Abstract
The host noncoding RNA 7SL is highly enriched in the virions of retroviruses. We examined the regions of 7SL that mediate packaging by HIV-1. Both the Alu domain and the S domain were sufficient to mediate specific packaging when expressed separately as truncations of 7SL. However, while the Alu domain competed with endogenous 7SL for packaging in proportion to Gag, the S domain was packaged additively, implying that the Alu and S domains are packaged via separate mechanisms and that the Alu domain is packaged by the same mechanism as endogenous 7SL. Further truncations of the Alu domain or mutation of the Alu domain helix 5c region significantly reduced packaging efficiency, implicating helix 5c as critical for packaging, reinforcing the finding that 7SL packaging is highly selective, and confirming that 7SL is not passively acquired. Surprisingly, when the Alu domain was mutated so that it no longer contained a binding site for the SRP protein heterodimer SRP9/14, it was no longer packaged in a competitive manner but instead was packaged additively with endogenous 7SL. These data support a model in which 7SL RNA is packaged via interactions between Gag and a 7SL RNA structure that exists transiently at a discrete stage of SRP biogenesis. Our data further indicate that a secondary “additive” pathway exists that can result in the packaging of certain 7SL derivatives in molar excess to endogenously packaged 7SL.
INTRODUCTION
Retroviruses, like all RNA viruses, are ribonucleoprotein (RNP) complexes. The primary protein component of retroviral particles is the structural polypeptide Gag, while the primary RNA component is the viral genomic RNA (gRNA). In vitro, nucleic acids or similar molecules are necessary to trigger the assembly of soluble Gag proteins into virus-like particles (9, 10). These RNA-protein interactions, some of which remain poorly understood, are critical during retroviral assembly.
The nucleocapsid (NC) domain is the primary region of Gag involved in interactions with RNA. Although it contributes to many viral functions, in simple terms NC contains RNA-binding motifs that either interact specifically with gRNA (the “zinc finger” motifs) or engage in nonspecific interactions with nucleic acids (the basic regions). Mutations in the latter dramatically reduce intact particle release, while mutations in the former do not (13, 55). Moreover, Gag can form VLPs in the absence of viral gRNA (2, 34, 48). Gag molecules from which NC has been deleted entirely form VLPs less efficiently than wild-type Gag; however, this phenotype can be rescued if NC is replaced with a sequence capable of forming protein-protein interactions, such as a leucine zipper motif (1, 14, 28, 61). Taken together, these data have led to a model in which RNA, but not necessarily the viral genome, promotes the multimerization of Gag through interactions with the NC domain during retroviral assembly.
Although the primary RNA component of retroviral particles is gRNA, retroviruses also package cellular RNAs, and as much as 30% of the RNA in a retroviral particle is host-derived (6, 7, 25, 35, 48). These RNAs are primarily small, noncoding, highly structured RNA polymerase III (Pol III) products, and some are packaged in molar excess over the genomic RNA (41, 42). One such Pol III product is the 301-nucleotide (nt) 7SL RNA, which has been recognized as a component of retroviral particles since the 1970s (7). Retroviruses for which RNA content has been studied in detail display replicable patterns of host RNA packaging, implying nonrandom acquisition of these RNAs. For example, in murine leukemia virus (MLV), the RNAs mY1, mY3, 7SL, and the murine 7SL derivative B1 are highly enriched in the virion (21, 41). Similarly, in human immunodeficiency virus type 1 (HIV-1), 7SL is highly enriched (17, 42). However, the human 7SL derivative scAlu RNA and the hY1 and hY3 RNAs are not present in appreciable amounts in HIV-1 (42, 54, 56). With the exception of the tRNAs that retroviruses use to prime reverse transcription (31, 38), any functions of these packaged host RNAs in viral replication remain unknown.
7SL RNA is well packaged by many retroviruses (7, 20). It is found at 3- to 4-fold molar excess over a monomer of MLV genomic RNA and at 6- to 7-fold molar excess over the genomic RNA of HIV (41, 42). 7SL has been implicated in the packaging of the cytidine deaminase APOBEC3G by HIV-1 (56–58, 60), although this finding remains controversial (3, 30, 51). Evidence for interactions between 7SL derivatives and retrotransposition machinery extends beyond exogenous retroviruses, since the highly repetitive Alu sequences found in primate and rodent genomes are derived from the Alu domain of 7SL (5, 47). Alu elements are retrotransposons, mobilized by retrovirus-like elements present in the genome. The origin of genomic Alu elements can be traced back at least to the origin of the Euarchontoglires lineage (32), which includes both primates and rodents and which diverged ∼85 million years ago. The association between 7SL and retroviruses can therefore be considered an ancient one.
The normal function of 7SL in the cell is as the RNA scaffold of the signal recognition particle (SRP), an RNP complex that is responsible for targeting nascent secretory and transmembrane polypeptides to the endoplasmic reticulum (ER) (for reviews, see references 15 and 37). 7SL RNA is composed of three domains (see Fig. 1): (i) the Alu domain, which binds the SRP proteins SRP9 and SRP14 and is responsible for arresting translation of the nascent polypeptide upon binding to the ribosome complex (24, 26, 52); (ii) the S domain, which binds the SRP proteins SRP19, SRP54, SRP68, and SRP72 and is responsible for targeting the arrested ribosome to the ER (22, 39, 49, 50); and (iii) the linker region, which separates the Alu and S domains. 7SL is also capable of acting as an enzyme, in the sense that a highly conserved region of the S domain, helix 8, is responsible for catalyzing the docking between SRP and the SRP receptor (44, 45). The fact that RNA (but not genomic RNA) is necessary for retroviral assembly, paired with the structural role of 7SL in the cell and its high abundance and selective packaging in at least most kinds of retroviral particles, appears consistent with the possibility that 7SL may play a structural role in retroviral assembly.
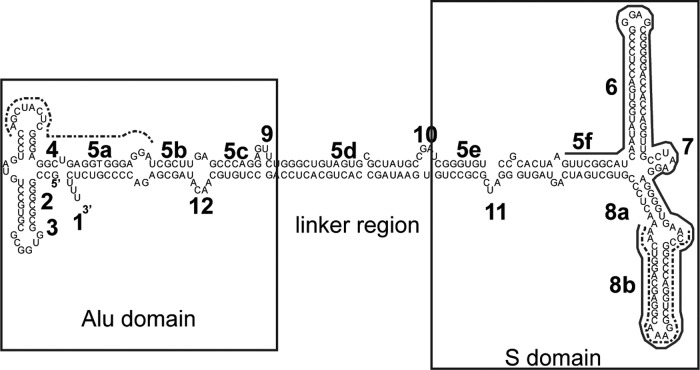
Fig 1.
