Abstract
PCNA is monoubiquitinated in response to DNA damage and fork stalling and then initiates recruitment of specialized polymerases in the DNA damage tolerance pathway, translesion synthesis (TLS). Since PCNA is reported to associate with Epstein-Barr virus (EBV) DNA during its replication, we investigated whether the EBV deubiquitinating (DUB) enzyme encoded by BPLF1 targets ubiquitinated PCNA and disrupts TLS. An N-terminal BPLF1 fragment (a BPLF1 construct containing the first 246 amino acids [BPLF1 1-246]) associated with PCNA and attenuated its ubiquitination in response to fork-stalling agents UV and hydroxyurea in cultured cells. Moreover, monoubiquitinated PCNA was deubiquitinated after incubation with purified BPLF1 1-246 in vitro. BPLF1 1-246 dysregulated TLS by reducing recruitment of the specialized repair polymerase polymerase η (Polη) to the detergent-resistant chromatin compartment and virtually abolished localization of Polη to nuclear repair foci, both hallmarks of TLS. Expression of BPLF1 1-246 decreased viability of UV-treated cells and led to cell death, presumably through deubiquitination of PCNA and the inability to repair damaged DNA. Importantly, deubiquitination of PCNA could be detected endogenously in EBV-infected cells in comparison with samples expressing short hairpin RNA (shRNA) against BPLF1. Further, the specificity of the interaction between BPLF1 and PCNA was dependent upon a PCNA-interacting peptide (PIP) domain within the N-terminal region of BPLF1. Both DUB activity and PIP sequence are conserved in the members of the family Herpesviridae. Thus, deubiquitination of PCNA, normally deubiquitinated by cellular USP1, by the viral DUB can disrupt repair of DNA damage by compromising recruitment of TLS polymerase to stalled replication forks. PCNA is the first cellular target identified for BPLF1 and its deubiquitinating activity.
INTRODUCTION
DNA damage prevents normal replicative synthesis of DNA at the replication fork. Lesions in DNA that cause stalling at the replication fork must be bypassed or avoided before replication can continue. DNA damage tolerance mechanisms collectively known as postreplication repair (PRR) allow for such lesion bypass and avoidance. There are two main subpathways of PRR, a translesion synthesis (TLS) mechanism that allows the DNA replication machinery to replicate damaged DNA templates and a template switch (TS) pathway that avoids damaged bases by using the undamaged nascent sister lagging strand as the template.
Many details of the TLS PRR pathway are well-established (46). TLS is initiated in response to replication fork stalling at sites of DNA damage. The resulting helicase-polymerase uncoupling at stalled replication forks generates long tracts of single-stranded DNA (ssDNA) that are coated by replication protein A (RPA) (9). RPA-coated ssDNA triggers the recruitment of a complex containing an E3 ubiquitin ligase termed “Rad18” and its binding partner DNA polymerase eta (Polη) to sites of DNA damage (12, 51). Once Rad18 has been recruited to sites of damage, it mediates monoubiquitination of proliferating cell nuclear antigen (PCNA) (a DNA polymerase processivity factor) at residue K164. Polη possesses a C-terminal PCNA-interacting polypeptide (PIP) box (containing the consensus PIP sequence Qxxhxxaa, where x is any amino acid, h represents a hydrophobic amino acid, and a represents an aromatic amino acid) and a ubiquitin-binding zinc finger UBZ motif which mediate its association with monoubiquitinated PCNA (2, 6, 49). Owing to a flexible active site that can accommodate certain bulky helix-distorting lesions (including UV-induced cyclobutane pyrimidine dimers or CPD), Polη can engage damaged DNA templates and execute error-prone replicative bypass of DNA lesions.
Polη is one of a family of specialized DNA polymerases that includes Polκ, Polι, and REV1 (46). Similar to Polη, other Y-family TLS DNA polymerases possess ubiquitin-binding motifs and are recruited to stalled replication forks via interactions with monoubiquitinated PCNA (2, 6). Different TLS polymerases have different lesion specificities, and thus collectively, TLS polymerases allow PRR of gaps resulting from diverse forms of DNA damage (31). Although TLS polymerase-mediated lesion bypass permits maintenance of DNA replication when the genome is damaged, the process is inherently error-prone and has the potential to induce mutations. An alternative repair pathway, template switch, is activated by polyubiquitination of K164-monoubiquitinated PCNA by Rad5 E3 ubiquitin ligase (16, 39). The cues for selection of error-prone TLS versus the error-free TS pathway for resolving stalled DNA replication remain obscure. Nevertheless, Rad18 activity is necessary for both TLS and TS.
Rad18 (as well as Rad5) mediated PCNA ubiquitination at K164 is antagonized by the cellular ubiquitin-specific protease 1 (USP1) (26). USP1 is the only known enzyme that deubiquitinates PCNA. However, the effects of USP1 are regulated. DNA damaging agents including UV irradiation induce degradation of USP1, thereby prolonging the PCNA monoubiquitinated state and recruitment of TLS polymerases (26). Thus, there is a dynamic equilibrium between activation of PCNA induced by ubiquitination and the supply of USP1 available in TLS.
In addition to its roles in DNA replication and PRR, PCNA appears to be involved in the viral lytic cycle of Epstein-Barr virus (EBV). EBV is a ubiquitous human gammaherpesvirus that causes infectious mononucleosis, and it is associated with Hodgkin's lymphoma, Burkitt's lymphoma, and nasopharyngeal carcinoma (42). Daikoku et al. (11) have reported that PCNA and several DNA repair proteins are assembled in EBV replication compartments, that PCNA is loaded onto newly synthesized viral DNA, and that levels of chromatin-bound PCNA increase during productive viral replication. However, the putative effects on viral replication itself and the mechanisms through which PCNA is involved in this context remain unknown.
BPLF1 is the large tegument protein of EBV (3,149 amino acids). It is classified as a late lytic cycle gene, but transcripts are detected as early as 6 to 8 h after infection (19, 56), and herpesvirus BPLF1 homologs have both early and late functions during infection, including virus entry, transport, and assembly (1, 4, 7, 13). Expression of short hairpin RNA (shRNA) against BPLF1 results in decreased EBV genome copy numbers (54).
