Abstract
Airborne viruses are expected to be ubiquitous in the atmosphere but they still remain poorly understood. This study investigated the temporal and spatial dynamics of airborne viruses and their genotypic characteristics in air samples collected from three distinct land use types (a residential district [RD], a forest [FR], and an industrial complex [IC]) and from rainwater samples freshly precipitated at the RD site (RD-rain). Viral abundance exhibited a seasonal fluctuation in the range between 1.7 × 106 and 4.0 × 107 viruses m−3, which increased from autumn to winter and decreased toward spring, but no significant spatial differences were observed. Temporal variations in viral abundance were inversely correlated with seasonal changes in temperature and absolute humidity. Metagenomic analysis of air viromes amplified by rolling-circle phi29 polymerase-based random hexamer priming indicated the dominance of plant-associated single-stranded DNA (ssDNA) geminivirus-related viruses, followed by animal-infecting circovirus-related sequences, with low numbers of nanoviruses and microphages-related genomes. Particularly, the majority of the geminivirus-related viruses were closely related to ssDNA mycoviruses that infect plant-pathogenic fungi. Phylogenetic analysis based on the replication initiator protein sequence indicated that the airborne ssDNA viruses were distantly related to known ssDNA viruses, suggesting that a high diversity of viruses were newly discovered. This research is the first to report the seasonality of airborne viruses and their genetic diversity, which enhances our understanding of viral ecology in temperate regions.
INTRODUCTION
The atmosphere is distributed over the Earth, and it is known to contain large numbers of chemical and biological particles. Airborne particles of biological origin or activity include atmospheric bacteria, fungal spores, pollen, and allergens (8–10, 26). These particles could be either the direct cause of epidemics and infectious diseases or the indirect cause of noninfectious diseases (e.g., hypersensitivity to aeroallergens, as with asthma) (70). The atmosphere is also known to act as a vehicle for the dispersal of unknown biological particulates, particularly viruses which are regarded as major environmental risk factors in complex disease pathogenesis (17, 23, 53).
Advances in molecular technologies have provided methods that facilitate sensitive and accurate resolution in environmental microbiological analysis, which overcomes the limitations of culture-dependent methods (21). However, very little is known about the microbiology of the atmosphere. Technical difficulties with low densities of airborne biological particles and the lack of air sampling standardization have limited research into the microbial ecology of the air (30, 73). In particular, viral genomes are so small that a large amount of viral particles must be obtained from air samples to be amplified, quantified, and sequenced. Furthermore, the absence of conserved genes and high genetic variation also make it difficult apply PCR assays to viral populations (5). Thus, there have been no previous metagenomic analyses of atmospheric viral ecology because great challenges are involved in determining the biodiversity and composition of airborne viruses. The recent development of “viral metagenomics” has provided a powerful tool for the exploration of viral diversity in a wide range of environments (13), and our knowledge of viral diversity has been totally reshaped by the discovery of numerous viruses in environmental and animal samples that are closely linked to public health (3, 11, 16, 42, 49, 56). A combination of viral metagenomics and air sampling will facilitate a new understanding of atmospheric viral ecology and the elucidation of new viruses.
The natural variation of airborne biological particles in response to meteorological parameters such as temperature, humidity, and light has also hindered research into atmospheric microbial ecology (60). Meteorological changes associated with anthropogenic influences, such as changing land use, are also known to affect the atmospheric microbial composition (9). In particular, recent studies of influenza virus have shown that virus transmission is enhanced by low temperatures and dry conditions (33, 55). However, our knowledge of how airborne viruses are affected by meteorological parameters is heavily biased toward known epidemiological agents, particularly influenza viruses. Thus, our current understanding of viruses found in the air is very limited, and the factors controlling the life cycles of atmospheric viruses remain elusive, which hampers our understanding of atmospheric viral ecology.
This study applied viral metagenomic approaches to the extensive characterization of airborne viral diversity and its composition in the near-surface atmosphere. Air samples were collected from three distinct land use sites (a residential district [RD], a forest [FR], and an industrial complex [IC]) while rainwater was also collected from one site (RD-rain). The airborne viromes were randomly amplified and analyzed by 454 pyrosequencing. The spatial and temporal variation of the airborne microbial population was determined by monitoring the viral and bacterial abundances at the RD site every month for 6 months. This allowed viral and bacterial variation to be correlated with meteorological parameters and land use types. This study provides novel insights into the temporal and spatial variation of airborne viruses and the diversity of airborne viruses in temperate regions.
MATERIALS AND METHODS
Sampling sites and sample collection.
Air samples were collected from three different land use types. Sampling sites were situated in the middle of Seoul city (site 1; RD), the central region of Korea (site 2; FR), and the west coast of Korea (site 3; IC). The three sites were representative of urban, forest, and coastal urban land use, respectively (see Fig. S1 in the supplemental material). Airborne particles smaller than 1 μm in diameter were collected and directed into 1 liter of phosphate-buffered saline (PBS) buffer (pH 7.4) (61) at 1 m above ground level for 48 h using a modified air sampling device referred to as a connector-linked direct precipitation air sampler with a water jet pump (Eyela) (flow rate, 19 liters min−1), which was different from other commercially available air sampling devices (66, 70) (Fig. 1). All sampling events were performed in biological replicates (n = 2) to minimize sampling and methodological variabilities. Sequences from biological replicates were combined for further analysis after pyrosequencing.
Fig 1.
Connector-linked direct precipitation air sampler.
The sampling program was conducted in two phases. The first phase conducted in August and September 2010 determined the genetic diversity of viruses in the near-surface atmosphere at the three sites. The second phase extended to every month from October 2010 to March 2011, and it enumerated the viruses and bacteria in the near-surface atmosphere of the RD site. Freshly precipitated rainfall (RD-rain) was also collected at the RD site during the rainy season to determine viral diversity in rain from the upper atmosphere. Meteorological data for each sampling time were downloaded from a standard onsite weather station (see Table S1 in the supplemental material).
Virus and bacteria enumeration.
Ten milliliters of the sampled mixture was used immediately for virus and bacteria enumerations, using protocols described by Patel et al. (41). Stained particles between 0.5 and 10 μm in diameter were regarded as airborne bacterial cells (8), whereas the particles smaller than 0.5 μm in diameter were presumed to be airborne viruses. At least eight fields per sample (30 to 150 cells per field) were counted at a magnification of ×1,000.
