Abstract
We found that Sweet potato feathery mottle virus (SPFMV) P1, a close homologue of Sweet potato mild mottle virus P1, did not have any silencing suppressor activity. Remodeling the Argonaute (AGO) binding domain of SPFMV P1 by the introduction of two additional WG/GW motifs converted it to a silencing suppressor with AGO binding capacity. To our knowledge, this is the first instance of the transformation of a viral protein of unknown function to a functional silencing suppressor.
TEXT
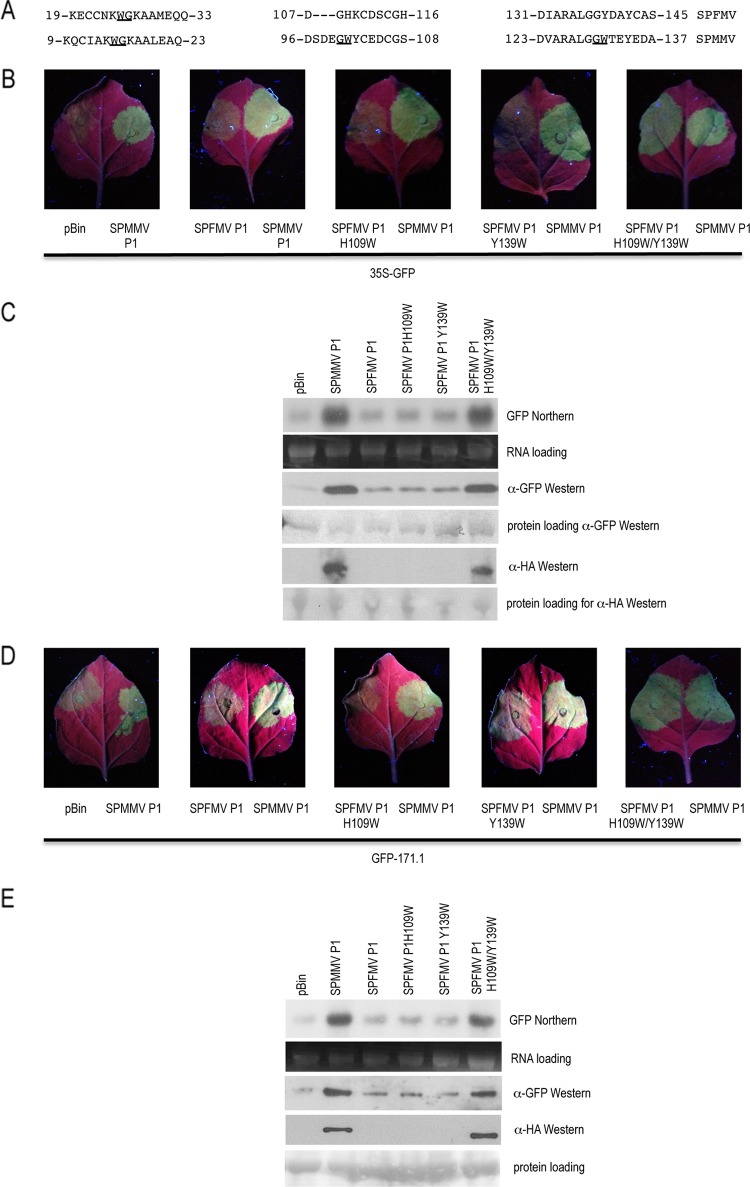
RNA silencing is a sequence-specific cellular process that leads to RNA degradation, inhibition of translation of mRNAs, or heterochromatin formation (10). RNA silencing has several functions; one of the most important is to counteract viruses and transposons. Viruses evolved silencing suppressors to inhibit RNA silencing (1). Cellular proteins possessing WG/GW domains are known to bind Argonaute (AGO) via their tryptophan (W) residues (3, 4, 6, 13, 15). The P1 protein of Sweet potato mild mottle virus (SPMMV) is a silencing suppressor that is able to counteract the active RNA-induced silencing complex (RISC) by binding AGO (5). Compared to the P1 proteins of members of the Potyvirus genus, ipomoviral SPMMV P1 has a long extension at its N-terminal end harboring 3 WG/GW domains spanning amino acid (aa) 1 to aa 140, and this region is absolutely necessary for suppressor activity and AGO binding (5). The closest homolog of SPMMV P1 is Sweet potato feathery mottle virus (SPFMV) P1 (14, 5). We sought to determine whether SPFMV P1 possesses RNA silencing suppressor activity. To do this, we prepared first-strand cDNA from RNA isolated from SPFMV-infected sweet potato plants commercially available at the German Collection of Microorganisms and Cell Cultures. Using degenerate primers (5′-AAGGATCCATGGCAWCYGYNATCBGYATYTGYGAA and 3′-AGAATTCTTTARWAYTGVDYGATRWATGGYARRRYRGAYCTACCAAG) designed according to available sequence data, we amplified the P1 open reading frame of a Nigerian SPFMV isolate (GenBank accession number JQ742091). Sequence analysis revealed a 689-aa protein that is 80 to 95% identical to known SPFMV P1 proteins. The N-terminal 193-aa region of SPFMV P1 was 41.5% identical to the corresponding part of SPMMV P1; the rest of the protein was 22% identical. However, the overall identity was 24.6%. Although the N-terminal 193-aa region of SPFMV P1 showed the highest homology to SPMMV P1, it contains only one WG/GW domain (Fig. 1A). To see if SPFMV P1 is able to inhibit RNA silencing, we performed the standard Agrobacterium coinfiltration assay (12). The coding region ending with a stop codon for SPFMV P1 was N terminally HA (hemagglutinin) tagged by being cloned into the binary vector pSanyi (7), transferred to agrobacteria, and then used with an agrobacterial strain harboring the 35S-labeled green fluorescent protein (GFP)-encoding reporter gene to coinfiltrate leaves of wild-type (WT) Nicotiana benthamiana. As a positive control, SPMMV P1 was used. Figure 1B and C show that 35S-GFP remained silenced in the presence of SPFMV P1; however, as was shown before, SPMMV P1 strongly inhibited silencing (5). Previously it was shown that the loss of any two of the WG/GW motifs of SPMMV P1 correlated with loss of silencing suppressor activity (5). We hypothesized, therefore, that the sole WG/GW motif in WT SPFMV P1 might be insufficient for silencing suppressor activity. Due to the high degree of amino acid identity between the SPFMV and SPMMV P1 proteins at their N-terminal ends, we determined amino acids in SPFMV P1 corresponding to the tryptophan residues of the second and third WG/GW motifs in SPMMV P1 (Fig. 1A). Using primers GATGTCCTGGATGGATGGAAGTGTGACAGCTGC and GCAGCTGTCACACTTCCATCCATCCAGGACATC along with primers CTAGAGCGTTAGGAGGGTGGGATGCATACTGTG and CACAGTATGCATCCCACCCTCCTAACGCTCTAG (nucleotide changes are underlined), the histidine residue at position 109 and the tyrosine residue at position 139 were changed to tryptophan using the QuikChange XL mutagenesis kit. Thus, three mutants, H109W, Y139W, and H109W/Y139W, were created, resulting in one additional WG/GW motif in the first two mutants, and two additional WG/GW motifs in the third. The mutants were tested for suppressor activity as described above. The H109W and Y139W mutants did not show silencing suppressor activity however, the H109W/Y139W mutant inhibited RNA silencing as well as SPMMV P1 (Fig. 1B). Our in vivo data were confirmed by Northern and Western blotting as described previously (5) (Fig. 1C).
Fig 1.
Analysis of SPFMV P1. (A) WG/GW motifs of SPFMV P1 and their corresponding SPMMV P1 parts. WG/GW amino acid duplets are underlined. Numbers indicate amino acid positions. In SPFMV P1, histidine 109 and tyrosine 139 are in bold. (B) Standard 35S-GFP agroinfiltration analysis of WT and mutant SPFMV P1 proteins. Infiltrated leaves at 3 days postinfection were illuminated with a hand-held UV lamp (Blak Ray B-100AP UV lamp; UVP), and pictures were taken with the camera of the iPhone 4S. (C) RNA and protein extracts of infiltrated patches were used to detect GFP expression by Northern and Western blotting. (D) Agroinfiltration analysis of WT and mutant SPFMV P1 proteins to inhibit active RISC. (E) Detection of GFP expression by Northern and Western blotting at 2 days postinfection.
The latter is a silencing suppressor that is able to inhibit active RISC (5). Next we checked if the highly homologous WT SPFMV P1 and/or any of the mutants we created interfere with active RISC. SPFMV strains expressing P1 and H109W, H139W, and H109W/Y139W mutant P1 were coinflitrated with the GFP1-171.1 reporter gene into N. benthamiana as described previously (5). Our results revealed that neither WT P1, containing one WG/GW motif, nor H109W or H139W mutant P1, having two WG/GW motifs, demonstrated silencing suppressor activity. However, H109W/Y139W mutant P1, having three WG/GW motifs, showed a potent ability to inhibit active RISC (Fig. 1D). Northern and Western blotting detected high GFP and suppressor expression by H109W/Y139W mutant SPFMV P1, as well as by the positive control (Fig. 1E). However, the WT and mutant SPFMV P1 proteins, which did not show suppressor activity, could be detected only when they coinfiltrated plants with Tobacco etch virus HC-Pro (9) (data not shown).