BPLF1 contains deubiquitinating (DUB) activity within the first 205 amino acids of the N-terminal region (49a, 49b). The deubiquitinating activity of BPLF1 and its catalytic triad (Cys-His-Asp) despite sequence diversity in this region are strictly conserved across the Herpesviridae, and mutation of the active site cysteine results in complete loss of enzymatic activity (20a, 33, 54). BPLF1 DUB activity cleaves both K63 and K48 polyubiquitin chains (54). A functionally active BPLF1 fragment has recently been shown to block degradation of cytosolic and endoplasmic reticulum (ER) proteins by removal of ubiquitin from substrates targeted to the proteasome (15). BPLF1 interacts with EBV ribonucleotide reductase (RR), deubiquitinates its large subunit (RR1), and downregulates RR activity (54). BPLF1 also has deneddylating activity that utilizes the same active site as the deubiquitinating activity. BPLF1 interacts with and deneddlylates Cullin ring ligases, which could potentially affect viral replication (19). In addition, the Kaposi's sarcoma-associated herpesvirus (KSHV) DUB homolog was recently shown to decrease retinoic acid-inducible gene I product (RIG-I) ubiquitination and reduce RIG-I-mediated interferon (IFN) signaling, suggesting that the viral DUB activity antagonizes RIG-I signaling during KSHV infection (27). Several herpesviruses whose deubiquitinating activity has been deleted have been constructed, and all result in approximately 10-fold decrease in viral infectivity (8, 21, 34, 53). A deubiquitinating activity knockout of Marek's disease virus, an alphaherpesvirus, reduced viral replication and severely impaired formation of T-cell lymphomas (29).
Given the likely critical roles of BPLF1 in viral infectivity, identification of relevant cellular targets of the viral DUB activity is of considerable interest. Because PCNA is associated with EBV DNA during its replication and mono- and polyubiquitination are critical for its functions in PRR, we asked whether PCNA is a target for EBV's deubiquitinating activity. In studies described here, we identify PCNA as a binding partner and substrate of BPLF1. Moreover, we demonstrate that BPLF1 inhibits the localization of Polη to DNA damage-induced nuclear repair foci. PCNA is the first cellular protein identified as a target for the EBV deubiquitinating enzyme. The cellular USP1 is established as the only known DUB that deubiquitinates PCNA. We show here that the EBV DUB, BPLF1, also specifically deubiquitinates PCNA and disrupts TLS repair. These findings identify a novel mechanism whereby herpesviruses can disrupt the cellular response to DNA damage through their DUB activity.
MATERIALS AND METHODS
Cell lines, growth, and transfection.
HEK293T, EBV293+, and H1299 cells were cultured as described previously (54). Transfections were performed with Lipofectamine 2000 (Invitrogen) (H129 cells) or Effectene (Qiagen) (HEK293T cells) according to the manufacturer's protocol.
Antibodies, immunoprecipitations, and Western blotting.
FLAG-tagged BPLF1 constructs (54) were immunoprecipitated with M2 affinity gel (Sigma) or with FLAG M2 antibody (Sigma) bound to Dynabeads magnetic beads (Invitrogen). PCNA was detected with PCNA (PC10) (Santa Cruz) and Polη with polyclonal Polη antibody (Abcam) (ab17725). Endogenous BPLF1 was detected with custom rabbit antibody against BPLF1 peptide (Thermo Scientific). Western blotting was performed as previously described (54).
Ub-AMC assay.
Assays were performed by incubating ubiquitin (Ub)–7-amido-4-methylcoumarin (AMC) (Enzo) with purified BPLF1 constructs essentially as described previously (54).
Genotoxin treatment and cellular fractionation.
HEK293T and H1299 cells were exposed to DNA damaging agents ultraviolet C (UVC) and hydroxyurea (HU). For the UV treatment, 24 to 48 h after transfection, medium was removed from the cells and replaced with a minimal volume of phosphate-buffered saline (PBS), the cells were subjected to 60 J/m2 UV, and the medium was replaced for 3 h before harvesting (5). For the HU treatment, the cells were treated with 2.5 mM HU for 3 h and then harvested. Lysates were fractionated into soluble and chromatin-containing fractions with the use of CSK buffer as described previously (28).
Immunofluorescence and visualization of Polη foci.
H1299 cells were transfected with YFP-tagged Polη (5), FLAG-tagged BPLF1 1-246 (a BPLF1 construct containing the first 246 amino acids), and inactive BPLF1 C61S constructs. Twenty-four hours after transfection, the samples were subjected to 60 J/m2 UV damage and incubated for 3 h. The cells were washed twice with PBS and fixed with 4% paraformaldehyde for 7 min. The cells were washed and fixed with 0.1% Triton X-100 for 10 min at room temperature (RT). The cells were washed and blocked with normal donkey serum at RT for 2 h and probed with primary FLAG antibody (1:1,000; mouse) (clone M2; Sigma) incubated with Alexa Fluor 594 mouse antibody, stained with 4′,6′-diamidino-2-phenylindole (DAPI), and examined with an Olympus IX 81-ZDC inverted fluorescence microscope and Hammatsu ORCA RC camera and QImaging RETIGA 4000R color camera at room temperature and ×40 magnification.
Construction of BPLF1 PIP mutant.
BPLF1 1-246 pFLAG-CMV-2 (CMV stands for cytomegalovirus) (54) served as the template. The PIP binding sequence QYTCVHLYF was converted to QYTCVHLAA (original and new amino acids shown in boldface type). The FWD (FWD stands for forward) primer was 5′ CAGTACACTTGCGTGCATCTCGCTGCTCTACCCGAGGCCTTTGAGACAG 3′, and the REV (REV stands for reverse) primer was 5′ CTGTCTCAAAGGCCTCGGGTAGAGCAGCGAGATGCACGCAAGTGTACTG 3′.
PCR was performed with Kapa HiFi Hot Start (Kapa Biosystems) polymerase according to the manufacturer's instructions. The entire sequence was verified.
Protein purification of PCNA and BPLF1 1-246 and in vitro interaction.