Viral morphology.
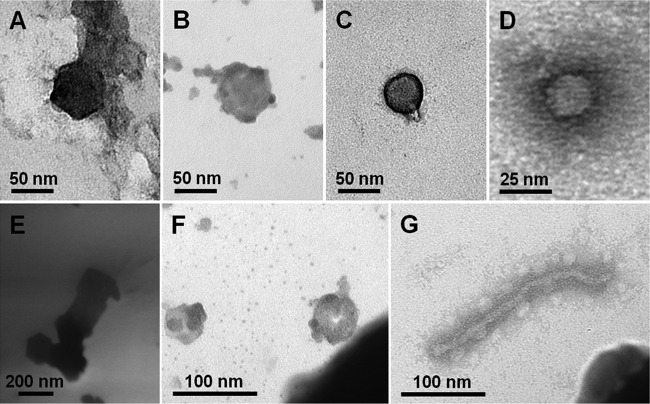
Repeated density gradient purification using ultracentrifugation was conducted before morphological analysis of airborne viruses using protocols described previously (40). In brief, air samples were passed sequentially through 3-μm-, 0.45-μm-, and 0.2-μm-pore-size filters (Advantec). Filtrates were then concentrated 40 times by tangential flow filtration (TFF) and loaded onto heavier CsCl layers (1.7, 1.5, and 1.2 g ml−1), which were prepared using 0.02-μm-pore-size-filtered PBS buffer and ultracentrifuged at 88,250 × g for 2 h at 4°C in a swinging bucket rotor (Beckman). Purified virus particles (VPs) were examined by transmission electron microscopy (TEM). Images were collected at magnifications of ×50,000 to ×80,000 using an energy-filtering transmission electron microscope (Carl Zeiss) (Fig. 2).
Fig 2.
Transmission electron micrographs showing different morphologies of virus particles found in the near-surface atmosphere.
Viral DNA extraction and sequencing.
VPs were concentrated 20 times by TFF as described above. After a further filtration with a 0.2-μm-pore-size filter, the filtrate was treated with 0.1 volume of chloroform for 10 min. The aqueous phase was collected and concentrated using centrifugal concentration filters (30 kDa; Ultra-15; Amicon). DNase I (25 U ml−1 at 37°C for 1 h) (TaKaRa Bio Inc.) was applied to the VP concentrates of each sample to remove any free DNA. The absence of microbial cells was verified by the amplification of the bacterial 16S rRNA gene using universal primers (8F and 518R) before viral DNA amplification. Viral DNA was extracted using a QIAamp MinElute Virus Spin Kit (Qiagen) and amplified with a REPLI-g Mini Kit (Qiagen), according to the manufacturer's instructions. This amplification step allowed preferential amplification of long strands of DNA or circular DNA genomes, excluding generation of a cDNA library for detection of RNA virus. Triplicate amplicons of each sample were subsequently pooled, purified with phenol-chloroform-isoamyl alcohol (25:24:1), and concentrated by ethanol precipitation. The resulting DNAs, purified from each of eight sets, were labeled with different multiplex identifiers (Roche) and sequenced by 454 GS-FLX titanium pyrosequencing (454 Life Sciences).
Bioinformatics. (i) Preprocessing.
To analyze the viral metagenomic sequences, denoised sequences were screened from raw sequence reads of viruses collected at the three sampling sites and rainwater as previously described by Kim et al. (29). Sequences with more than one ambiguous base call (Ns), an average quality score less than 25, and those shorter than 101 bp were excluded first, and exact duplicate sequences with 100% sequence identity and a maximum 1-bp difference in length were also excluded.
(ii) Viral taxonomic identification.
Viral sequence reads for the four combined viromes were compared against the CAMERA databases, i.e., CAMERA's nonidentical (nondredundant [nr]) peptide sequence and viral protein databases (http://camera.calit2.net). The NCBI Reference data sets in CAMERA contain sequence data updated on 16 January 2012 from NCBI Refseq release 51 and GenBank release 187. To perform taxonomic assignment, the MEGAN program (version 4.62.7) (24) was used for acceptance of the output of a BLAST search based on the lowest-common-ancestor algorithm with one minimal support hit. Viral sequences were annotated using BLASTx, and matches with expected values (E values) of less than 10−3 for the CAMERA nr peptide sequences and 10−3 for the CAMERA viral proteins were used for identification. A cross-BLAST analysis was conducted to compare the viromes from the three land use types and rainwater using BLASTn, which compared common sequences among viromes. Pooled air viromes were used for cross-BLAST analysis, and overlapping sequences were assembled into contigs based on 98% minimal identity with a minimal 35-bp overlap (72). Sequences assigned as RNA viruses (less than 3%) with significant similarities were excluded because only DNA was used for sequencing in this study.
(iii) Phylogenetic diversity of ssDNA viruses.
To determine the phylogenetic diversity of ssDNA viruses, viral sequences assigned as geminiviruses, circoviruses, nanoviruses, and microphage-related genomes were assembled using a meta-assembler with the default setting provided on the CAMERA server. Of the assembled contigs, large contigs (>500 bp) were used to predict open reading frames (ORFs) with the FragGeneScan method (45), and predicted ORFs were compared to the GenBank nonredundant (nr) database (BLASTp; E value of <10−5). A phylogenetic tree of geminiviruses was constructed based on replication initiator protein (Rep) sequences (Gemini_AL1) of the ORFs related to geminiviruses, which were aligned with sequences from pfam00799, rice paddy soil (28), Phytoplasma plasmids (4), and a geminivirus-related mycovirus (74). A phylogenetic tree was constructed based on replication initiator protein sequences of the ORFs related to circoviruses and nanoviruses, which were aligned with sequences from pfam02407, rice paddy soil (28), an Antarctic lake (32), a dragonfly (48), Entamoeba histolytica (54), Giardia intestinalis (1), canarypox virus (64), and a Bifidobacterium pseudocatenulatum plasmid (19) and from circo-like sequences from reclaimed water and marine environments (47). A phylogenetic tree was constructed based on partial capsid (Vp1) protein sequences of the ORFs related to microphages, as previously described (12), which were aligned with 13 sequences from the Sargasso Sea (3), 8 sequences from microbialites (12), 3 sequences from an Antarctic lake (32), and 15 cultured microphage sequences. Conserved regions of porcine circovirus-1 (amino acids 14 to 84 of the Rep proteins) and beet curly top virus (amino acids 7 to 117 of the Gemini_AL1 proteins) were used in phylogenetic analyses. The predicted protein sequences of large contigs from the four viromes were aligned with the reference sequences using MUSCLE (http://www.ebi.ac.uk/Tools/muscle) with default settings. Phylogenetic trees were constructed by performing 1,000 randomly replicated bootstraps based on neighbor joining (52) with the p-distance (pairwise distance) model, using MEGA5 (59).