Secondary structure of human 7SL RNA, allele B. Alu and S domains are boxed. Boldface numbers indicate previously established nomenclature for 7SL helices and loops (58). These designations are preserved throughout the present study to indicate the origins of specific 7SL segments. The dashed lines represent the probes used for Northern blotting, and the solid line represents the probe used in the RNase protection assays.
Previously, we identified the trans-acting determinants of 7SL packaging (29). We demonstrated that even minimal assembly-competent forms of HIV Gag that retain only the C-terminal domain (CTD) of capsid (CA) and spacer protein 1 (sp1) package 7SL RNA as efficiently as intact Gag, but only a fragment of 7SL was observed in particles that lacked NC. This finding indicates that an interaction with CA/sp1 is sufficient for 7SL acquisition but that the Alu domain of 7SL is protected from endonucleolytic processing when NC is present (29). We generated 7SL mutants and tested their packaging to define the cis-acting sequences that mediate specific 7SL packaging by HIV-1.
MATERIALS AND METHODS
Plasmids.
Most 7SL-derivative expression plasmids were made by overlap extension PCR templated by the plasmid p7SL30.1 (62), which contains human 7SL allele B with its endogenous promoter, and inserting the PCR products into the backbone of p7SL30.1 in place of the wild-type (WT) 7SL coding region. 7SL derivatives expressed by the U6 promoter were made synthetically by filling out overlapping 60-nt primers using Taq DNA polymerase. These products were inserted into the multiple cloning site of the plasmid pBluescript KS(+). HIV-1 virions were produced by transfecting 293T or ET cells (46) with the plasmid pHIVpuro, a ΔEnv, ΔVif, ΔVpr version of HIV-1 with the env coding region replaced by a gene coding for puromycin resistance (33). Templates for riboprobes that recognize HIV-1 genomic RNA and 7SL (pSRK1520-1 and pBRU-7SL, respectively) were previously described (36, 42).
Cells and transfections.
Virus was produced in 293T or ET cells (a 293T derivative that constitutively expresses ecotropic envelope [46]), which were cultured and maintained in Dulbecco modified Eagle medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Gemini) at 37°C under 5% CO2. Target cells were D17/pJET cells (canine cells that constitutively express the ecotropic receptor [40]), which were cultured and maintained in DMEM supplemented with 10% bovine serum (CS; Gemini). Transfection was carried out on 100-mm plates using polyethyleneimine (PEI; Polysciences, Inc.), as described previously (29). For each cotransfection of a plasmid that expressed a 7SL derivative with pHIVpuro (the plasmid that expressed the HIV virus-like particles), 8 μg of each plasmid was used. Plasmid DNA was replaced by purified herring sperm DNA (Roche) in the mock samples and the samples that expressed HIV without exogenous 7SL so that the total amount of DNA and PEI used remained constant. The medium-transfection mixture was removed and replaced with 10 ml of fresh medium 24 h after transfection.
Virus processing and RNA isolation.
Tissue culture media was harvested 48 h after transfection and filtered through 0.22-μm-pore-size filters. Virus was quantified by a reverse transcription (RT) assay as described previously (53). Virus was concentrated from 10 ml of medium by centrifugation through a 2-ml 20% sucrose cushion in phosphate-buffered saline for 2 h at 4°C and 25,000 rpm using a Sorvall Surespin 630 rotor in a Sorvall Discovery 90 ultracentrifuge. The pellets were resuspended in 500 μl of TRIzol (Invitrogen), and RNA was isolated according to the manufacturer's instructions. Cells were harvested 48 h after transfection by scraping cells into 2 ml of TRIzol per 100-mm plate, and RNA was isolated according to the manufacturer's instructions.
Northern blotting.
Northern blotting was carried out as described previously (29). Loading was normalized so that RNA from equivalent amounts of virus was loaded in each lane. For cellular RNA, 0.6% of the total RNA isolated from a confluent 100-mm plate was loaded into each lane. The locations recognized by the oligonucleotide probes used are indicated by dashed lines in Fig. 1 and were as follows: to detect the Alu domain (see Fig. 2b, 3b, and 5a) we used 5′-ATCCTCCAGCCTCAGCCTCCCGAGTAGCTG-3′, and to detect the S domain (Fig. 4b) we used 5′-TTTTGACCTGCTCCGTTTCCGACCT-3′. Images were acquired by scanning with a Typhoon Trio variable mode imager, and quantification was performed using the one-dimensional gel imaging feature of ImageQuant TL (v7.0).
Fig 2.
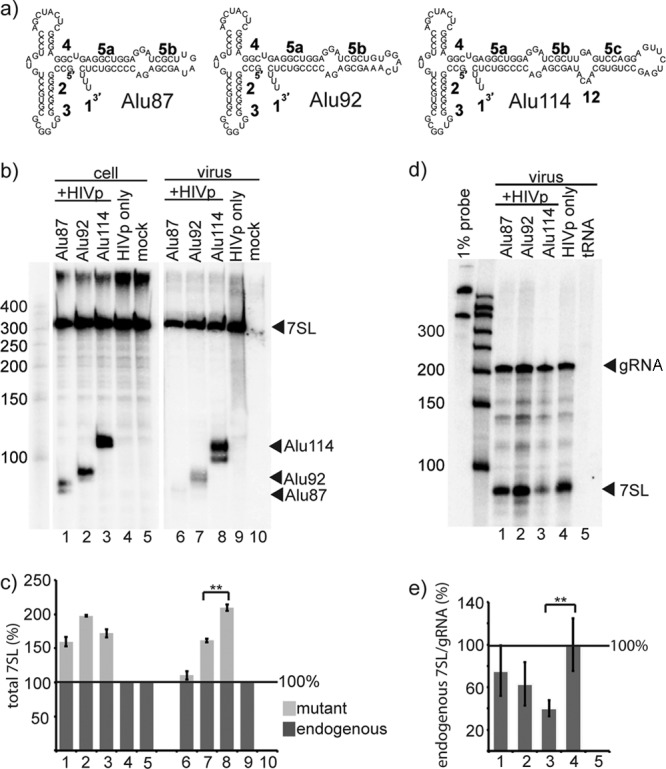
The Alu domain is sufficient for packaging. (a) Schematics of the Alu domain derivatives used. Alu114 corresponded to the entire Alu domain, and Alu92 and Alu87 were successively shorter truncations. (b) Northern blot of RNA from cotransfected cells and HIVp virus, showing the expression and packaging of the Alu domain derivatives. The probe used was against the Alu domain of 7SL. The multiple bands observed for the Alu derivatives likely reflect alternate 3′ end modifications (11); virion encapsidation of subsets of Pol III processing products has been described previously (20). (c) Quantification of mutant (light gray) and endogenous (dark gray) 7SL RNA from Northern blots. Column numbers correspond to the lane numbers in panel B. The mean amount of total 7SL is shown; endogenous 7SL was set to 100% for cells and virus. (d) RPA of RNA from HIVp virus. Riboprobes used were against the S domain of 7SL and HIV genomic RNA (gRNA). (e) Quantification of RPAs of HIVp virus RNA. Column numbers correspond to the lane numbers in panel D. gRNA, genomic RNA. The mean amount of endogenous 7SL RNA per gRNA is shown; the 7SL/gRNA value of the HIVp-only sample was set at 100%. Error bars indicate ± the standard deviation. **, P < 0.01 (n = 3 independent experiments).