His-tagged PCNA (His-PCNA) and His-tagged BPLF1 (His-BPLF1) constructs (54) were grown in BL21 Codon Plus DE3-RIPL cells (Stratagene) and were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 to 4 h. The cultures were centrifuged, and the pellet was resuspended in HLB buffer (50 mM Na2HPO4 [pH 8.0], 20 mM imidazole, 10% glycerol, 0.02% NP-40, 5 mM β-mercaptoethanol [BME], 150 mM NaCl). The cells were lysed by sonication, and cleared lysates were incubated with nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) overnight. The beads were eluted with LB plus 300 mM imidazole and dialyzed against S100 buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 10% glycerol, 0.02% NP-40, 5 mM BME, plus protease inhibitors). Purified His-BPLF1 1-246 and His-PCNA were incubated together for 30 min at 25°C.
Cell viability assay.
H1299 cells were transfected with BPLF1 1-246, BPLF1 with the active site cysteine mutated (BPLF1 C61S), and BPLF1 PIP. Forty-eight hours posttransfection, cells were UV irradiated with 0, 1, 2, 5, and 10 J/m2 and were grown for 12 days. The cells were washed with PBS and counted.
RESULTS
PCNA interacts with BPLF1 in vivo.
Since PCNA is monoubiquitinated following DNA damage and reported to bind to EBV DNA during viral replication (11), we evaluated whether BPLF1 and PCNA can interact. A BPLF1 construct containing the first 246 amino acids (BPLF1 1-246) that retains DUB activity was ectopically expressed in 293T cells (54). Mutation of the active site cysteine (C61) in the backbone of BPLF1 1-246 (BPLF1 C61S) results in loss of deubiquitinating activity (54). Cells were transfected with FLAG-tagged versions of BPLF1 1-246 and BPLF1 C61S, and extracts were immunoprecipitated with FLAG antibody (Sigma), then separated by SDS-PAGE, and probed for PCNA. As shown in Fig. 1A, PCNA was immunoprecipitated specifically with BPLF1 1-246, but it associated less efficiently with BPLF1 C61S. These data demonstrate that BPLF1 interacts with PCNA in vivo either directly or through a complex involving both PCNA and BPLF1. It is noteworthy that the enzymatically active form of BPLF1 interacts more strongly with PCNA, which suggests that functional BPLF1 is required for its interaction with PCNA.
Fig 1.
BPLF1 interacts with PCNA both in vivo and in vitro. (A) In an in vivo interaction, immunoprecipitations (IPs) were performed on lysates from 293T cells expressing BPLF1 1-246 followed by Western blotting (WB) for endogenous levels of PCNA. Western blotting of lysates detected PCNA, BPLF1 1-246, and BPLF1 C61S. The relative amounts of constituents are shown by densitometry beneath each row. Approximately 10% of BPLF1 C61S bound to PCNA (when total expression levels of BPLF1 1-246 and BPLF1 C61S are factored in) with WT BPLF1 binding set at 100% (1.0 is equivalent to 100%). (B) In an in vivo interaction, FLAG-tagged BPLF1 1-246 and HA-tagged PCNA were overexpressed in 293T cells, and IP was performed with FLAG antibody. HA-PCNA and FLAG-BPLF1 1-246 were present in the reaction mixtures as shown in the middle and bottom panels. Mixing of lysates from samples represented by lanes 1 to 4 demonstrate that interaction occurred in the cells and not in the lysates (lane 5). Lysates from the indicated samples (lanes 5 to 8) were mixed as indicated prior to IP (e.g., lane 5 contains the samples from lanes 1 and 2 mixed together). The middle and bottom panels show the presence of BPLF1 and PCNA. The position of the IgG band is indicated by black arrows to the right of the panels. (C) In an in vitro interaction, BPLF1 1-246 and PCNA were purified from E. coli and incubated together for 30 min at 25°C. IPs were performed with BPLF1 antiserum and probed for PCNA. PCNA and BPLF1 were present in the reaction mixtures as shown in the middle and bottom panels.
In addition, immunoprecipitations (IPs) with hemagglutinin (HA)-tagged PCNA (pCMV-HA-PCNAwt [wt stands for wild type]) (57) and FLAG-tagged BPLF1 1-246 revealed that PCNA and BPLF1 1-246 interact within the cell (Fig. 1B, lanes 1 to 4). Lysates from reaction mixtures overexpressing BPLF1 and PCNA from separate samples that were mixed prior to IP demonstrate that interactions were not artifacts that occurred after cell lysis under IP conditions (Fig. 1B, lane 5).
PCNA interacts with BPLF1 in vitro.
To determine whether the interaction between BPLF1 and PCNA is direct, in vitro reactions were performed. BPLF1 1-246 and PCNA were purified from Escherichia coli and incubated together for 30 min at 25°C. Immunoprecipitations were performed with BPLF1 antibodies and probed for the presence of PCNA (Fig. 1C). The results indicate that BPLF1 1-246 interacts directly with PCNA.
Deubiquitinating activity of BPLF1 removes ubiquitin from PCNA in vivo.
To determine whether PCNA is a substrate for BPLF1, we examined whether its ubiquitination was affected by the viral DUB activity. 293T cells were transfected with an expression vector encoding myc-Rad18 (to enhance PCNA monoubiquitination) (44) alone or in combination with a BPLF1 expression plasmid. Cells expressing Rad18 and BPLF1 were treated with hydroxyurea (HU) or UV irradiated to increase PCNA ubiquitination and activation of translesion synthesis. After treatment, cell lysates were separated into soluble (combined cytosol and nucleosol) and insoluble (chromatin) fractions. Monoubiquitinated PCNA should be detected mainly in the chromatin fraction, since the monoubiquitinated form is almost exclusively associated with the chromatin (31, 32). As expected, Rad18 and genotoxin treatments induced ubiquitination of chromatin-bound PCNA (as evidenced by a PCNA mobility shift of ∼8 kDa) (Fig. 2A). Interestingly, both Rad18 and genotoxin-induced PCNA ubiquitination were nullified in cells expressing BPLF1 1-246 (Fig. 2A). These data indicate that expression of BPLF1 results in the removal of ubiquitin from PCNA and suggests that the viral enzyme will influence the translesion synthesis response to DNA damage.
Fig 2.