Statistical analysis.
All data were checked for normality before statistical tests. One-way analysis of variance (ANOVA) was used to test differences between viral/bacterial abundances and land use types/months. P values in pairwise comparisons were corrected using Duncan's method. Parametric Pearson's correlation coefficients were calculated to examine trends between viral/bacterial abundances and meteorological parameters (temperature, absolute humidity, and relative humidity). All analyses were conducted using the SAS program.
Sequencing data accession number.
The viromes of the three different land use types and rainwater are accessible in the NCBI Sequence Read Archive under accession number SRP007810.1.
RESULTS
Spatial viral and bacterial abundance in the near-surface atmosphere above different land use sites.
The abundance of viruses and bacteria in the air collected from the three distinct land use types (RD, FR, and IC) was determined using a fluorescence staining method with confocal laser scanning microscopy (see Fig. S2 in the supplemental material). Viral abundance in the near-surface atmosphere above three land use types ranged from 1.7 × 106 to 4.0 × 107 viruses m−3, while bacterial abundance ranged from 8.6 × 105 to 1.1 × 107 bacteria m−3. Virus-to-bacterium ratios ranged from 1.6 to 3.6 (average, 2.2). Although spatial variations in viruses and bacteria in the near-surface atmosphere were investigated by air sampling conducted at the three different land use types, no significant differences in viral or bacterial abundances were found between them (see Fig. S3A and B).
Temporal variation in airborne microbiota and its relationship to meteorological parameters.
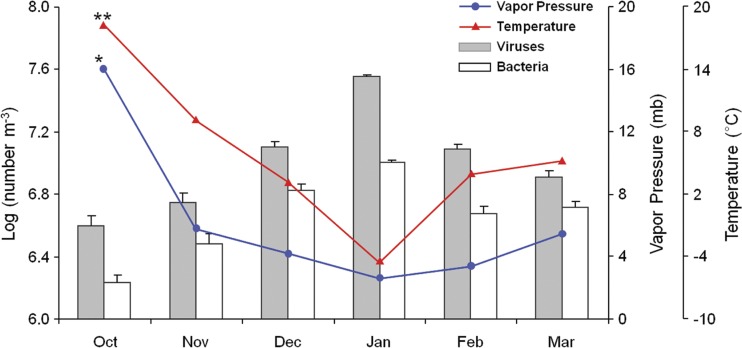
Temporal variations in viruses and bacteria in the near-surface atmosphere were investigated using monthly sampling campaigns between October 2010 and March 2011 at the RD site. Although no significant differences in viral and bacterial abundances between different land use types were observed, there were highly significant differences in viral and bacterial abundances at monthly time scales (viruses, F = 69.07, P < 0.001, one-way ANOVA; bacteria, F = 39.54, P < 0.001, one-way ANOVA) (Fig. 3; see also Fig. S3C and D in the supplemental material). Viral and bacterial abundances in the near-surface atmosphere at the RD site increased from autumn to winter and decreased toward spring.
Fig 3.
Temporal abundance of airborne viruses and bacteria and correlations with meteorological parameters. The graph shows total viral and bacterial abundances in samples collected in the residential district during 6 months (monthly time scales), as determined by direct microscopy (viruses, P < 0.001; bacteria, P < 0.001 [one-way ANOVA]). Temporal variations in total viral and bacterial abundances are significantly negatively correlated with temperature (**) (viruses, ρ = −0.78, P < 0.001; bacteria, ρ = −0.82, P < 0.001) and vapor pressure (*) (viruses, ρ = −0.70, P < 0.001; bacteria, ρ = −0.78, P < 0.001) on the basis of parametric Pearson's correlation coefficients. Error bars represent standard errors of means.
Possible correlations among viral and bacterial abundances in the near-surface atmosphere and meteorological factors (temperature, relative humidity, and vapor pressure) were investigated at the RD site. Vapor pressure was used as a measure of absolute humidity (55). Temperature and vapor pressure gradually declined toward January in winter and increased from March in the spring, whereas relative humidity was relatively constant throughout seasons (see Fig. S4 in the supplemental material). Viral and bacterial abundances exhibited seasonal variation at the RD site, where they were significantly inversely correlated with temperature (viruses, ρ = −0.78, P < 0.001; bacteria, ρ = −0.82, P < 0.001) and vapor pressure (viruses, ρ = −0.70, P < 0.001; bacteria, ρ = −0.78, P < 0.001) (Fig. 3). However, there were no significant correlations with relative humidity. Thus, temperature and absolute humidity are major factors influencing viral and bacterial abundance in the near-surface atmosphere.
Metagenomic diversity of single-stranded DNA (ssDNA) viruses in the near-surface atmosphere.
A metagenomic approach with high-throughput sequencing was used in the extensive characterization of viral assemblages present in air samples collected from the three land use types and from rainwater collected at the RD site. Low-quality sequences were excluded, and the following sequences were obtained from each site: 17,943 sequences from RD, 20,691 sequences from FR, 14,332 sequences from IC, and 15,776 sequences from RD-rain (Table 1; see also Table S2 in the supplemental material). Sequences were compared to the CAMERA's NCBI reference sequence data sets (viral proteins and CAMERA's nr peptide sequences) using BLASTx (E < 10−3) and were assigned best matches based on their BLAST similarity. No significant similarities (E > 10−3) were found for 59.0% of RD, 49.7% of FR, 80.0% of IC, and 79.9% of RD-rain sequences when the viromes were compared with the CAMERA's viral proteins data set. The remaining sequences (41.0% of RD, 50.3% of FR, 20.0% of IC, and 20.1% of RD-rain) were identified as viruses (E < 10−3) compared to the CAMERA's viral protein database (see Table S2).