Fig 3.
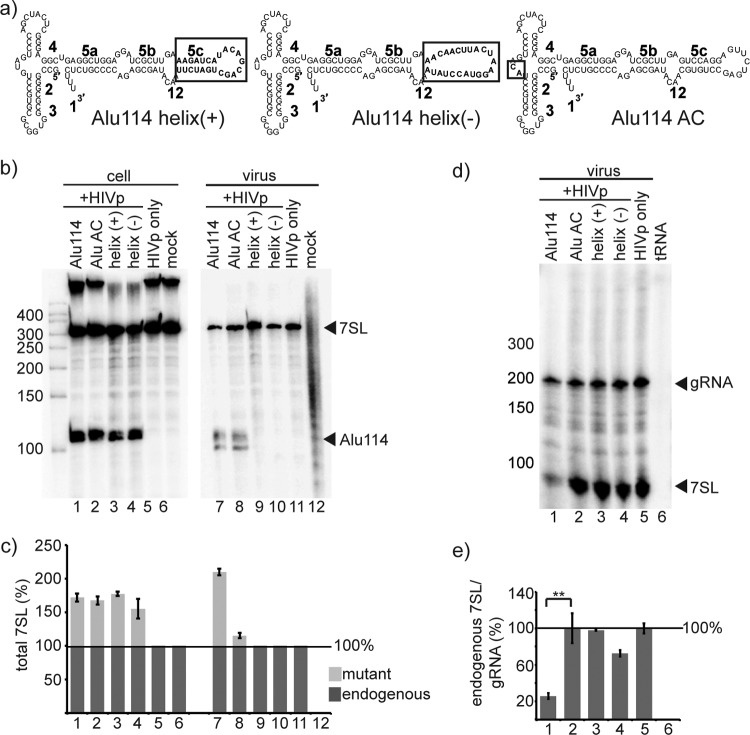
Disruption of helix 5c, but not interference with SRP9/14 binding, abrogates packaging of the Alu domain. (a) Schematics of mutants of the Alu domain derivative Alu114. The mutated regions are boxed. (b) Northern blot of RNA from transfected cells and HIVp virus, showing the expression and packaging of Alu114 and the Alu domain mutants. The probe used recognized the Alu domain. (c) Quantification of mutant (light gray) and endogenous (dark gray) 7SL RNA from Northern blots. Column numbers correspond to the lane numbers in panel B. The mean amount of total 7SL is shown; endogenous 7SL was set to 100% for cells and virus. (d and e) RNase protection assay (d) and quantification of RNA from HIVp virus based on RPAs like than in panel d (e). Column numbers correspond to the lane numbers in panel d. The mean amount of endogenous 7SL RNA per genomic RNA is shown; the value of endogenous 7SL/gRNA of the HIVp-only sample was set at 100%. Error bars indicate ± the standard deviation. **, P < 0.01 (n = 3 independent experiments).
Fig 5.
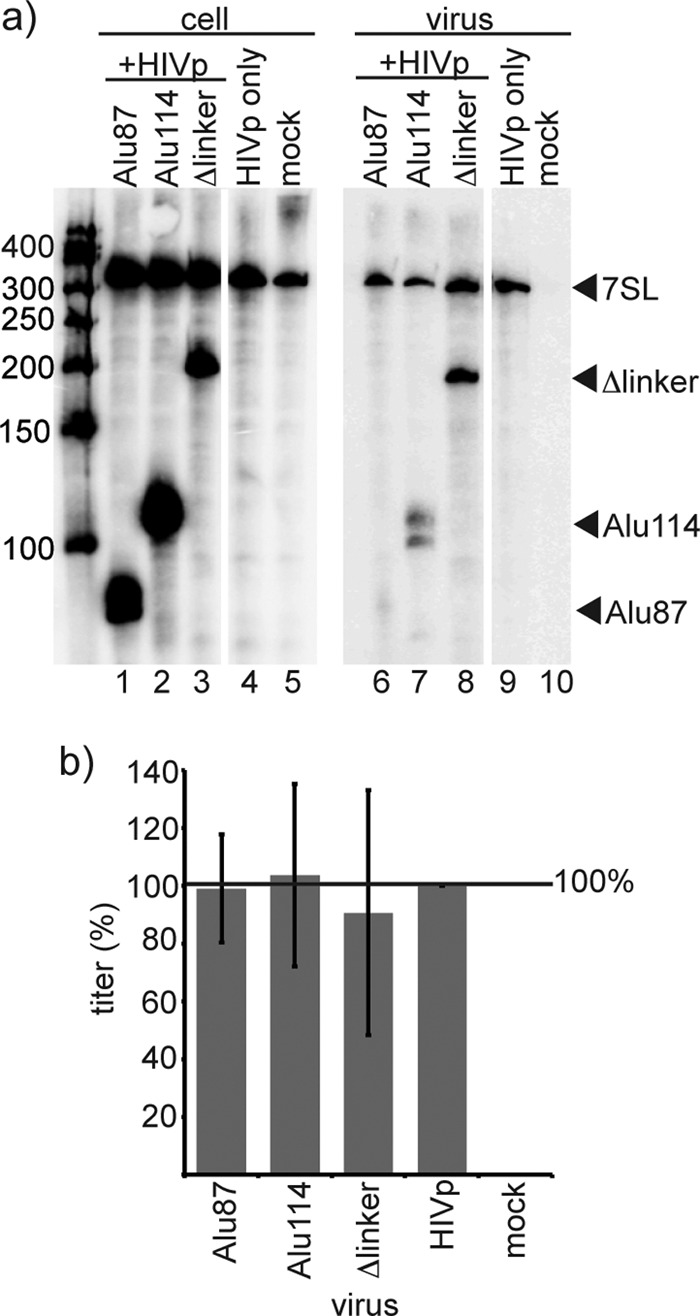
Packaging of 7SL derivatives does not have detectable effects on viral infectivity. (a) Northern blot of RNA from transfected cells and HIV-1 virus, showing the expression and packaging of 7SL derivatives, visualized with an Alu domain probe. (b) Titer of 7SL derivative-containing HIVpuro virus, normalized for virion content by an RT assay. Error bars indicate ± the standard deviation. n = 3 independent experiments.