BPLF1 1-246 deubiquitinates PCNA in vivo and in vitro. (A) In vivo, 293T cells were transfected with BPLF1 and Rad18 expression plasmids. Rad18, PCNA's natural ubiquitin ligase, was utilized to enhance PCNA monoubiquitination. Cells were treated with hydroxyurea (HU) or UV irradiated and fractionated into chromatin-containing and soluble fractions. Samples were probed with PCNA and Rad18 antibody. (B) In vitro, H1299 cells were transfected with Rad18 and treated with 2 μM hydroxyurea for 2 h. Fractions containing ubiquitinated PCNA were evenly divided from a single ubiquitinated lysate and incubated with (+) or without (−) purified 1 μM BPLF1 1-246 and placed at 37°C overnight. Reactions were performed with lysates from 1 × 107 H1299 cells. Western blots were probed with antibody against PCNA. The relative density of PCNA-Ub band is indicated below the blot (1.0 is equivalent to 100%).
Although the levels of soluble Rad18 were diminished in BPLF1-expressing cells, BPLF1 did not affect the levels of Rad18 in the chromatin compartment (where PCNA ubiquitination occurs). Therefore, the reduced PCNA ubiquitination we observed in BPLF-expressing cells is most likely due to PCNA deubiquitination.
DUB activity of BPLF1 removes ubiquitin in vitro from PCNA.
The reduced levels of ubiquitinated PCNA observed in BPLF1-expressing cells (Fig. 2A) could have resulted from reduced Rad18-mediated E3 ligase activity or through BPLF1-mediated deubiquitination of PCNA (or both mechanisms). To distinguish between these possibilities, we performed in vitro PCNA deubiquitination assays. A chromatin fraction containing ubiquitinated PCNA from HU-treated and Rad18-overexpressing H1299 cells was evenly divided and incubated with or without active recombinant bacterial BPLF1 1-246 (purified from E. coli as described previously [54]). As shown in Fig. 2B, chromatin-associated PCNA was deubiquitinated by purified BPLF1 1-246. The levels of unmodified PCNA in chromatin were unaffected by incubation with BPLF1 1-246. The results of these in vitro experiments confirm that BPLF1 has PCNA-directed DUB activity.
BPLF1 DUB activity results in reduced recruitment of DNA polymerase η.
Monoubiquitinated PCNA interacts much more strongly with Polη than unmodified PCNA (32). Monoubiquitinated PCNA is necessary for the DNA damage-inducible recruitment of Polη to sites of replication stalling (6, 32). Therefore, we predicted that deubiquitination of PCNA by BPLF1 DUB would compromise damage-induced recruitment of Polη to chromatin. To test this prediction, 293T cells were transfected with a Rad18 expression plasmid to increase the levels of ubiquitinated PCNA and treated with UV or hydroxyurea. As shown in Fig. 3A, ectopically expressed BPLF1 1-246 reduced the levels of chromatin-associated Polη by 84% and 69% in UV- and HU-treated cells, respectively. A catalytically inactive BPLF1 mutant did not affect PCNA ubiquitination or chromatin-association of Polη. The chromatin association of Polη is dependent on its interaction with ubiquitinated PCNA. Thus, the levels of chromatin-associated Polη are reduced concomitant with loss of monoubiquitinated PCNA in BPLF1-expressing cells. The amount of Polη does not decrease upon expression of BPLF1, but the amount of Polη recruited to the chromatin is decreased. Therefore, the inhibitory effect of BPLF1 on Polη recruitment is most likely mediated via PCNA-directed DUB activity.
Fig 3.
BPLF1 DUB activity disrupts polymerase η recruitment and focus formation. Polymerase η is shown as pol eta in the figure. (A) DUB activity decreases Polη recruitment to chromatin. 293T cells were transfected with BPLF1 1-246, exposed to UV or HU treatment, fractionated, and probed for Polη. The relative density indicates the percentage of Polη recruited to the chromatin. The bottom panels show expression of PCNA and ubiquitinated PCNA (PCNA-Ub) in the chromatin. (B) H1299 cells were transfected with YFP-tagged Polη and FLAG-tagged BPLF1 1-246 and BPLF1 C61S constructs and irradiated with UV. Cells were fixed, permeabilized, and incubated with fluorescent antibody against FLAG. Nuclei were detected with DAPI staining. (C) Foci from transfected immunofluorescence samples were counted and quantitated as the average number of foci per cell. The foci from 20 cells were counted. (D) Samples in which Polη foci formation was abolished. PCNA was also deubiquitinated by BPLF1 DUB activity. In samples transfected in tandem with those for immunofluorescence, H1299 cells were harvested after UV treatment and probed for PCNA by Western blotting. The bottom panel shows expression of BPLF1 1-246 and BPLF1 C61S.
DNA polymerase η focus formation is disrupted by BPLF1 DUB activity.
A hallmark of activation of the TLS pathway is formation of nuclear foci containing Polη localized at sites of DNA damage. Polη focus formation depends on PCNA ubiquitination (6, 32). To investigate further the effects of BPLF1 on TLS pathway activation, we expressed yellow fluorescent protein (YFP)-tagged Polη (YFP-Polη) alone or together with BPLF1 1-246 or BPLF1 C61S in H1299 cells. We used fluorescence microscopy to examine the effects of BPLF1 on the UV-inducible redistribution of Polη to nuclear foci. UV treatment induces an increase in the number of cells containing YFP-Polη foci (48). In cultures coexpressing Polη and active BPLF1 1-246, but not inactive BPLF1 C61S, there was a striking reduction of focus formation (Fig. 3B). Polη focus formation in cells expressing inactive BPLF1 C61S was essentially the same as that observed in untransfected cells after UV damage (6, 32, 48). Formation of Polη foci in cells expressing BPLF1 1-246 was almost completely abolished, whereas cells expressing the inactive mutant had little effect on foci (approximately 25-fold reduction as quantitated in Fig. 3C). Figure 3D shows the results of Western blotting of samples treated at the same time as samples used for immunofluorescence. Loss of ubiquitinated PCNA is evident when BPLF1 is expressed and demonstrates the correlation between PCNA ubiquitination and formation of Polη foci. Thus, DUB activity of BPLF1 alters translesion synthesis by inhibiting Polη recruitment to sites of DNA damage.
The PCNA-interacting peptide is conserved across herpesviral deubiquitinating enzymes.
