] [PubMed] [Google Scholar]
- 15. Bohan CA, et al. 1992. Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 2:391–407 [PMC free article] [PubMed] [Google Scholar]
- 16. Boller K, et al. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89:567–572 [DOI] [PubMed] [Google Scholar]
- 17. Buonaguro L, Buonaguro FM, Giraldo G, Ensoli B. 1994. The human immunodeficiency virus type 1 Tat protein transactivates tumor necrosis factor beta gene expression through a TAR-like structure. J. Virol. 68:2677–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buscher K, et al. 2006. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec, and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 16:223–234 [DOI] [PubMed] [Google Scholar]
- 19. Caldwell RL, lane KB, Shepherd VL. 2006. HIV-1 Tat interaction with cyclin T1 represses mannose receptor and the bone morphogenetic protein receptor-2 transcription. Arch. Biochem. Biophys. 449:27–33 [DOI] [PubMed] [Google Scholar]
- 20. Caputo A, Sodroski JG, Haseltine WA. 1990. Constitutive expression of HIV-1 tat protein in human Jurkat T cells using a BK virus vector. J. Acquir. Immune Defic. Syndr. 3:372–379 [PubMed] [Google Scholar]
- 21. Chu WM, Ballard R, Carpick BW, Williams BR, Schmid CW. 1998. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 18:58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Contreras-Galindo R, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. 2007. Comparative longitudinal studies of HERV-K and HIV-1 RNA titers in HIV-1-infected patients receiving successful versus unsuccessful highly active antiretroviral therapy. AIDS Res. Hum. Retrovir. 23:1083–1086 [DOI] [PubMed] [Google Scholar]
- 23. Contreras-Galindo R, et al. 2006. A new Real-Time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J. Virol. Methods 136:51–57 [DOI] [PubMed] [Google Scholar]
- 24. Contreras-Galindo R, et al. 2008. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol. 82:9329–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retrovir. 22:979–984 [DOI] [PubMed] [Google Scholar]
- 26. Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. 2007. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res. Hum. Retrovir. 23:116–122 [DOI] [PubMed] [Google Scholar]
- 27. Contreras-Galindo RA, et al. 2011. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J. Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dandekar DH, Ganesh KN, Mitra D. 2004. HIV-1 Tat directly binds to NFκB enhancer sequence: role in viral and cellular gene expression. Nucleic Acids Res. 32:1270–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Das AT, Harwig A, Berkhout B. 2011. The HIV-1 Tat protein has a versatile role in activating viral transcription. J. Virol. 85:9506–9516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Demarchi F, d'Adda di Fagagna F, Falaschi A, Giacca M. 1996. Activation of transcription factor NF-κB by the Tat protein of human immunodeficiency virus type 1. J. Virol. 70:4427–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Denne M, et al. 2007. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J. Virol. 81:5607–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Denner J, Phelps RC, Lower J, Lower R, Kurth R. 1995. Antibody response of pregnant women, tumor and AIDS Patients against the human endogenous retrovirus HERV-K. J. Cancer Res. Clin. Oncol. 121:5 [Google Scholar]
- 33. Dong QG, et al. 2002. The function of multiple IκB: NF-κB complexes in the resistance of cancer cells to taxol-induced apoptosis. Oncogene 21:6510–6519 [DOI] [PubMed] [Google Scholar]
- 34. Dunlap KA, et al. 2006. Endogenous retroviruses regulate peri-implantation placental growth and differentiation. Proc. Natl. Acad. Sci. U. S. A. 103:14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunn CA, Medstrand P, Mager DL. 2003. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. U. S. A. 100:12841–12846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ehlhardt S, et al. 2006. Human endogenous retrovirus HERV-K(HML-2) Rec expression and transcriptional activities in normal and rheumatoid arthritis synovia. J. Rheumatol. 33:16–23 [PubMed] [Google Scholar]
- 37. Ensoli B, Barillari G, Salahuddin SZ, Gallo RC, Wong-Staal F. 1990. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature 345:84–86 [DOI] [PubMed] [Google Scholar]