Fig 4.
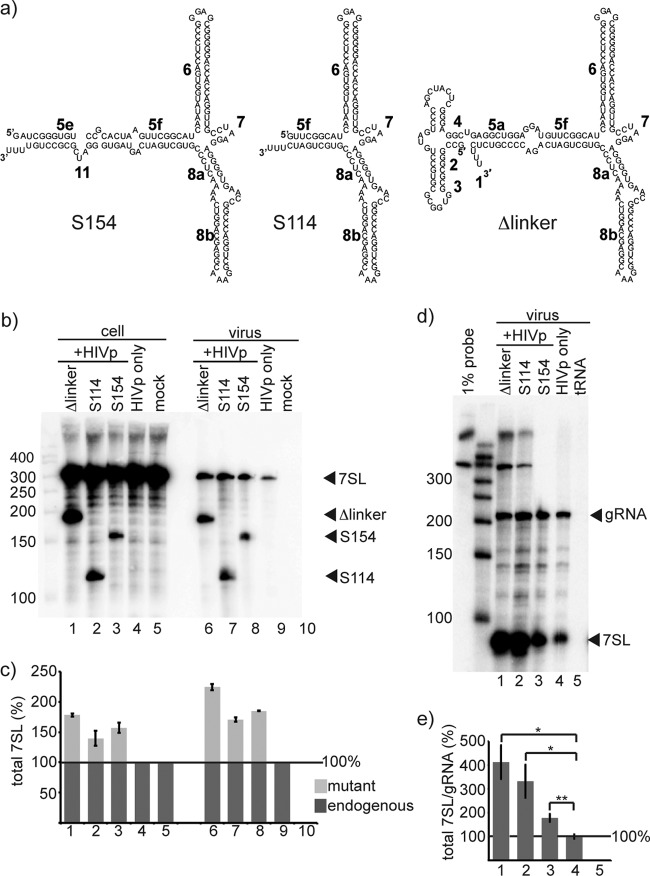
The S domain mediates additive packaging. (a) Schematics of the S domain derivatives used. (b) Northern blot of RNA from transfected cells and HIVp virus, showing the expression and packaging of the S domain derivatives. The probe used recognizes the S domain. (c) Quantification of mutant (light gray) and endogenous (dark gray) 7SL RNA from Northern blots. Column numbers correspond to the lane numbers in panel b. The mean amount of total 7SL is shown; endogenous 7SL was set to 100% for cells and virus. (d) RPA of RNA from HIVp virus. (e) Quantification of RPAs of RNA from HIVp virus. Column numbers correspond to the lane numbers in panel d. The mean amount of endogenous 7SL RNA per genomic RNA is shown; the value of 7SL/gRNA of the HIVp-only sample was set at 100%. Error bars indicate ± the standard deviation. *, P < 0.05; **, P < 0.01 (n = 3 independent experiments).
RPAs.
RNase protection assays (RPAs) were performed as previously described (41). The riboprobes used recognized either a portion of the HIV-1 gag gene (CA/sp/NC) or the S domain of 7SL from C116 to C217 (indicated as a solid line in Fig. 1).
Titer.
HIVpuro virus for infection was produced by transfection of ET cells as described above. Medium was harvested after 48 h and filtered through 0.22-μm-pore-size filters. Then, 100-mm plates of D17/pJET cells were infected with 2 ml of DMEM plus 10% CS, 5 μg of Polybrene (Sigma)/ml, and serial dilutions of HIV-containing medium. Two hours later, the medium was discarded, and 10 ml of fresh medium was added to each plate. After 2 days incubation, the medium was replaced with DMEM plus 10% CS plus 2 μg of puromycin (Sigma)/ml, which was changed every 2 days until colonies were visible at 12 to 14 days after infection. The colonies were stained with trypan blue and counted.
Statistical analysis.
Statistical significance was analyzed by performing two-tailed, unpaired Student t test on pairs of data, using the statistical functions of Microsoft Excel. Differences were considered significant if the P value was <0.05.
RESULTS
An isolated Alu domain competes with endogenous 7SL for packaging.
To determine the minimal region of 7SL capable of mediating specific packaging, truncations of 7SL were created that corresponded to successively shorter regions of the Alu domain. As diagrammed in Fig. 2a, Alu114 contained an internal deletion in 7SL from G82 to C268, thus yielding a 114-nt RNA that corresponded to the entire Alu domain, while Alu92 and Alu87 contained larger deletions and thus encoded smaller RNAs. Alu114 corresponds to the same region of 7SL that is the ancestor of the murine 7SL derivative B1, which is packaged by MLV (41), and of the human 7SL derivative scAlu. The Alu domain derivatives were cotransfected with HIVpuro (33)—a plasmid that expresses HIV virus-like particles that contain all viral proteins except Env, Vif, and Vpr and will henceforth be referred to as HIVp—and the RNA content of cotransfected cells and HIVp virions produced from these cells was examined by Northern blotting (Fig. 2b). Although expression of all three Alu domain truncations was readily detectable in cotransfected cells (Fig. 2b, lanes 1 to 3), only Alu114 was packaged in high quantities in virions (lane 8), with Alu92 and Alu87 packaged less well (lanes 6 and 7). Quantification of these blots revealed that Alu114 comprised ca. 50% of the total 7SL (endogenous 7SL and Alu114) in HIVp virions from cotransfected cells (Fig. 2c), whereas Alu92 comprised ca. 35% and Alu87 comprised ca. 10% of the total 7SL. The amount of Alu114 and Alu92 that was packaged by HIVp virions was significantly different (P < 0.01). Another way of addressing packaging selectivity is to determine how enriched specific RNAs are, by comparing their levels in virions to their levels in cells (41). If the enrichment factor of endogenous 7SL was set to 1, Alu114 was found to be enriched 1.5-fold in the virus over its concentration in the cell.