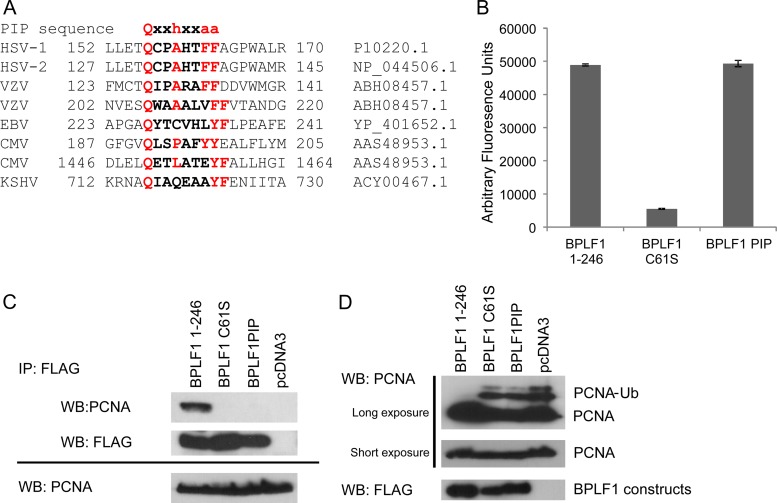
Many proteins, including Polη, that interact with PCNA contain a PCNA-interacting peptide (PIP domain). The conserved sequence of the PIP domain is Qxxhxxaa where h represents a hydrophobic amino acid, a represents an aromatic amino acid, and x represents any amino acid. A large number of proteins that interact with PCNA, including many DNA replication and repair factors, contain this interacting motif, but it is not required. We found a PIP motif in BPLF1 at amino acids 227 to 234. Compared with a canonical PIP box, the putative viral PIP sequence contains an insertion of an extra amino acid between the hydrophobic and aromatic residues (Qxxhxxaa versus Cxxhxxxaa). From sequence analysis, all members of the herpesvirus family that we investigated have potential PIP domains within their N-terminal regions (Fig. 4A) which suggests that deubiquitination of PCNA and subsequent inhibition of translesion repair is conserved across the Herpesviridae.
Fig 4.
BPLF1 contains a PIP domain that is conserved and is essential for interaction with PCNA. (A) PIP domain is conserved across herpesviral DUBs. The PCNA-interacting peptide sequence is shown at the top of the panel, and h stands for a hydrophobic amino acid, a stands for an aromatic amino acid, and x is any amino acid. The accession numbers are shown on the right. In some cases, 3 amino acids are present between the hydrophobic and aromatic amino acids. The sequences for herpes simplex virus 1 (HSV-1) and 2 (HSV-2), vesicular stomatitis virus (VSV), EBV, cytomegalovirus (CMV), and Kaposi's sarcoma-associated herpesvirus (KSHV) are shown. (B) Mutation of the PIP domain does not affect DUB activity. Fluorogenic ubiquitin-AMC was incubated with BPLF1 constructs purified by IP. Protein expression was normalized for the BPLF1 constructs. (C) Mutation of the PIP domain in BPLF1 abolishes interaction with PCNA. 293T cells were transected with FLAG-tagged constructs: BPLF1 1-246, BPLF1 C61S, and BPLF1 PIP. Immunoprecipitations were performed on whole-cell lysates with FLAG antibody and probed for endogenous PCNA (top two panels). Expression of BPLF1 constructs and PCNA levels were detected by Western blotting of total lysates, shown in the bottom panel. (D) The PIP domain is essential for PCNA deubiquitination. H1299 cells were transfected with BPLF1 constructs as noted and subjected to UV damage. Cells were fractionated into chromatin-containing fractions, and Western blotting was performed for PCNA. The top panel shows both ubiquitinated and unmodified PCNA. Only WT BPLF1 removed ubiquitin from PCNA. Unmodified PCNA expression levels are shown as a short exposure in the middle panel, and the levels of BPLF1 constructs are shown in the in bottom panel.
BPLF's PIP domain is essential for its interaction with PCNA.
The BPLF1 sequence from amino acids 227 to 235 closely resembles the published PIP consensus sequence. To determine whether this PIP-like (or “putative PIP”) domain is involved in the interaction with PCNA, immunoprecipitations were performed with 293T cell extracts overexpressing FLAG-tagged BPLF1 1-246, BPLF1 C61S, and BPLF1 PIP (the wild-type [WT] sequence of QYTCVHLYF was converted to QYTCVHLAA). The aromatic region in the PIP domain is commonly mutated to alanine (YF to AA) to disrupt protein interactions involving the PIP domain (2, 22, 25, 52). Incubation of purified BPLF1 constructs with fluorogenic Ub-AMC (54) demonstrated that the BPLF1 PIP construct maintains enzymatic activity (Fig. 4B). After immunoprecipitation with FLAG antibody, Western blots were probed with antibody against endogenous PCNA (Fig. 4C). While PCNA was immunoprecipitated efficiently with WT BPLF1 1-246, its associations with the PIP mutant BPLF1 and catalytically inactive BPLF1 C61S were largely abolished. The results suggest that BPLF1 binding to PCNA involves at least two sites including a PIP-like sequence. These results demonstrate that the specificity of the BPLF1 PIP domain and the DUB active sites are intrinsic to its interaction with PCNA.
We expected that deubiquitination of PCNA by BPLF1 PIP would be compromised due to its inability to bind PCNA. To test this prediction, cells were transfected with BPLF1 1-246, BPLF1 C61S, and BPLF1 PIP and UV irradiated to induce PCNA monoubiquitination (Fig. 4D). As expected, monoubiquitinated PCNA was not detected in BPLF1 1-246-expressing cells, but it was present in cells expressing similar levels of BPLF1 C61S and PIP mutants. Therefore, abolishing the interaction of PCNA and BPLF1 abolishes deubiquitination of PCNA by BPLF1. These results underscore that BPLF1-mediated PCNA deubiquitination is a specific, nonpromiscuous activity.
BPLF1 DUB activity results in decreased cell viability after UV damage as reflected by cell numbers.
To assess phenotypic changes due to inhibition of TLS, we measured cell viability after UV damage in the presence of BPLF1. We hypothesized that cells with the ability to repair DNA through TLS would be able to recover and survive DNA damage, whereas cells that express BPLF1, and therefore have an inhibited TLS pathway, would not survive and would consequently result in reduced cell numbers. H1299 cells were transfected with BPLF1 1-246, BPLF1 C61S, and BPLF1 PIP and irradiated with graduated doses of UV. Adherent cells were washed and counted 12 days after UV. Figure 5 demonstrates that cells overexpressing BPLF1 resulted in decreased cell numbers after exposure to 5 and 10 J/m2, whereas both enzymatically inactive and PIP mutants had no effect. It thus appears that EBV BPLF1, by deubiquitinating PCNA and presumably interrupting TLS, resulted in the loss of the ability to repair DNA damage and subsequent cell death.