- 38. Ensoli B, et al. 1993. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J. Virol. 67:277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans LH, et al. 2009. Mobilization of endogenous retroviruses in mice after infection with an exogenous retrovirus. J. Virol. 83:2429–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farre D, et al. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31:3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Flockerzi A, et al. 2008. Expression patterns of transcribed human endogenous retrovirus HERV-K (HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics 9:354 doi:10.1186/1471-2164-9-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frankel AD, Young JA. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1–25 [DOI] [PubMed] [Google Scholar]
- 43. Freimanis G, et al. 2010. A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: investigating mechanisms of pathogenesis. Clin. Exp. Immunol. 160:340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frendo JL, et al. 2003. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 23:3566–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fuchs NV, et al. 2011. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J. Virol. 85:3436–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujioka S, et al. 2004. Stabilization of p53 is a novel mechanism for proapoptotic function of NF-κB. J. Biol. Chem. 279:27549–27559 [DOI] [PubMed] [Google Scholar]
- 47. Fujioka S, et al. 2003. Inhibition of constitutive NF-κB activity by IκBα M suppresses tumorigenesis. Oncogene 22:1365–1370 [DOI] [PubMed] [Google Scholar]
- 48. Galli UM, et al. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223–3228 [DOI] [PubMed] [Google Scholar]
- 49. Garber ME, et al. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garrison KE, et al. 2007. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 3:e165 doi:10.1371/journal.ppat.0030165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gatignol A, Duarte M, Daviet L, Chang YN, Jeang KT. 1996. Sequential steps in Tat trans-activation of HIV-1 mediated through cellular DNA, RNA, and protein binding factors. Gene Expr. 5:217–228 [PMC free article] [PubMed] [Google Scholar]
- 52. Gross H, et al. 2010. The NP9 protein encoded by the human endogenous retrovirus HERV-K (HML-2) negatively regulates gene activation of the Epstein-Barr virus nuclear antigen 2 (EBNA2). Int. J. Cancer 129:1105–1115 [DOI] [PubMed] [Google Scholar]
- 53. Herrmann M, Neidhart M, Gay S, Hagenhofer M, Kalden JR. 1998. Retrovirus-associated rheumatic syndromes. Curr. Opin. Rheumatol. 10:347–354 [DOI] [PubMed] [Google Scholar]
- 54. Herve CA, Lugli EB, Brand A, Griffiths DJ, Venables PJ. 2002. Autoantibodies to human endogenous retrovirus-K are frequently detected in health and disease and react with multiple epitopes. Clin. Exp. Immunol. 128:75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hohenadl C, et al. 1999. Transcriptional activation of endogenous retroviral sequences in human epidermal keratinocytes by UVB irradiation. J. Invest. Dermatol. 113:587–594 [DOI] [PubMed] [Google Scholar]
- 56. Howcroft TK, Strebel K, Martin MA, Singer DS. 1993. Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science 260:1320–1322 [DOI] [PubMed] [Google Scholar]
- 57. Hsiao FC, Lin M, Tai A, Chen G, Huber BT. 2006. Cutting edge: Epstein-Barr virus transactivates the HERV-K18 superantigen by docking to the human complement receptor 2 (CD21) on primary B cells. J. Immunol. 177:2056–2060 [DOI] [PubMed] [Google Scholar]
- 58. Hughes JF, Coffin JM. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. U. S. A. 101:1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ishida T, et al. 2008. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun. 8:15. [PMC free article] [PubMed] [Google Scholar]
- 60. Jang KL, Collins MK, Latchman DS. 1992. The human immunodeficiency virus tat protein increases the transcription of human Alu repeated sequences by increasing the activity of the cellular transcription factor TFIIIC. J. Acquir. Immune Defic. Syndr. 5:1142–1147 [PubMed] [Google Scholar]
- 61. Jeang KT, Xiao H, Rich EA. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837–28840 [DOI] [PubMed] [Google Scholar]
- 62. Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709–732 [DOI] [PubMed] [Google Scholar]