Table 1.
Overview of the total number of viral sequences and contigs for air samples
| Samplea | No. of raw reads | No. of high quality, nonredundant sequences | No. of assembled contigs (>100 bp) | No. of singletons | % of reads assembled into contigs | No. of large contigs (>500 bp) | No. of known contigs |
|---|---|---|---|---|---|---|---|
| RD | 24,721 | 17,943 | 190 | 3,082 | 83 | 86 | 78 |
| FR | 23,316 | 20,691 | 160 | 10,806 | 48 | 84 | 62 |
| IC | 20,578 | 14,332 | 151 | 3,934 | 73 | 73 | 64 |
| RD-rain | 18,829 | 15,776 | 252 | 1,438 | 91 | 109 | 54 |
RD, residential district; RD-rain, rainwater precipitated at the RD site; FR, forest; IC, industrial complex.
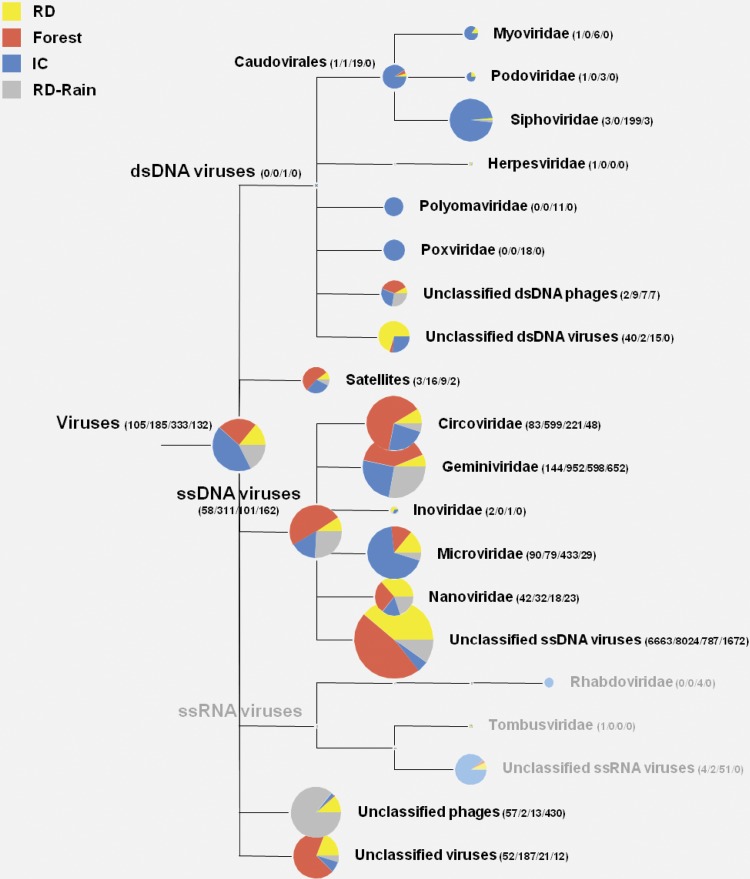
The identified sequences present in the four viromes were classified into DNA viruses, mostly ssDNA viruses (75.3 to 97.6%). Sequences from the four viromes were distributed across 12 viral families, which were composed of six double-stranded DNA (dsDNA) viral families (Myo-, Podo-, Sipho-, Herpes-, Polyoma-, and Poxviridae), five ssDNA viral families (Circo-, Gemini-, Ino-, Micro-, and Nanoviridae), and one ssDNA satellites (Fig. 4; see also Table S3 in the supplemental material). Of the identified sequences, the majority of viral sequences in the four viromes were characterized as geminivirus-related viruses accounting for 92% of RD, 86% of FR, 29% of IC, and 71% of RD-rain. On average, 71% of the geminivirus-related sequences were identified as Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 (SsHADV-1)-related viruses; i.e., geminivirus-related sequences comprised 98% of RD, 89% of FR, 28% of IC, and 71% of RD-rain sequences. Sequences related to circoviruses were the next most abundant after geminivirus-related sequences, representing an average of 10% of sequences identified in the four viromes (2% of RD, 6% of FR, 27% of IC, and 4% of RD-rain). On average, 1% of the sequences in the four viromes were nanovirus-related sequences, and 4% were microphage-related sequences. The taxonomic profile of the RD virome was similar to the profiles of FR and RD-rain at the viral family level but dissimilar to that of IC, which contained a high abundance of dsDNA siphophage-, ssDNA circovirus-, and microphage-related sequences (Fig. 4). The recent study of Lysholm et al. (34) raised the possibility that some of sequences were likely to be derived from reagents used for nucleic acid treatment. Although it is unlikely that contamination accounts for much of current data because of differences of the viromes from place to place, DNAs from reagent or nucleic acids extraction kits possibly account for some of the data.
Fig 4.
Overview of the viral families from the three land use types and rainwater based on best-match analysis. Viral metagenomic sequences were compared to the viral proteins data set in CAMERA's NCBI reference set comprising data released from GenBank (release 187) and Refseq (release 51). Comparison was performed with the BLASTx algorithm with an E-value cutoff 10−3. The MEGAN program (version 4.62.7) was used for acceptance of the output of a BLAST search based on the lowest-common-ancestor algorithm. The number of assigned sequence reads of each virome is provided in parentheses (RD/FR/IC/RD-rain). The branches shown in gray indicated the assignment to RNA viruses, which considered false matches of the sequences as described in Materials and Methods.
Genotypic differences in viral assemblages from the three land use types and rainwater.
Cross-BLASTn (E < 10−3; ≥98% identity and ≥35-bp overlap) was used to compare the viral genotypes of airborne viral assemblages between the land use types and rainwater. It was assumed that the viral genotype of RD was very similar to that of RD-rain because the viral genotype of the RD virome should coincide with that of the RD-rain virome, which reflected viruses present in the upper and lower troposphere of the RD site. The RD-rain virome shared 52.9% of its sequences with the RD virome, 2.4% with the IC virome, and 1.3% with the FR virome, suggesting that the RD viral assemblage was much more similar to that of IC than of FR and supporting the assumption that the viral assemblage of RD-rain was most similar to that of RD (Table 2).