Fig 5.
BPLF1 results in reduced viability of UV-irradiated cells. H1299 cells were transfected with BPLF1 constructs and irradiated with UV. After 12 days, cells were counted.
Endogenous levels of BPLF1 expressed during EBV reactivation result in diminished ubiquitination of PCNA.
To determine whether the levels of BPLF1 expressed endogenously during EBV infection could result in deubiquitination of PCNA, we induced the viral lytic cycle with the viral transactivator BZLF1 with and without expression of shRNA against BPLF1. We have shown that shRNA 893 produces knockdown of BPLF1 transcripts and results in reduced viral genome copy numbers (54). When shRNA against endogenously expressed full-length BPLF1 was expressed, ubiquitinated PCNA increased by 61% after DNA damage, whereas the total levels of PCNA remained fairly constant (94% versus 100%) (Fig. 6A). These experiments were repeated after three different doses of UV (30, 50, and 70 J/m2). All exposures resulted in similar levels of PCNA ubiquitination and relative deubiquitination and are quantitated in Fig. 6B. The ratios obtained are virtually identical to those in Fig. 6A. Importantly, we demonstrate detection of both PCNA monoubiquitination and deubiquitination during the course of endogenous infection. While these data do not demonstrate that endogenous BPLF1 DUB activity is directly responsible for PCNA deubiquitination, since the entire gene is silenced and not just the DUB activity, the results do show that with endogenous levels found during viral replication, ubiquitination of PCNA is reduced, presumably due to BPLF1's DUB activity. Whether EBV infection itself induces ubiquitination of PCNA and activates postreplication repair is under study.
Fig 6.
EBV lytic induction and endogenous expression of BPLF1 results in decreased PCNA ubiquitination. (A) EBV293+ cells were induced with the viral trans activator BZLF1 (+). shRNA against BPLF1 was expressed where indicated. Forty-eight hours after transfection, the cells were irradiated with 50 J/m2 UV and incubated for 3 h before harvesting. Lysates were separated into chromatin-containing fractions and were probed for endogenous PCNA and endogenous levels of BPLF1. Increased PCNA monoubiquitination is observed when BPLF1 levels are suppressed with shRNA (top and bottom panels). These results suggest that PCNA deubiquitination by BPLF1 occurs under endogenous expression of full-length BPLF1 during EBV infection. (B) Quantitation of the results of 3 PCNA ubiquitination experiments of EBV293+ cells after induction and shRNA against BPLF1. Samples were prepared as above for panel A and treated with 30, 50, and 70 J of UV irradiation. The relative amount of PCNA ubiquitination were quantitated and graphed. Relative changes in PCNA ubiquitation were normalized to total expression of PCNA. The value for untreated samples (no shRNA) was set at 1. Average BPLF1 knockdown was approximately 70% as determined by densitometry. Total PCNA ubiquitination was similar over the 30- to 70-J/m2 range. The error bar indicates the standard deviation.
DISCUSSION
Despite the discovery of herpesvirus gene-encoded deubiquitinating enzymes in 2005 (33), information on their virological and biochemical functions remains scanty. The conservation of DUB activity across all the Herpesviridae, human and nonhuman, underscores their importance. DUB activity is expressed from the distal-end terminal regions of the large tegument proteins. Remarkably, there is less than 20% homology in the N-terminal regions of these proteins, but the enzymatic active site is strictly conserved and similarly localized in all of them. Herpes simplex virus (HSV) VP1/2 DUB protein has been largely analyzed from the perspective of its function in viral maturation as a component in virion assembly, with little attention to its distinctive enzymatic activity.
Herpesviral DUBs contribute to viral replication; an approximately 10-fold reduction in viral titer results from deleting or suppressing DUB expression (8, 21, 34, 53). However, the role of DUB activity in promoting viral replication is unknown. Since BPLF1 is expressed during the EBV replicative cycle, and not during latent infection, lytic cycle proteins were anticipated to be primary targets of DUB activity. Indeed, the only target of BPLF1 DUB activity that has been identified is the EBV ribonucleotide reductase (RR) (54). One of the two subunits of the EBV RR is ubiquitinated, similar to cellular RR where a single subunit is also ubiquitinated, and deubiquitination of EBV RR by BPLF1 leads to inhibition of its activity. EBV RR ubiquitination may be required for its function in resting B lymphocytes where deoxynucleoside triphosphate (dNTP) pools are low (17).
In this paper, we identify PCNA as the first cellular target of EBV DUB activity. PCNA is primarily recognized for its processivity function during cellular replication, but it is also essential for DNA repair and other functions, including chromatin remodeling, sister chromatid cohesion, and cell cycle control (50). In its monoubiquitinated state, PCNA recruits TLS polymerases and most likely other ubiquitin-binding proteins (55) to sites of DNA damage. We show here that BPLF1 interacts directly with PCNA and antagonizes Rad18-dependent PCNA monoubiquitination.
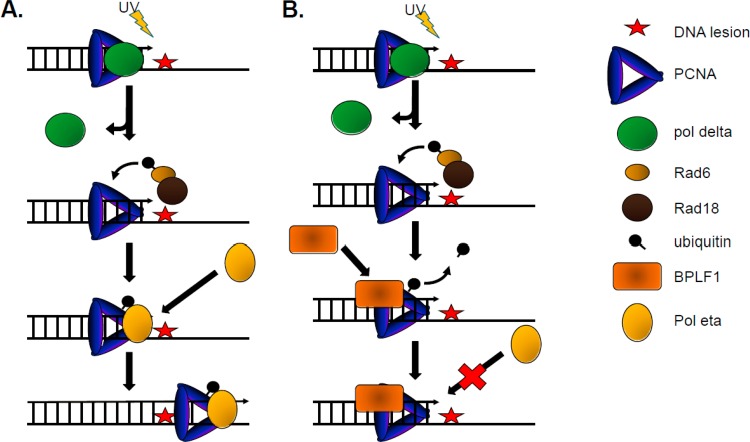
Finally, we show that the inhibitory effect of BPLF1 on PCNA monoubiquitination resulted in impaired recruitment of Polη to chromatin and virtually abolished formation of nuclear foci containing Polη. Consequently, EBV's DUB activity results in inhibition of TLS (Fig. 7). It is likely that other events that depend on PCNA monoubiquitination, including recruitment of other Y-family DNA polymerases and SNM1A, will also be influenced by BPLF1.