The amount of endogenous 7SL per virion was subsequently quantified by performing a RNase protection assay (RPA) using two riboprobes: one to the 7SL S domain that recognized endogenous 7SL, but not Alu114, and one that recognized viral genomic RNA (Fig. 2d). RT assays were performed on the viral samples to quantify viral proteins, and the amount of genomic RNA per sample was found to be proportional to the RT for each sample. Thus, detection of genomic RNA allowed for normalization to the number of virions. Comparing the endogenous 7SL signal for the HIVp-only virus (lane 4) to that for the Alu114-containing virus (lane 3) showed that the ratio of endogenous 7SL to gRNA in virions was ∼2-fold lower in HIVp virions produced by cotransfected cells in which ∼40% of the total 7SL-type RNA was Alu114 than it was in virions produced in the absence of Alu114 (Fig. 2e). This difference in endogenous 7SL packaging was statistically significant (P < 0.01). Similarly, the amount of endogenous 7SL was reduced ca. 35% in HIVp virions containing Alu92, and ca. 14% in HIVp virions containing Alu87. Paired with Fig. 2c data, these findings indicated that the total amount of 7SL-type RNAs (7SL and Alu114, Alu92, or Alu87) in virions remained constant, implying that the Alu domain derivatives competed for packaging with endogenous 7SL. Based on previously determined endogenous 7SL packaging levels (12 to 14 copies) (42), these data indicate that under the conditions here, six to seven molecules of Alu114 replaced the corresponding number of endogenous 7SL molecules per viral particle, or approximately four molecules of Alu92 or one molecule of Alu87 was packaged per virion when these later derivatives were coexpressed.
Helix 5c, but not binding to SRP9/14, is required for packaging of the Alu domain.
Next, the difference between the well-packaged Alu114 and the poorly packaged truncations was dissected to define the packaging signals in the Alu domain. The primary difference between these RNAs was the presence of helix 5c (H5c) in Alu114 (Fig. 1 and a). To investigate whether the length, secondary structure, flexibility, or sequence of H5c accounted for the increased packaging of Alu114, H5c of Alu114 was mutated to produce Alu114 helix(+) and Alu114 helix(−) (Fig. 3a). In Alu114 helix(+), the sequence of H5c was mutated in a way predicted to maintain the H5c helix-loop conformation; in Alu114 helix(−), both the sequence and the predicted secondary structure of H5c were altered.
To assess the effects of H5c modification on RNA packaging, Northern blotting was performed on RNA from cells cotransfected with HIVp and the Alu domain derivatives, and on RNA from virions produced from these cells (Fig. 3b). Although expression of both constructs was detectable in cells (lanes 3 and 4), neither Alu derivative was present in the virus above the threshold of detection. By quantifying the background signal and the signal of packaged Alu114, we determined that both helix(+) and helix(−) RNAs were packaged at least 6-fold less efficiently than Alu114 (compare lane 7 to lanes 9 and 10). Quantification of these Northern data is presented in Fig. 3c. These results suggest that the specific sequence of H5c, rather than its length or the basic features of its secondary structure, was crucial for packaging.
To further address Alu packaging determinants, the roles of this RNA's protein binding partners were assessed. During SRP biogenesis, the first proteins that bind nascent 7SL are the protein heterodimer SRP9/14, which forms a complex with the Alu domain (12). In initial work not shown, the SRP9/14 protein content of virions was examined by Western blotting to determine whether SRP9/14 was packaged by HIVp. Although the results indicated that the SRP9/14 in virions was at least 8-fold de-enriched compared to cells when normalized to 7SL content, this sensitivity was not high enough to conclusively exclude SRP9/14 packaging in virions because SRP9/14 exists in 20-fold excess over 7SL in the cell (8).
Therefore, the possible role of SRP9/14 binding was examined indirectly, by creating a derivative of Alu114 in which the binding site for SRP9/14 was abolished by changing the nucleotides G24 and U25 to A and C, respectively (Fig. 3a, henceforth referred to as Alu AC). In RNA/protein cocrystals, G24 and U25 are the only nucleotides of 7SL that are specifically recognized by SRP9/14 (59), and mutations to G24 and U25 reduce SRP9/14 binding up to 50-fold in vitro (11, 18). To assess the influence of an intact SRP9/14 binding site on the packaging of Alu114, cells were cotransfected with HIVp and the Alu AC expression construct, and Northern blotting was performed on the RNA content of these cells and the virions produced by them (Fig. 3b). Similar to observations with the unmutated Alu114 derivative (lanes 1 and 7), the Alu AC mutant was detectable in both cells (lane 2) and virus (lane 8). These results are quantified in Fig. 3c. The relatively efficient packaging of Alu AC suggested that the binding of SRP9/14 was not required for the packaging of the Alu domain.
We subsequently quantified the amount of endogenous 7SL packaged in the presence of the Alu AC mutant by RPA (Fig. 3d). Unlike the significant decrease in endogenous 7SL observed in virions containing Alu114 (lane 1), the endogenous 7SL signal in virions containing the Alu AC mutant remained unchanged compared to gRNA (compare lane 2 to lane 5). Quantification of the RPA confirmed that the endogenous 7SL to gRNA ratio decreased in Alu114-containing virions, but not in Alu AC-containing virions (Fig. 3e). The difference in the packaging of endogenous 7SL between the Alu114-containing virions and the Alu AC-containing virions was statistically significant (P < 0.01). These data indicate that while binding to SRP9/14 was not required for packaging of the Alu domain, the mechanism by which the Alu domain was packaged was no longer competitive with endogenous 7SL when its interaction with SRP9/14 was ablated.
S domain-only RNAs are packaged in addition to endogenous 7SL.