Table 2.
A cross-BLAST analysis based on the contig assembly process
| Source | % Sequence identity by land use typea |
|||
|---|---|---|---|---|
| RD | FR | IC | RD-Rain | |
| RD | 2.6 | 14.4 | 52.9 | |
| FR | 0.7 | 0 | 1.3 | |
| IC | 21.9 | 4.2 | 2.4 | |
Parameters for the search were an E value of <10−3 and 98% minimal identity with minimal 35-bp overlap. RD, residential district; RD-rain, rainwater precipitated at the RD site; FR, forest; IC, industrial complex.
Phylogenetic diversity of airborne ssDNA viruses in the near-surface atmosphere.
Single-stranded DNA viruses formed the majority in viral assemblages collected from the land use types (Fig. 4). The genetic diversity of these putative ssDNA viruses was determined by comparing the ORFs of assembled contigs (>500 bp) for each virome with sequences in the GenBank nr database (BLASTp, E < 10−3). ORFs that contained a conserved protein sequence for ssDNA viruses (Rep protein for gemini-, circo-, and nanoviruses; Cap protein for microphages) were analyzed further. A total of 52 contigs from the four viromes had significant similarities (E < 10−5) to conserved protein sequences from gemini-, circo-, and nanoviruses and microphages (RD, 17 contigs; FR, 15 contigs; IC, 14 contigs; and RD-rain, 7 contigs). Overall, amino acid identities of the putative airborne ssDNA viral sequences had less than 50% sequence identity to known conserved protein sequences, according to BLASTp searches (Table 3). Phylogenetic analyses were conducted using the 52 contigs to determine genotypic diversity of putative ssDNA viruses with previously known viruses.
Table 3.
The features of putative ssDNA viral genome elements in the near-surface atmosphere
| Virome | Contig no. | Length (bp) | Nonanucleotide (position) | Description | BLASTp |
|||
|---|---|---|---|---|---|---|---|---|
| Rep sequence |
Amino acid profile |
|||||||
| Best hit (E value) | Coverage (%) | Identity (%) | Length (aa)a | |||||
| RD | 7 | 1,433 | Geminiviridae | Sri Lankan cassava mosaic virus (2e−06) | 73 | 25 | 295 | |
| 11 | 727 | Virus NG10 (3e−17) | 73 | 36 | 242 | |||
| 13 | 865 | Virus NG10 (5e−16) | 59 | 38 | 261 | |||
| 35 | 3,057 | TAGTATTAC (1792–1800) | Circovirus-like genome RW-C (1e−56) | 59 | 46 | 409 | ||
| 40 | 1,841 | Geminiviridae | Eragrostis curvula streak virus (1e−33) | 94 | 31 | 395 | ||
| 42 | 2,463 | Geminiviridae | African cassava mosaic virus (2e−15) | 75 | 31 | 285 | ||
| 56 | 1,188 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (3e−78) | 99 | 43 | 374 | |||
| 61 | 408 | Geminiviridae | Rhynchosia golden mosaic virus (3e−14) | 85 | 37 | 135 | ||
| 99 | 426 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (1e−31) | 66 | 71 | 141 | |||
| 101 | 438 | Virus NG19 (3e−12) | 64 | 41 | 145 | |||
| 154 | 504 | Virus NG10 (1e−14) | 74 | 41 | 167 | |||
| 164 | 1,835 | TAGTATTAC (1677–1685) | Trichoplax adhaerens (6e−21) | 68 | 31 | 345 | ||
| 173 | 2,134 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (1e−69) | 83 | 45 | 396 | |||
| 174 | 2,148 | TGTTATTAC (1937–1945) | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (5e−60) | 96 | 40 | 365 | ||
| 176 | 1,835 | Trichoplax adhaerens (2e−26) | 81 | 33 | 298 | |||
| 182 | 2,359 | TAATACTAA (2077–2085) | Geminiviridae | Tomato mottle leaf curl virus (1e−13) | 41 | 32 | 482 | |
| FR | 4 | 1,727 | CAGTATTAC (807–815) | Giardia intestinalis (6e−22) | 82 | 32 | 251 | |
| 13 | 2,467 | Sewage associated circovirus (4e−13) | 80 | 32 | 220 | |||
| 26 | 2,364 | TAGTATTAC (1101–1109) | Circoviridae | Porcine circovirus 2 (8e−23) | 65 | 31 | 449 | |
| 127 | 582 | Circoviridae | Circovirus NGchichken38/NGA/2009 (8e−23) | 90 | 39 | 193 | ||
| 128 | 2,415 | Circoviridae | Cyclovirus PK5510 (1e−27) | 97 | 31 | 279 | ||
| 129 | 579 | Circoviridae | Circovirus NGchichken38/NGA/2009 (3e−22) | 89 | 39 | 192 | ||
| 130 | 2,421 | Circoviridae | Cyclovirus PK5510 (1e−27) | 97 | 31 | 279 | ||
| 133 | 2,958 | TAGTATTAC (1332–1340) | Circovirus-like genome RW-A (4e−25) | 87 | 32 | 318 | ||
| 134 | 2,010 | TAGTATTAC (921–929) | Circoviridae | Circovirus NGchichken38/NGA/2009 (3e−22) | 42 | 40 | 665 | |
| 138 | 2,370 | TAATATTAC (1182–1190) | Geminiviridae | Velvet bean severe mosaic virus (3e−34) | 97 | 33 | 316 | |
| 139 | 2,549 | CTATATTAC (2054–2062) | Circoviridae | Chimpanzee stool avian-like circovirus Chimp17 (7e−22) | 64 | 30 | 434 | |