Fig 7.
BPLF1 DUB activity disrupts translesion synthesis. (A) Translesion synthesis overview. DNA replication proceeds until it stalls at the sites of DNA damage. PCNA is ubiquitinated at K164 by the Rad18/Rad6 complex. The TLS polymerase Polη (shown as Pol eta in the figure) is recruited to the site of DNA damage, replaces the replicative polymerase, and permits bypass of lesions. (B) BPLF1 DUB activity inhibits Polη recruitment to sites of DNA damage. PCNA is ubiquitinated in response to DNA damage. BPLF1 then removes ubiquitin from PCNA and blocks recruitment and binding of Polη to damaged DNA. Polη interacts with only the ubiquitinated form of PCNA.
We emphasize that the inhibitory effects of BPLF1 on ubiquitinated PCNA and distal events in TLS are specific, since they were abolished as a result of mutations in the PIP region of BPLF1 that specifically prevent BPLF1/PCNA binding. Many of the human herpesvirus PIP domains (including EBV's) contain an extra amino acid between the hydrophobic and aromatic residues. However, noncanonical PIP domains also exist (such as in Polη and Polι) (25). In addition, regions flanking the PIP domain are important for protein binding and subsequent signaling (23). Since PIP sequences are conserved in all the herpesviral DUB proteins, the effects on PCNA and TLS are likely evolutionarily conserved and may be generalized to all the Herpesviridae.
Deubiquitination of PCNA by USP1 is thought to operate as a safeguard against error-prone translesion synthesis. The levels of PCNA monoubiquitination can be regulated positively through cellular E3 ligase (i.e., Rad18) or negatively by cellular DUB (i.e., USP1) activities. We introduce here another avenue for negative regulation of PCNA ubiquitination and TLS, namely, deubiquitination by EBV BPLF1 activity. This is the first example of a viral enzyme regulating PCNA ubiquitination and only the second known PCNA-directed DUB.
In addition to its role in PCNA deubiquitination and TLS, USP1 is involved in attenuation of another DNA repair process, namely, the Fanconi anemia (FA) pathway (18, 41). The FA pathway is activated in response to various replication fork-stalling DNA lesions and leads to PCNA-dependent monoubiquitination of FANCD2 (20, 38, 45). Monoubiquitinated (activated) FANCD2 associates with chromatin and directs replication-coupled repair of interstrand cross-links (ICL) (35, 38, 47) and may also stimulate homologous recombination (HR) (40). Monoubiquitinated FANCD2 is a direct substrate for USP1, and genotoxin-induced degradation of USP1 constitutes a major mechanism for FA pathway activation. We have considered the possibility that BPLF1 might (similar to USP1) deubiquitinate PCNA and FANCD2, thereby inhibiting both TLS and FA pathways. In preliminary results, genotoxin-induced recruitment of FANCD2 to chromatin was indeed reduced in BPLF1-expressing cells. Therefore, in addition to perturbing TLS, it is very likely that BPLF1 compromises the FA pathway and perhaps other DNA repair mechanisms.
Finally, how might deubiquitination and PCNA abet replication of Epstein-Barr Virus? BPLF1 is classified as a late lytic cycle gene. However, BPLF1 protein is incorporated in the virion tegument and therefore is already present at the onset of infection. BPLF1 RNA is detected as early as 6 to 8 h postinfection (19, 56). Since PCNA associates with EBV DNA during its replication, BPLF1 may affect viral replication. Whether PCNA is ubiquitinated in this context is unknown. We are currently investigating the potential effects of PCNA and its ubiquitination state on EBV replication and infection.
EBV replication shuts down replication of cellular DNA, producing an S-phase-like cellular environment that is likely advantageous for viral lytic replication (36, 37). It has been proposed that newly synthesized viral DNA is sensed as foreign and induces a DNA damage response, and cell cycle checkpoints are activated (37). Arrest of cell cycle provides an interval for repair of damaged DNA. Checkpoint signaling is regulated by two major kinases, ATM (ataxia-telangiectasia mutated) and ATR (ataxia-telangiectasia and Rad3-related). ATM responds to double-strand breaks (3), and ATR responds primarily to stalled replication forks (10). Induction of checkpoint pathways during EBV infection results in inhibition of cellular DNA replication, while permitting viral replication to proceed. Deubiquitination of PCNA by BPLF1 stalls DNA repair processes and likely renders a state in which cellular replication cannot proceed, allowing cellular factors to be utilized by the virus. In addition, BPLF1 during this time may interact with PCNA and pirate it from sites of cellular DNA damage to sites of viral DNA replication where it might function as an accessory factor during processing of replicating viral DNA. In this scenario, BPLF1's DUB activity could act simultaneously to inhibit cellular DNA replication and enhance viral replication.
EBV, in addition to its lytic replicative cycle, is known for its ability to transform latently infected human cells, and was in fact the first human tumor virus identified (14, 24, 42, 43). In this capacity, the virus is known to cause genetic instability of host DNA (30), possibly through the virus triggering activation of repair pathways and co-opting DNA and replication repair factors for its own replication in the early lytic phase before transition to the latent phase of the infection. In this scenario, when triggered by the DNA damage response, EBV may also engage cellular factors such as PCNA during its replication. Whether such events occur, the sequence of events, and the underlying mechanism present intriguing new areas for exploration.
ACKNOWLEDGMENTS
We thank P. Kannouche for the HA-tagged PCNA construct (pCMV-HA-PCNAwt).
This work was supported by NIH grants T32-CA09156-35 (C.B.W.), P01-CA19014-29 (J.S.P.), and R01-ES09558 (C.V.).