- 63. Johnston JB, et al. 2001. Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann. Neurol. 50:434–442 [DOI] [PubMed] [Google Scholar]
- 64. Jones KA, Peterlin BM. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:717–743 [DOI] [PubMed] [Google Scholar]
- 65. Jones RB, et al. 2012. Human endogenous retrovirus K(HML-2) Gag- and Env-specific T-cell responses are infrequently detected in HIV-1-infected subjects using standard peptide matrix-based screening. Clin. Vaccine Immunol. 19:288–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Katsumata K, et al. 1999. Cytokine regulation of env gene expression of human endogenous retrovirus-R in human vascular endothelial cells. Clin. Immunol. 93:75–80 [DOI] [PubMed] [Google Scholar]
- 67. Kaufmann S, et al. 2010. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J. Gen. Virol. 91:1494–1502 [DOI] [PubMed] [Google Scholar]
- 68. Khan AS, Muller J, Sears JF. 2001. Early detection of endogenous retroviruses in chemically induced mouse cells. Virus Res. 79:39–45 [DOI] [PubMed] [Google Scholar]
- 69. Kinoshita S, et al. 1997. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6:235–244 [DOI] [PubMed] [Google Scholar]
- 70. Kobori M, et al. 2004. Wedelolactone suppresses LPS-induced caspase-11 expression by directly inhibiting the IKK complex. Cell Death Differ. 11:123–130 [DOI] [PubMed] [Google Scholar]
- 71. Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL. 2002. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate-early protein 0. Virus Res. 86:93–100 [DOI] [PubMed] [Google Scholar]
- 72. Laderoute MP, et al. 2007. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS 21:2417–2424 [DOI] [PubMed] [Google Scholar]
- 73. Leghmari K, Bennasser Y, Bahraoui E. 2008. HIV-1 Tat protein induces IL-10 production in monocytes by classical and alternative NF-κB pathways. Eur. J. Cell Biol. 87:947–962 [DOI] [PubMed] [Google Scholar]
- 74. Leghmari K, Contreras X, Moureau C, Bahraoui E. 2008. HIV-1 Tat protein induces TNF-alpha and IL-10 production by human macrophages: differential implication of PKC-βII and -δ isozymes and MAP kinases ERK1/2 and p38. Cell. Immunol. 254:46–55 [DOI] [PubMed] [Google Scholar]
- 75. Lewis N, Williams J, Rekosh D, Hammarskjold ML. 1990. Identification of a cis-acting element in human immunodeficiency virus type 2 (HIV-2) that is responsive to the HIV-1 Rev and human T-cell leukemia virus types I and II rex proteins. J. Virol. 64:1690–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu J, Perkins ND, Schmid RM, Nabel GJ. 1992. Specific NF-κB subunits act in concert with Tat to stimulate human immunodeficiency virus type 1 transcription. J. Virol. 66:3883–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Löwer R, Löwer J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 93:5177–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. 1995. Identification of a Rev.-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Magin C, Lower R, Lower J. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mallet F, et al. 2004. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl. Acad. Sci. U. S. A. 101:1731–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mameli G, et al. 2007. Regulation of the syncytin-1 promoter in human astrocytes by multiple sclerosis-related cytokines. Virology 362:120–130 [DOI] [PubMed] [Google Scholar]
- 82. Mann DL, et al. 1989. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res. Hum. Retrovir. 5:253–255 [DOI] [PubMed] [Google Scholar]
- 83. Mayer J, et al. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257–258 [DOI] [PubMed] [Google Scholar]
- 84. Medstrand P, Landry JR, Mager DL. 2001. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 276:1896–1903 [DOI] [PubMed] [Google Scholar]
- 85. Messeguer X, et al. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18:333–334 [DOI] [PubMed] [Google Scholar]
- 86. Minghetti L, et al. 2004. Multiple actions of the human immunodeficiency virus type-1 Tat protein on microglial cell functions. Neurochem Res. 29:965–978 [DOI] [PubMed] [Google Scholar]
- 87. Mingyan Y, Xinyong L, De Clercq E. 2009. NF-κB: the inducible factors of HIV-1 transcription and their inhibitors. Mini Rev. Med. Chem. 9:60–69 [DOI] [PubMed] [Google Scholar]
- 88. Molle D, et al. 2007. A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Muster T, et al. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 63:8735–8741 [PubMed] [Google Scholar]