| 141 | 2,980 | Circovirus-like genome RW-A (5e−28) | 85 | 34 | 321 | |||
| 149 | 1,786 | TAATATTAA (1528–1536) | Geminiviridae | Sida Brazil virus (2e−15) | 66 | 32 | 305 | |
| 150 | 1,816 | TAGTATTAG (1351–1359) | Geminiviridae | Rhynchosia golden mosaic virus (4e−17) | 83 | 29 | 291 | |
| 157 | 1,769 | Geminiviridae | Beet curly top virus (2e−12) | 50 | 35 | 246 | ||
| IC | 2 | 3,877 | Circovirus-like genome CB-A (1e−40) | 40 | 40 | 666 | ||
| 7 | 1,098 | Circovirus-like genome CB-A (3e−28) | 76 | 37 | 308 | |||
| 104 | 2,079 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (5e−33) | 73 | 47 | 205 | |||
| 119 | 2,310 | TAGTATTAC (664–672) | Nanoviridae | Banana bunchy top virus (3e−21) | 90 | 32 | 315 | |
| 123 | 2,797 | Circoviridae | Columbid circovirus (1e−38) | 69 | 34 | 459 | ||
| 126 | 1,891 | TAGTATTAC (174–182) | Circoviridae | Barbel circovirus (1e−32) | 92 | 31 | 289 | |
| 132 | 2,008 | CAGTATTAC (1154–1162) | Circoviridae | Dragonfly cyclovirus (1e−46) | 75 | 42 | 312 | |
| 138 | 940 | CAGTATTAC (292–300) | Circoviridae | Raven circovirus (2e−33) | 84 | 39 | 311 | |
| 139 | 1,930 | CAGTATTAC (696–704) | Nanoviridae | Faba bean necrotic yellows virus (2e−19) | 14 | 53 | 640 | |
| 141 | 2,806 | Circovirus-like genome CB-A (2e−33) | 46 | 34 | 668 | |||
| RD-rain | 15 | 1,675 | Geminiviridae | Tomato leaf curl karnataka virus (4e−13) | 66 | 31 | 243 | |
| 16 | 1,523 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (1e−08) | 82 | 32 | 154 | |||
| 23 | 2,551 | Virus TN1 (2e−15) | 30 | 38 | 434 | |||
| 194 | 1,083 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (7e−66) | 98 | 43 | 359 | |||
| 209 | 2,038 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (2e−15) | 87 | 34 | 199 | |||
| 216 | 2,316 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (3e−41) | 93 | 44 | 204 | |||
| 232 | 2,148 | Sclerotinia sclerotiorum hypovirulence-associated DNA virus (2e−33) | 80 | 51 | 179 | |||
aa, amino acids.
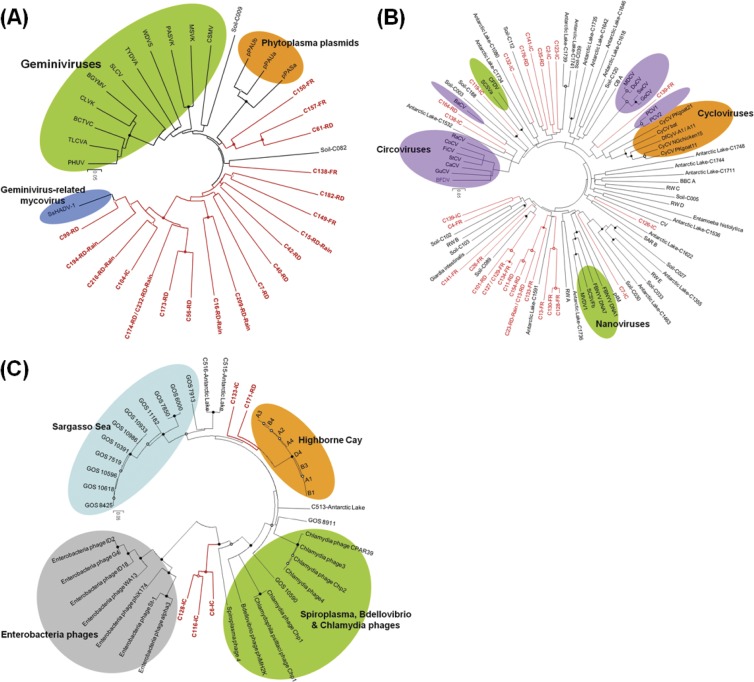
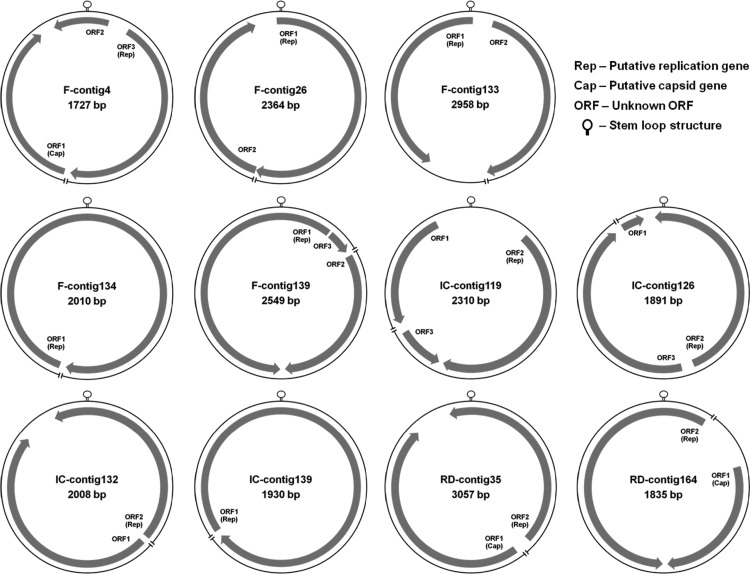
Phylogenetic analysis based on Rep protein sequences (Gemini_AL1, pfam00799) showed that 20 contigs (RD, 9 sequences; FR, 4 sequences; IC, 1 sequence; RD-rain, 6 sequences) related to geminiviruses were distantly related to previously known geminiviruses, whereas most of the contigs were closely related to SsHADV-1 (Fig. 5A). Phylogenetic analysis of Rep protein sequences (pfam02407) showed that 24 contigs were related to circoviruses, while 4 contigs related to nanoviruses were distantly related to previously known circoviruses, nanoviruses, and environmental circovirus-like genomes (Fig. 5B). However, many were closely related to environmental sequences from rice paddy soil (28) and an Antarctic lake (32). Repeated sequences both sides of a conserved nonanucleotide sequence in stem-loop structure are typically found in the viral families Circoviridae and Nanoviridae, where the sequences initiate rolling-circle replication (62, 67). Stem-loop structures with slight sequence variations of nonanucleotides were found in 9 contigs related to circoviruses (RD, 2 contigs; FR, 5 contigs; and IC, 3 contigs) and 2 contigs related to nanoviruses (IC) from the 24 contigs related to circoviruses and nanoviruses (see Table S4 in the supplemental material). Eleven putative genome elements were constructed based on stem-loop structures, putative Rep genes, putative Cap genes, or unknown ORFs in the assembled contigs, which are genomic features of the viral families Circoviridae and Nanoviridae (Fig. 6). One RD contig and four IC contigs had putative microphage Cap protein sequences (pfam02305) with significant similarities to the capsid protein sequences of chlamydia phages, spiroplasma phages, and Bdellovibrio microphages in the viral family Microviridae (E < 10−5). Based on partial Vp1 protein sequences, the airborne microphages belonged to distant genotypes compared with cultured isolates and environmental sequences (Fig. 5C). Amino acid sequences of viruses in the near-surface atmosphere of the three land use types and rainwater had low sequence identity with conserved protein sequences, and there was a diverse range of novel ssDNA viruses detected, including genomes distantly related to geminiviruses, circoviruses, nanoviruses, and microphages.