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Abaitua F, Daikoku T, Crump CM, Bolstad M, O'Hare P. 2011. A single mutation responsible for temperature-sensitive entry and assembly defects in the VP1-2 protein of herpes simplex virus. J. Virol. 85:2024–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acharya N, et al. 2008. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 105:17724–17729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakkenist CJ, Kastan MB. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506 [DOI] [PubMed] [Google Scholar]
- 4. Batterson W, Furlong D, Roizman B. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bi X, et al. 2006. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol. Cell. Biol. 26:3527–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bienko M, et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310:1821–1824 [DOI] [PubMed] [Google Scholar]
- 7. Bottcher S, et al. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403–13411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bottcher S, et al. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 82:6009–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cimprich KA, Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daikoku T, et al. 2006. Postreplicative mismatch repair factors are recruited to Epstein-Barr virus replication compartments. J. Biol. Chem. 281:11422–11430 [DOI] [PubMed] [Google Scholar]
- 12. Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 29:625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet i:702–703 [DOI] [PubMed] [Google Scholar]
- 15. Ernst R, et al. 2011. Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS Biol. 8:e1000605 doi:10.1371/journal.pbio.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fox JT, Lee KY, Myung K. 2011. Dynamic regulation of PCNA ubiquitylation/deubiquitylation. FEBS Lett. 585:2780–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao WY, Cara A, Gallo RC, Lori F. 1993. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc. Natl. Acad. Sci. U. S. A. 90:8925–8928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcia-Higuera I, et al. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249–262 [DOI] [PubMed] [Google Scholar]
- 19. Gastaldello S, et al. 2010. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat. Cell Biol. 12:351–361 [DOI] [PubMed] [Google Scholar]
- 20. Geng L, Huntoon CJ, Karnitz LM. 2010. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J. Cell Biol. 191:249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a. González CM, Wang L, Damania B. 2009. Kaposi's sarcoma-associated herpesvirus encodes a viral deubiquitinase. J. Virol. 83:10224–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gredmark-Russ S, et al. 2009. A gammaherpesvirus ubiquitin-specific protease is involved in the establishment of murine gammaherpesvirus 68 infection. J. Virol. 83:10644–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haracska L, et al. 2005. A single domain in human DNA polymerase iota mediates interaction with PCNA: implications for translesion DNA synthesis. Mol. Cell. Biol. 25:1183–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Havens CG, Walter JC. 2009. Docking of a specialized PIP box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henderson E, Miller G, Robinson J, Heston L. 1977. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology 76:152–163 [DOI] [PubMed] [Google Scholar]
- 25. Hishiki A, et al. 2009. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 284:10552–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang TT, et al. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8:339–347 [DOI] [PubMed] [Google Scholar]
- 27. Inn KS, et al. 2011. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 85:10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Izumi M, Yatagai F, Hanaoka F. 2001. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 276:48526–48531 [DOI] [PubMed] [Google Scholar]
- 29. Jarosinski K, Kattenhorn L, Kaufer B, Ploegh H, Osterrieder N. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. U. S. A. 104:20025–20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamranvar SA, Gruhne B, Szeles A, Masucci MG. 2007. Epstein-Barr virus promotes genomic instability in Burkitt's lymphoma. Oncogene 26:5115–5123 [DOI] [PubMed] [Google Scholar]
- 31. Kannouche PL, Lehmann AR. 2004. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle 3:1011–1013 [PubMed] [Google Scholar]
- 32. Kannouche PL, Wing J, Lehmann AR. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14:491–500 [DOI] [PubMed] [Google Scholar]
- 33. Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547–557 [DOI] [PubMed] [Google Scholar]
- 34. Kim ET, Oh SE, Lee YO, Gibson W, Ahn JH. 2009. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 83:12046–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Knipscheer P, et al. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326:1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kudoh A, et al. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 77:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kudoh A, et al. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163 [DOI] [PubMed] [Google Scholar]
- 38. Montes de Oca R, et al. 2005. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 105:1003–1009 [DOI] [PubMed] [Google Scholar]
- 39. Motegi A, et al. 2006. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 175:703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakanishi K, et al. 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. U. S. A. 102:1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nijman SM, et al. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17:331–339 [DOI] [PubMed] [Google Scholar]
- 42. Pagano J. 2009. EBV diseases, p 794 In Damania B, Pipas J. (ed), DNA tumor viruses. Springer, New York, NY [Google Scholar]
- 43. Pagano JS. 1995. Epstein-Barr virus: therapy of active and latent infection, p 155–195 In Jeffries DJ, De Clercq E. (ed), Antiviral chemotherapy. John Wiley & Sons Ltd., London, United Kingdom [Google Scholar]
- 44. Palle K, Vaziri C. 2011. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA topoisomerase 1 inhibition. Cell Cycle 10:1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park HK, Wang H, Zhang J, Datta S, Fei P. 2010. Convergence of Rad6/Rad18 and Fanconi anemia tumor suppressor pathways upon DNA damage. PLoS One 5:e13313 doi:10.1371/journal.pone.0013313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prakash S, Johnson RE, Prakash L. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317–353 [DOI] [PubMed] [Google Scholar]
- 47. Raschle M, et al. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134:969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sabbioneda S, et al. 2008. Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases. Mol. Biol. Cell 19:5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sabbioneda S, et al. 2009. Ubiquitin-binding motif of human DNA polymerase eta is required for correct localization. Proc. Natl. Acad. Sci. U. S. A. 106:E20–E21 (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a. Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. 2005. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 79:15582–15585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49b. Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gaudet R, Ploegh HL. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stoimenov I, Helleday T. 2009. PCNA on the crossroad of cancer. Biochem. Soc. Trans. 37:605–613 [DOI] [PubMed] [Google Scholar]
- 51. Ulrich HD. 2004. How to activate a damage-tolerant polymerase: consequences of PCNA modifications by ubiquitin and SUMO. Cell Cycle 3:15–18 [PubMed] [Google Scholar]
- 52. Vidal AE, et al. 2004. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota. J. Biol. Chem. 279:48360–48368 [DOI] [PubMed] [Google Scholar]
- 53. Wang J, Loveland AN, Kattenhorn LM, Ploegh HL, Gibson W. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Whitehurst CB, et al. 2009. The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J. Virol. 83:4345–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang K, Moldovan GL, D'Andrea AD. 2010. RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J. Biol. Chem. 285:19085–19091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yuan J, Cahir-McFarland E, Zhao B, Kieff E. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 80:2548–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zlatanou A, et al. 2011. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell 43:649–662 [DOI] [PubMed] [Google Scholar]