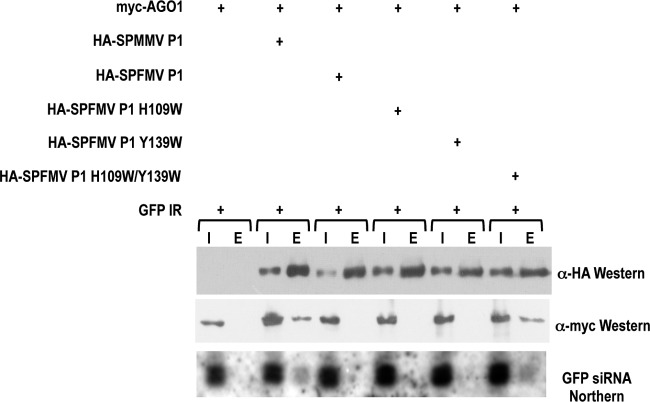
To test AGO binding capacity, the HA-tagged WT and mutant forms of SPFMV P1 were used for coinfiltration with 6×mycAGO1 (16) and a 35S-GFP inverted repeat. To avoid the effect of RNA silencing, the GFP16c/RDR6i N. benthamiana line was used in this experiment, which does not have silencing against (even transiently expressed) transgenes (5, 11). Western and Northern analyses of inputs and eluates of immunoprecipitations of the HA-tagged WT and mutant SPFMV and SPMMV P1 proteins were carried out. Small-RNA-loaded AGO binding ability was detected in the SPFMV H109W/Y139W mutant and WT SPMMV P1 proteins but not in the SPFMV WT or H109W or Y139W mutant P1 protein (Fig. 2), thus indicating that silencing suppressor activity strongly correlated with the capability of AGO binding. Any of the two WG/GW motifs was sufficient for silencing of suppressor activity and AGO binding in the case of SPMMV P1 (5). However, three WG/GW motifs were absolutely required for SPFMV P1 to gain silencing suppressor activity.
Fig 2.
Physical interaction of AGO1 and WT and mutant SPFMV P1 proteins. Western and Northern analyses of inputs (I) and eluates (E) of immunoprecipitations carried out with extracts of infiltrated leaves. IR, inverted repeat.
Bioinformatic analysis showed a close evolutionary relationship between the P1 proteins of SPMMV and SPFMV (14). Although WT SPFMV P1 did not show any silencing suppressor activity, remodeling of the AGO hook by changing only two amino acids to tryptophan resulted in a protein that inhibits active RISC by the same mechanism as the SPMMV P1 prototype (5). Thus, the close evolutionary relationship between the P1 proteins of SPMMV and SPFMV was further proven by our functional analyses. SPFMV, in a synergistic interaction with Sweet potato chlorotic stunt virus (SPCSV), causes the very severe sweet potato virus disease (8). RNase 3 protein, the silencing suppressor of SPCSV, was found to mediate viral synergism between SPCSV and SPFMV (2). In such a synergistic interaction, one powerful suppressor can support the spread of two viruses and that might explain why SPFMV P1 did not bear silencing suppressor activity.
Finally, to our knowledge, this is the first instance in which a viral protein of unknown function was turned into a functional RNA silencing suppressor.
ACKNOWLEDGMENTS
This research was supported by a grant from the Hungarian Scientific Research Fund (OTKA K81591) and grants TÁMOP-4.2.1/B-09/1/KONV.-2010-0005 and TÁMOP-4.2.2/B-10/1-2011-0012. Lóránt Lakatos was a recipient of the Bólyai János fellowship.
We are grateful to Andrew McDowell for critical reading of the manuscript.
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Burgyán J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci. 16:265–272 [DOI] [PubMed] [Google Scholar]
- 2. Cuellar WJ, et al. 2009. Elimination of antiviral defense by viral RNase III. Proc. Natl. Acad. Sci. U. S. A. 106:10354–10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. 2001. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20:2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. El-Shami M, et al. 2007. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21:2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Giner A, Lakatos L, Garcia-Chapa M, Lopez-Moya JJ, Burgyán J. 2010. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6:e1000996 doi:10.1371/journal.ppat.1000996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. 2005. RNA polymerase IV directs silencing of endogenous DNA. Science 308:118–120 [DOI] [PubMed] [Google Scholar]
- 7. Kertész S, et al. 2006. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34:6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kreuze JF, et al. 2008. RNA silencing-mediated resistance to a crinivirus (Closteroviridae) in cultivated sweet potato (Ipomoea batatas L.) and development of sweet potato virus disease following co-infection with a potyvirus. Mol. Plant Pathol. 9:589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lakatos L, et al. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25:2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Melnyk CW, Molnar A, Baulcombe DC. 2011. Intercellular and systemic movement of RNA silencing signals. EMBO J. 30:3553–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwach F, Vaistij FE, Jones L, Baulcombe DC. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138:1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silhavy D, et al. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Till S, et al. 2007. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 14:897–903 [DOI] [PubMed] [Google Scholar]
- 14. Valli A, Lopez-Moya JJ, Garcia JA. 2007. Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 88:1016–1028 [DOI] [PubMed] [Google Scholar]
- 15. Zhang L, et al. 2007. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell 28:598–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X, et al. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20:3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]