Fig 5.
Genetic diversity of airborne viruses characterized as ssDNA gemini- (A), circo- and nanoviruses (B), and microphages (C). Phylogenetic trees based on neighbor-joining with a p-distance model were constructed with MEGA5 using 1,000 randomly replicated bootstraps. Airborne viral sequences (RD, FR, IC, and RD-rain) are shown in red. The filled and empty circles at the internal nodes indicate bootstrap values of >50% and >90%, respectively. (A) Phylogenetic tree of replication initiator protein sequences (Gemini_AL1) for airborne geminivirus-like viruses with sequences from pfam00799 (accession numbers: PHUV, Q06923; TLCVA, P36279; BCTVC, P14991; CLVK, P14982; BGYMV, P05175; SLCV, P29048; TYDVA, P31617; WDVS, CAC84660; PASVK, Q00338; MSVK, P03568; CSMV, P18921), rice paddy soil (Soil-C009, ABQX01000044; Soil-C082, ABQX01000029), Phytoplasma plasmids (pPAUa, Q0QLC1; pPAUb, Q0QLC5; pPASa, Q2NIE5), and a geminivirus-related mycovirus (SsHADV-1, YP_003104796). Conserved regions of beet curly top virus (amino acids 7 to 117 of the Gemini_AL1 proteins) were used for phylogenetic analysis. The scale bar represents 0.05 amino acid substitutions per site. (B) Topology of the phylogenetic tree based on replication initiator protein sequences (Rep) for airborne circo- and nanovirus-like viruses using sequences from pfam02407 (BFDV, NP_047275; GuCV, YP_803546; FiCV, YP_803549; StCV, YP_610960; CoCV, NP_059527; CaCV, NP_573442; PCV1, NP_065678; PCV2, NP_937956; CyCV PKgoat21, YP_004152327; CyCV bat, YP_004152331; CyCV PKgoat11, YP_004152329; SwCV, ABU48445; GoCV, NP_150368; MDCV, YP_164517; DuCV, YP_271918; BaCV, YP_004376332; SCSVF, Q87009; FBNYV DNA1, Q66862; FBNYV DNA7, O91250; SCSVFb, Q87013; and MVDV1, Q9Z0D3), rice paddy soil (Soil-C89, ABQX01000039; Soil-C039, ABQX01000087; Soil-C003, ABQX01000007; Soil-C188, ABQX01000068), an Antarctic lake, a dragonfly (DfCyV-A1, ADY18020; DfCyV-A11, ADY18004), Entamoeba histolytica (XP_648754), Giardia intestinalis (AAF28772), canarypox virus (CV, NP_955176), a plasmid of Bifidobacterium pseudocatenulatum (p4M, NC_003527), and circovirus-like sequences from reclaimed water (RW_E, ACQ78164; RW_A, ACQ78155; RW_B, ACQ78158; RW_C, ACQ78160; RW_D, ACQ78163) and marine environments (BBC_A, ACQ78175; CB_A ACQ78166; SAR_B, ACQ78174). Conserved regions of porcine circovirus-1 (amino acids 14 to 84 of the Rep proteins) were used for phylogenetic analysis. (C) Phylogenetic tree of major capsid protein sequences (Vp1) from airborne microphages using Antarctic lake, Sargasso Sea, Highborne Cay, and cultured isolates (pfam02305). Partial capsid protein sequences were used for phylogenetic analysis. Enterobacteria|phages were used as outgroups. The scale bar represents 0.05 amino acid substitutions per site. Conserved regions of chlamydia phage Chp1 (amino acids 341 to 583 of the Cap proteins) were used for phylogenetic analysis. PHUV, pepper huasteco yellow vein virus; TLCVA, tomato leaf curl virus; BCTVC, beet curly top virus; CLVK, African cassava mosaic virus; BGYMV, bean golden yellow mosaic virus; SLCV, squash leaf curl virus; TYDVA, tobacco yellow dwarf virus; WDVS, wheat dwarf virus; PASVK, panicum streak virus; MSVK, maize streak virus; CSMV, chloris striate mosaic virus; BFDV, beak and feather disease virus; GuCV, gull circovirus; FiCV, finch circovirus; StCV, starling circovirus; CoCV, columbid circovirus; CaCV, canary circovirus; PCV1, porcine circovirus 1; PCV2, porcine circovirus 2; CyCV PKgoat21, cyclovirus PKgoat21/PAK/2009; CyCV bat, cyclovirus bat/USA/2009; CyCV PKgoat11, cyclovirus PKgoat11/PAK/2009; SwCV, swan circovirus; GoCV, goose circovirus; MDCV, muscovy duck circovirus; DuCV, duck circovirus; BaCV, YP_004376332; SCSVF, subterranean clover stunt virus; FBNYV DNA1, faba bean necrotic yellows virus; FBNYV DNA7, faba bean necrotic yellows virus; SCSVFb, subterranean clover stunt virus; and MVDV1, milk vetch dwarf virus.
Fig 6.