Truncations of 7SL corresponding to isolated S domains were also created to determine the influence of the S domain on 7SL packaging. The S domain is the most highly conserved portion of SRP RNAs, and orthologs of the S domain RNA are found in all kingdoms of life (15). Because the 7SL promoter contains intragenic promoter elements within the Alu domain (19), an isolated S domain could not be expressed under the WT 7SL promoter. Therefore, the S domain derivatives S154 and S114 were expressed using the U6 promoter, a Pol III type III promoter that contains only extragenic promoter elements (43) (Fig. 4a). S154 corresponded to the entirety of the S domain, while S114 corresponded to the 111-nt fragment of the S domain that is retained in minimal VLPs (29). To exclude the possibility that any phenotypes of the S domain RNAs resulted from use of the heterologous U6 promoter, a third S domain derivative, termed Δlinker, was constructed in which the S domain was expressed by the native 7SL promoter. The Δlinker mutant contained two deletions in 7SL, from C61 to A119 and from A231 to G286, which removed the 7SL linker region RNA stem and parts of the Alu and S domains (Fig. 4a). The left-hand 75 nt of Δlinker, corresponding to part of the Alu domain (helices 2, 3, 4, and 5a), contained the intragenic promoter elements. However, as determined in Fig. 3 and data not shown, this portion of 7SL was incapable of mediating packaging (Fig. 2). The right-hand 111 nt of Δlinker corresponded to the S114 derivative described above (helices 5f, 6, 7, and 8).
The RNA content of virions produced by cells cotransfected with expression constructs for these S domain derivatives and HIVp was examined by Northern blotting (Fig. 4b). S154 and S114 were detected in both cells (lanes 2 and 3) and HIVp virus (lanes 7 and 8). The mutant Δlinker was also packaged by HIVp (Fig. 4b, lane 6), confirming that the S domain was capable of mediating packaging regardless of its promoter. These results are quantified in Fig. 4c. If the enrichment factor of endogenous 7SL was set to 1, S114 was found to be enriched 1.8-fold in the virus over its concentration in the cell, S154 was found to be enriched 1.5-fold in the virus over its concentration in the cell, and Δlinker was found to be enriched 1.6-fold in the virus over its concentration in the cell. We also quantified the total 7SL RNA content (S domain derivatives and endogenous 7SL) compared to the genomic RNA content of the HIVp virions by an RPA (Fig. 4d). Viral protein quantification by RT activity confirmed that genomic RNA was proportional to the number of virions present in each sample (data not shown). When normalized to encapsidated gRNA, the total 7SL signal in the S domain derivative-containing virions was 2- to 4-fold higher than that for the HIVp-only virus (compare lanes 1 to 3 to lane 4). The amount of total 7SL packaged in the S domain derivative-containing virions was significantly different than the amount of total 7SL packaged in the HIVp-only virions (P < 0.05). Quantified in Fig. 4e, these results confirm that the S domain derivatives were packaged in addition to endogenous 7SL. Based on 12 to 14 copies of endogenous 7SL per viral particle, these data indicate that there were ∼40 copies of Δlinker per viral particle, ∼30 copies of S114 per viral particle, or ∼10 copies of S154 per viral particle in addition to the endogenous 7SL.
Packaging of exogenous 7SL, or Alu or S domain derivatives, does not affect viral infectivity detectably.
Because HIVp genomic RNAs are competent for reverse transcription and integration and contain a puromycin-resistant selectable marker, HIVp puromycin-resistant colony-forming titer was used to determine whether incorporating 7SL derivatives was detrimental to viral infectivity in a single-cycle replication assay. HIVp virions that packaged Alu domain or S domain derivatives were used to infect target cells, and the puromycin-resistant titers from a single round of replication were determined. In these experiments, virions were pseudotyped with ecotropic Env and titered on cells that constitutively expressed the ecotropic receptor. Producer cells were cotransfected with HIVp and the Alu domain derivatives Alu87 or Alu114 or the S domain derivative Δlinker. Northern blotting of the RNA content of producer cells and virus was performed to verify exogenous 7SL derivative expression and packaging levels (Fig. 5a). Under the transfection conditions used here, these virions contained either 50% of the endogenous 7SL replaced with Alu114, or (in the Δlinker S domain-containing virions) additional 7SL derivatives in 1- to 1.5-fold molar excess of the encapsidated endogenous 7SL.
The puromycin-resistant CFU titers of these viruses are presented in Fig. 5b. When normalized to input virus as quantified by RT activity, no significant difference between the titer of virus containing Alu114 or Δlinker (Fig. 5b, columns 2 and 3) and those of HIVpuro-only virus (column 4) was detected (P > 0.05). Similarly, the expression of the nonpackaged 7SL derivative Alu87 by producer cells had no effect on the titer of HIVpuro (column 1). These data indicate that the packaging of exogenous 7SL derivatives did not detectably affect viral infectivity under the conditions tested here.
DISCUSSION
We examined the determinants of 7SL packaging by HIV-1 by establishing packaging properties of overexpressed 7SL truncations. 7SL RNA consists of three domains: the left-hand Alu domain, the right-hand S domain, and the linker region that separates the two (Fig. 1). The results here demonstrated that both the Alu and S domains were capable of mediating packaging independently. They also showed that the Alu domain competed with endogenous 7SL for packaging, while the amount of endogenous 7SL remained unchanged when excess S domain was packaged. Some 7SL derivatives that were highly expressed were not packaged at detectable levels, providing information on the determinants of 7SL packaging and confirming that the packaging of 7SL does not result from a random encapsidation of cellular RNAs. These data indicate that there are two mechanisms by which 7SL can become encapsidated: a “competitive” pathway and an “additive” pathway.
A truncation of 7SL corresponding to the entire Alu domain, called Alu114, was found to replace some of the endogenous 7SL in HIVp particles. This competition for packaging between Alu114 and endogenous 7SL implies that endogenous 7SL is packaged via the same packaging pathway as Alu114. This competitive pathway is therefore likely to be the endogenous pathway by which HIV-1 recruits 7SL. This suggests that the Gag-proportionate packaging of endogenous 7SL that is characteristic of several retroviruses (41, 42, 54) involves an interaction of 7SL's Alu domain with Gag. Furthermore, the decreased packaging of the shorter Alu domain truncations, Alu87 and Alu92, and of the Alu114 derivatives in which helix 5c was mutated, suggests that H5 of the Alu domain is crucial in the endogenous pathway. The competitive packaging of Alu RNAs that lacked the S domain suggests that the endogenous pathway does not require recognition of the S domain by Gag.