- 90. Nelson PN, et al. 2003. Demystified. Human endogenous retroviruses. Mol. Pathol. 56:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nguyen KL, Mllano, et al. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319:163–175 [DOI] [PubMed] [Google Scholar]
- 92. Noguchi H, et al. 2004. A new cell-permeable peptide allows successful allogeneic islet transplantation in mice. Nat. Med. 10:305–309 [DOI] [PubMed] [Google Scholar]
- 93. Ono A, Freed EO. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U. S. A. 98:13925–13930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ono M, Kawakami M, Ushikubo H. 1987. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J. Virol. 61:2059–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ott M, et al. 1997. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science 275:1481–1485 [DOI] [PubMed] [Google Scholar]
- 96. Padow M, et al. 2000. Analysis of human immunodeficiency virus type 1 containing HERV-K protease. AIDS Res. Hum. Retrovir. 16:1973–1980 [DOI] [PubMed] [Google Scholar]
- 97. Page KA, Landau NR, Littman DR. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64:5270–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Raney A, Kuo LS, Baugh LL, Foster JL, Garcia JV. 2005. Reconstitution and molecular analysis of an active human immunodeficiency virus type 1 Nef/p21-activated kinase 2 complex. J. Virol. 79:12732–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ratner L, et al. 1987. Complete nucleotide sequences of functional clones of the virus associated with the acquired immunodeficiency syndrome, HTLV-1II/LAV. Haematol Blood Transfus. 31:404–406 [DOI] [PubMed] [Google Scholar]
- 100. Ratner L, et al. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 3:57–69 [DOI] [PubMed] [Google Scholar]
- 101. Reiche J, Pauli G, Ellerbrok H. 2010. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res. 20:435–440 [DOI] [PubMed] [Google Scholar]
- 102. Romani B, Engelbrecht S, Glashoff RH. 2010. Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J. Gen. Virol. 91:1–12 [DOI] [PubMed] [Google Scholar]
- 103. Ruda VM, et al. 2004. Tissue specificity of enhancer and promoter activities of a HERV-K (HML-2) LTR. Virus Res. 104:11–16 [DOI] [PubMed] [Google Scholar]
- 104. Ruprecht K, et al. 2008. Human endogenous retrovirus family HERV-K(HML-2) RNA transcripts are selectively packaged into retroviral particles produced by the human germ cell tumor line Tera-1 and originate mainly from a provirus on chromosome 22q11.21. J. Virol. 82:10008–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sadaie MR, Benter T, Wong-Staal F. 1988. Site-directed mutagenesis of two trans-regulatory genes (tat-III,trs) of HIV-1. Science 239:910–913 [DOI] [PubMed] [Google Scholar]
- 106. Sauter M, et al. 1996. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 56:4362–4365 [PubMed] [Google Scholar]
- 107. Sauter M, et al. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69:414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Scala G, et al. 1994. The expression of the interleukin 6 gene is induced by the human immunodeficiency virus 1 TAT protein. J. Exp. Med. 179:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Schaeffer E, Geleziunas R, Greene WC. 2001. Human immunodeficiency virus type 1 Nef functions at the level of virus entry by enhancing cytoplasmic delivery of virions. J. Virol. 75:2993–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Schanab O, et al. 2011. Expression of human endogenous retrovirus K is stimulated by ultraviolet radiation in melanoma. Pigment Cell Melanoma Res. 24:656–665 [DOI] [PubMed] [Google Scholar]
- 111. Seifarth W, et al. 1998. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J. Virol. 72:8384–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sekigawa I, et al. 2010. A new hypothesis of the possible mechanisms of gender differences in systemic lupus erythematosus. Clin. Exp. Rheumatol. 28:419–423 [PubMed] [Google Scholar]
- 113. Sekigawa I, Ogasawara H, Kaneko H, Hishikawa T, Hashimoto H. 2001. Retroviruses and autoimmunity. Intern. Med. 40:80–86 [DOI] [PubMed] [Google Scholar]
- 114. Serafino A, et al. 2009. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 315:849–862 [DOI] [PubMed] [Google Scholar]
- 115. Shi J, Qin X, Zhao L, Wang G, Liu C. 2011. Human immunodeficiency virus type 1 Tat induces B7-H1 expression via ERK/MAPK signaling pathway. Cell. Immunol. 271:280–285 [DOI] [PubMed] [Google Scholar]