The endogenous pathway appears to limit the number of 7SL molecules acquired. Multiples studies have shown that a fixed number (12 to 16) of 7SL molecules are acquired by HIV-1, and that the packaging of endogenous 7SL occurs in proportion to Gag and independent of the packaging of gRNA (42, 48, 54). The evidence here that Alu domain derivatives replaced endogenous 7SL in the virion implies that exogenously expressed Alu domains serve as substrates in the endogenous pathway, which involves a counting mechanism by which an assembling viral particle acquires a certain number of 7SL molecules. In contrast to this, the additive pathway does not appear to involve a counting mechanism because the total number of RNAs in the virus appeared to increase when additive 7SL derivatives were packaged.
The change in phenotype from competitive to additive packaging when the binding site of SRP9/14 was ablated in Alu114 AC suggests that SRP9/14 binding is involved in the endogenous 7SL packaging pathway. Binding of SRP9/14 to 7SL occurs early in SRP biogenesis and is responsible for maintaining 7SL in the nucleolus for subsequent steps in SRP complex formation (12, 27). Thus, the evidence here that 7SL derivatives that cannot bind SRP9/14 were packaged via the additive pathway suggests that location and timing are crucial to the mechanisms of 7SL packaging.
The data that the SRP9/14 binding capability is crucial for packaging via the endogenous pathway suggests a possible model for 7SL packaging. In this model, the endogenous, competitive pathway may involve acquisition of a form of 7SL that ordinarily exists transiently, early in its biogenesis, whereas the additive pathway may involve acquisition of 7SL later during SRP biogenesis and in a different location in the cell. This model suggests that the regulated trafficking of endogenous 7SL through the cell may prevent the packaging of endogenous 7SL through the additive pathway. Observations with the SRP9/14 binding site mutant suggested that the binding of SRP9/14 may transiently produce a three-dimensional structure in helix 5c of the Alu domain that is recognized by Gag but that is altered upon binding of the remaining SRP proteins. In this model, Alu114 was packaged exclusively by the endogenous pathway because it could not bind further SRP proteins and therefore was locked in a packaging-competent form, while Alu AC and the S domain truncations were never in the correct conformation.
This model raises the possibility that HIV-1 acquires 7SL in the nucleus. Although fluorescence microscopy visualization of Gag during HIV assembly and evidence that HIV-1 replication does not require Crm1-dependent nuclear trafficking appears inconsistent with this notion, existing work does not rule out Crm1-independent nuclear trafficking or the possibility that a small subset of HIV-1 Gags may traffic through the nucleus (4, 23). It is also possible that HIV-1 Gag acquires 7SL in the cytoplasm but before SRP54 has bound 7SL, that expression of HIV-1 changes the subcellular localization of 7SL, or that a fraction of nuclear SRP assembly intermediates intersect with retroviral assembly in some other way.
The effect of 7SL derivative packaging on virus titer was examined to determine whether alterations to the 7SL content of virions affected infectivity. Under the conditions tested here—that is, between approximately 6 and 24 replaced or additional 7SL-type molecules per virion—the packaging of 7SL derivatives had no discernible effect on titer. This was true whether the packaged 7SL derivative was acquired via the endogenous pathway (Alu114) or the additive pathway (Δlinker). These findings suggest that the normal complement of 7SL is not required for HIV-1 infectivity. If 7SL does contribute to viral replication, the lack of a detectable effect on infectivity when Alu114 replaced some of the endogenous 7SL implies either that the Alu domain alone is capable of fulfilling any 7SL-specific roles, or that the remaining endogenous 7SL is capable of fulfilling that role. Similarly, the lack of a detectable effect on infectivity when Δlinker was packaged implies that the packaging of extra 7SL did not destabilize the virion or interfere with viral replication cycle. This lack of evidence for a role of 7SL in the viral life cycle may instead indicate that 7SL packaging results because 7SL is a parasite of retroviruses, much like how Alu elements parasitize the L1 retrotransposition machinery (16). However, it is noteworthy that all of the experiments here were performed under overexpression conditions in 293T cells: conditions known to mask some forms of restriction and other forms of virus-host interactions. In addition, we did not produce virions that completely lacked 7SL or 7SL derivatives and cannot rule out a role for 7SL in viral replication.
In summary, these studies establish two pathways by which 7SL can be acquired by HIV-1: the endogenous pathway, which involves the Alu domain and SRP9/14 binding, and an additive pathway, which occurs in the absence of SRP9/14 binding and which may be dependent on alternative trafficking of 7SL. The additive pathway can result in the packaging of additional 7SL RNA, and the studies here provide evidence that the nature of these additionally packaged RNAs can be artificially manipulated. Our demonstrated ability to manipulate the RNA content of retroviral virions has potentially far-reaching applications as a delivery mechanism for modified RNAs.
ACKNOWLEDGMENTS
This study was supported by NIH grant R21 AI080276 and a Gates Grand Challenges Exploration Grant to A.T. and NIH grant T32 GM 07544 to S.E.K.
We thank Kathy Spindler and Adewunmi Onafuwa-Nuga for their helpful comments on the manuscript.