- 116. Stevens RW, et al. 1999. Antibody to human endogenous retrovirus peptide in urine of human immunodeficiency virus type 1-positive patients. Clin. Diagn. Lab. Immunol. 6:783–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sune C, Garcia-Blanco MA. 1995. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 69:6572–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Swanson MD, Winter HC, Goldstein IJ, Markovitz DM. 2010. A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 285:8646–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Tada H, et al. 1990. Trans-activation of the JC virus late promoter by the tat protein of type 1 human immunodeficiency virus in glial cells. Proc. Natl. Acad. Sci. U. S. A. 87:3479–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tai AK, Luka J, Ablashi D, Huber BT. 2009. HHV-6A infection induces expression of HERV-K18-encoded superantigen. J. Clin. Virol. 46:47–48 [DOI] [PubMed] [Google Scholar]
- 122. Takeda S, Sugimoto K, Otsuki H, Hirochika H. 1998. Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Mol. Biol. 36:365–376 [DOI] [PubMed] [Google Scholar]
- 123. Tandon R, et al. 2011. Identification of human endogenous retrovirus-specific T cell responses in vertically HIV-1-infected subjects. J. Virol. 85:11526–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. 1992. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 6:1457–1465 [DOI] [PubMed] [Google Scholar]
- 125. Toufaily C, Landry S, Leib-Mosch C, Rassart E, Barbeau B. 2011. Activation of LTRs from different human endogenous retrovirus (HERV) families by the HTLV-1 tax protein and T-cell activators. Viruses 3:2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Towler EM, et al. 1998. Functional characterization of the protease of human endogenous retrovirus, K10: can it complement HIV-1 protease? Biochemistry 37:17137–17144 [DOI] [PubMed] [Google Scholar]
- 127. Turcanova VL, Bundgaard B, Hollsberg P. 2009. Human herpesvirus-6B induces expression of the human endogenous retrovirus K18-encoded superantigen. J. Clin. Virol. 46:15–19 [DOI] [PubMed] [Google Scholar]
- 128. Turner G, et al. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535 [DOI] [PubMed] [Google Scholar]
- 129. Vacca A, et al. 1994. Human immunodeficiency virus type-1 tat enhances interleukin-2 promoter activity through synergism with phorbol ester and calcium-mediated activation of the NF-AT cis-regulatory motif. Biochem. Biophys. Res. Commun. 205:467–474 [DOI] [PubMed] [Google Scholar]
- 130. van der Kuyl AC. 2012. HIV infection and HERV expression: a review. Retrovirology 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Villarreal L. 2012. Viruses and host evolution: virus-mediated self identity. Adv. Exp. Med. Biol. 738:185–217 [DOI] [PubMed] [Google Scholar]
- 132. Wang-Johanning F, et al. 2012. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J. Natl. Cancer Inst. 104:189–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Weissman JD, et al. 1998. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc. Natl. Acad. Sci. U. S. A. 95:11601–11606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Williams RD, Lee BA, Jackson SP, Proudfoot NJ. 1996. Activation domains of transcription factors mediate replication-dependent transcription from a minimal HIV-1 promoter. Nucleic Acids Res. 24:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Yang J, et al. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. U. S. A. 96:13404–13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yang S, et al. 1999. Generation of retroviral vector for clinical studies using transient transfection. Hum. Gene Ther. 10:123–132 [DOI] [PubMed] [Google Scholar]
- 137. Yedavalli VS, Benkirane M, Jeang KT. 2003. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 278:6404–6410 [DOI] [PubMed] [Google Scholar]
- 138. Yezid H, Konate K, Debaisieux S, Bonhoure A, Beaumelle B. 2009. Mechanism for HIV-1 Tat insertion into the endosome membrane. J. Biol. Chem. 284:22736–22746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zsiros J, Jebbink MF, Lukashov VV, Voute PA, Berkhout B. 1998. Evolutionary relationships within a subgroup of HERV-K-related human endogenous retroviruses. J. Gen. Virol. 79(Pt 1):61–70 [DOI] [PubMed] [Google Scholar]