Footnotes
Published ahead of print 16 May 2012
REFERENCES
- 1. Accola MA, Strack B, Gottlinger HG. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aldovini A, Young RA. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bach D, et al. 2008. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology 5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baluyot MF, Grosse SA, Lyddon TD, Janaka SK, Johnson MC. 2012. CRM1-dependent trafficking of retroviral Gag proteins revisited. J. Virol. 86:4696–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bennett EA, et al. 2008. Active Alu retrotransposons in the human genome. Genome Res. 18:1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177–218 [DOI] [PubMed] [Google Scholar]
- 7. Bishop JM, et al. 1970. The low-molecular-weight RNAs of Rous sarcoma virus. II. The 7S RNA. Virology 42:927–937 [DOI] [PubMed] [Google Scholar]
- 8. Bovia F, Fornallaz M, Leffers H, Strub K. 1995. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol. Biol. Cell 6:471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell S, Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campbell S, Vogt VM. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang DY, Newitt JA, Hsu K, Bernstein HD, Maraia RJ. 1997. A highly conserved nucleotide in the Alu domain of SRP RNA mediates translation arrest through high-affinity binding to SRP9/14. Nucleic Acids Res. 25:1117–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Sinha K, Perumal K, Gu J, Reddy R. 1998. Accurate 3′ end processing and adenylation of human signal recognition particle RNA and Alu RNA in vitro. J. Biol. Chem. 273:35023–35031 [DOI] [PubMed] [Google Scholar]
- 13. Cimarelli A, Sandin S, Hoglund S, Luban J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crist RM, et al. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cross BC, Sinning I, Luirink J, High S. 2009. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell. Biol. 10:255–264 [DOI] [PubMed] [Google Scholar]
- 16. Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41–48 [DOI] [PubMed] [Google Scholar]
- 17. Didierlaurent L, et al. 2011. Role of HIV-1 RNA and protein determinants for the selective packaging of spliced and unspliced viral RNA and host U6 and 7SL RNA in virus particles. Nucleic Acids Res. 39:8915–8927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emde G, Frontzek A, Benecke BJ. 1997. Secondary structure of the nascent 7S L RNA mediates efficient transcription by RNA polymerase III. RNA 3:538–549 [PMC free article] [PubMed] [Google Scholar]
- 19. Englert M, Felis M, Junker V, Beier H. 2004. Novel upstream and intragenic control elements for the RNA polymerase III-dependent transcription of human 7SL RNA genes. Biochimie 86:867–874 [DOI] [PubMed] [Google Scholar]
- 20. Faras AJ, Garapin AC, Levinson WE, Bishop JM, Goodman HM. 1973. Characterization of the low-molecular-weight RNAs associated with the 70S RNA of Rous sarcoma virus. J. Virol. 12:334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garcia EL, et al. 2009. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J. Virol. 83:12526–12534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gowda K, Chittenden K, Zwieb C. 1997. Binding site of the M-domain of human protein SRP54 determined by systematic site-directed mutagenesis of signal recognition particle RNA. Nucleic Acids Res. 25:388–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grewe B, et al. 2012. Cytoplasmic Utilization of Human Immunodeficiency Virus type 1 Genomic RNA Is Not Dependent on a Nuclear Interaction with Gag. J. Virol. 86:2990–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halic M, et al. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427:808–814 [DOI] [PubMed] [Google Scholar]
- 25. Houzet L, et al. 2007. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 35:2695–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huck L, et al. 2004. Conserved tertiary base pairing ensures proper RNA folding and efficient assembly of the signal recognition particle Alu domain. Nucleic Acids Res. 32:4915–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jacobson MR, Pederson T. 1998. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 95:7981–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson MC, Scobie HM, Ma YM, Vogt VM. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177–11185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keene SE, King SR, Telesnitsky A. 2010. 7SL RNA is retained in HIV-1 minimal virus-like particles as an S-domain fragment. J. Virol. 84:9070–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khan MA, et al. 2007. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleiman L. 2002. tRNALys3: the primer tRNA for reverse transcription in HIV-1. IUBMB Life 53:107–114 [DOI] [PubMed] [Google Scholar]
- 32. Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. 2007. Evolutionary history of 7SL RNA-derived SINEs in supraprimates. Trends Genet. 23:158–161 [DOI] [PubMed] [Google Scholar]
- 33. Laurent LC, Olsen MN, Crowley RA, Savilahti H, Brown PO. 2000. Functional characterization of the human immunodeficiency virus type 1 genome by genetic footprinting. J. Virol. 74:2760–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Levin JG, Grimley PM, Ramseur JM, Berezesky IK. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Linial ML, Miller AD. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125–152 [DOI] [PubMed] [Google Scholar]
- 36. Lu K, Heng X, Summers MF. 2011. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 410:609–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luirink J, Sinning I. 2004. SRP-mediated protein targeting: structure and function revisited. Biochim. Biophys. Acta 1694:17–35 [DOI] [PubMed] [Google Scholar]
- 38. Mak J, Kleiman L. 1997. Primer tRNAs for reverse transcription. J. Virol. 71:8087–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Menichelli E, Isel C, Oubridge C, Nagai K. 2007. Protein-induced conformational changes of RNA during the assembly of human signal recognition particle. J. Mol. Biol. 367:187–203 [DOI] [PubMed] [Google Scholar]
- 40. O'Reilly L, Roth MJ. 2000. Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses. J. Virol. 74:899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Onafuwa-Nuga AA, King SR, Telesnitsky A. 2005. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J. Virol. 79:13528–13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Onafuwa-Nuga AA, Telesnitsky A, King SR. 2006. 7SL RNA, but not the 54-kDa signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA 12:542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paule MR, White RJ. 2000. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28:1283–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peluso P, et al. 2000. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science 288:1640–1643 [DOI] [PubMed] [Google Scholar]
- 45. Peluso P, Shan SO, Nock S, Herschlag D, Walter P. 2001. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry 40:15224–15233 [DOI] [PubMed] [Google Scholar]
- 46. Pfeiffer JK, Topping RS, Shin NH, Telesnitsky A. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rowold DJ, Herrera RJ. 2000. Alu elements and the human genome. Genetica 108:57–72 [DOI] [PubMed] [Google Scholar]
- 48. Rulli SJ, et al. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 81:6623–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siegel V, Walter P. 1988. Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein-RNA “footprinting.” Proc. Natl. Acad. Sci. U. S. A. 85:1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Siegel V, Walter P. 1986. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature 320:81–84 [DOI] [PubMed] [Google Scholar]
- 51. Strebel K, Khan MA. 2008. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology 5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Strub K, Moss J, Walter P. 1991. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol. Cell. Biol. 11:3949–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Telesnitsky A, Blain S, Goff SP. 1995. Assays for retroviral reverse transcriptase. Methods Enzymol. 262:347–362 [DOI] [PubMed] [Google Scholar]
- 54. Tian C, Wang T, Zhang W, Yu XF. 2007. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 35:7288–7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang SW, Aldovini A. 2002. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76:11853–11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang T, et al. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 81:13112–13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. 2008. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 375:1098–1112 [DOI] [PubMed] [Google Scholar]
- 58. Wang T, et al. 2008. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology 377:71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weichenrieder O, Wild K, Strub K, Cusack S. 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408:167–173 [DOI] [PubMed] [Google Scholar]
- 60. Zhang W, et al. 2010. Association of potent human antiviral cytidine deaminases with 7SL RNA and viral RNP in HIV-1 virions. J. Virol. 84:12903–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zwieb C, Ullu E. 1986. Identification of dynamic sequences in the central domain of 7SL RNA. Nucleic Acids Res. 14:4639–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]