Genome organizations of the 11 putative circular genome elements assembled from viral metagenomes collected from FR, RD, and IC. Each genome element consists of a putative stem-loop structure with a conserved nonanucleotide sequence and a putative Rep gene sequence. Where there was a putative Cap gene sequence and an unidentified ORF, either or both are included, except for FR contig 134 and IC contig 139. Slashes in the genome elements indicate that the genome elements were not generated from fully sequenced genomes.
DISCUSSION
The diversity and community composition of viruses in the atmosphere were previously uncharacterized and remain as “one of the last frontiers of biological exploration on Earth” (50). Thus, this study explored, for the first time, the diversity of viruses in the near-surface atmosphere at the three distinct land use types. The study of spatial variations in viral and bacterial abundances at the three land use sites indicated no significant differences. However, temporal variations in viral and bacterial abundances were inversely correlated with temperature and absolute humidity at the RD site. Lowen et al. showed that temperature and relative humidity, the actual water vapor pressure of the air at a specific temperature and expressed in percentage, constrained the transmission and viability of the influenza virus (33). However, no distinct variation in relative humidity was observed to occur with seasonal change in the current study. Variation in relative humidity was not significantly correlated with total viral and bacterial abundances. This result was in agreement with the study of Shaman et al. and suggests that absolute humidity, the actual water vapor content of air irrespective of temperature, rather than relative humidity, might be a strong modulator of air's viral content (55). Seasonal cycles in temperature and absolute humidity, rather than relative humidity, may be the critical factors controlling seasonal variations in viruses and bacteria in the atmosphere. Thus, the high incidence of respiratory infections caused by viruses in the winter, such as respiratory syncytial virus, human rhinovirus, human bocavirus, and influenza virus, might be due to a correlation between high viral abundance and meteorological characteristics (39, 68, 71). Moreover, the lack of any significant difference in viral and bacterial abundances among sites may be attributable to similar meteorological conditions at the three sites (see Table S1 in the supplemental material). Viral and bacterial abundances in the atmosphere of Caribbean ranged from 104 to 105 viruses or bacteria m−3 under a fluorescence staining method (20). The difference in viral and bacterial abundances compared with the current study may be attributable to different meteorological characteristics in the two regions. UV radiation is also well known to be a primary factor influencing the survival of viruses and bacteria (69). Thus, the difference in UV exposures between the two different latitudes may also be responsible for the discrepancy in viral and bacterial abundances.
A high percentage of unknown sequences has been reported in other viral metagenomic studies (40 to 90% unknown sequences) (3, 40), and the current study also found that greater than 50% of the sequences of the three land use types and rainwater were uncharacterized. Taxonomy-independent analysis of the four viromes using a cross-BLAST search showed that the RD virome was more different from the FR virome than the IC virome, which differed from the comparison of viral taxonomy profiles, suggesting that a large number of novel viruses may be present in the atmosphere. Moreover, high similarity in viral genotypes between viromes at the same site (RD versus RD-rain virome) compared to the viromes at other land use types (RD versus FR or IC) suggests that the diversity and composition of airborne viruses may be dependent on land use type.
Most viromes characterized to date are dominated by bacteriophages that infect bacteria, especially dsDNA phages (sipho-, podo-, and myophages) and ssDNA microphages. The airborne viral assemblages described in this study exhibited distinct compositions that were dominated by a diverse range of ssDNA viral families (Gemini-, Circo-, Nano-, and Microviridae). The employment of multiple-displacement amplification (MDA) with phi29 polymerase has enhanced the investigation of ssDNA viruses as well as dsDNA viruses in a variety of environments (13). Single-stranded DNA viruses are known as an abundant viral class capable of infecting vertebrates, plants, and bacteria (25, 58, 62, 67), and the high genotypic diversity of ssDNA viruses has recently been reported from many environments, including rice paddy soil (28), microbialites (12), the sea (3), an Antarctic lake (32), human feces (44), and insects (36, 37, 48). Despite the consideration of preferential amplification of MDA toward a circular genome (27, 28), the detection of a large proportion of eukaryote-infecting ssDNA viruses with a small proportion of ssDNA microphages suggested distinct airborne viral assemblages compared with other viromes such as rice paddy soil (28), reclaimed water (49), marine (3), fresh water (51), an Antarctic lake (32), and human feces (44). The overwhelming majority of the ssDNA viruses detected in air viromes have not been found in previous studies, except for an Antarctic lake dominated by ssDNA viruses, where metabolic activity is restricted in the spring. The ice cover of the lake melts in the summer, and the higher transmission of radiation permits photosynthesis resulting in a prokaryote bloom, which is followed by a peak in dsDNA virus abundance. The oligotrophic air ecosystem has basically low moisture and nutrient content, and low abundance and density of airborne bacteria attributed to metabolic limitation have been considered (9). Thus, the abundance of ssDNA viruses infecting plants and mammals might become relatively dominant in airborne viral assemblages.
Atmospheric conditions during the sampling period (August to September) were typical summer conditions at the three different land use types. Most of the characterized sequences (28.8 to 92.0%) were identified as geminivirus-related viruses. A small number of sequences related to nanoviruses (0.3 to 0.7%) were also detected, suggesting a high abundance of plant-associated viruses in the near-surface atmosphere. Plant-derived particulates, such as plant debris and pollen, are largely abundant in summer libraries (18, 26), and plant-associated bacteria dominate the microbial community composition of aerosols (8, 9, 15, 18) including Sphingobacterium (a leaf surface indicator) and Sphingomonadales in the phylum Proteobacteria. Therefore, it is logical to assume that local terrestrial sources, especially plant-associated materials, may be a potential source of airborne viral assemblages. Geminiviruses, their satellites, and nanoviruses are generally known to infect crops and weeds worldwide. These viruses are spread by the infected plants or viruliferous insect vectors, such as whiteflies (35, 57, 65). However, a large number of sequences related to geminiviruses were detected, and phylogenetic analyses based on Rep protein indicated that their contigs were distantly related to previously known geminiviruses (Fig. 5A and B), which suggests a lack of knowledge regarding the ecology of geminiviruses and the possibility of airborne geminivirus transmission. Interspecies genetic recombination is known to frequently occur in ssDNA viruses, especially geminiviruses (38). Thus, it is assumed that genetic recombination might produce chimeric genotypes in the predominant ssDNA viruses found in the air virome which were related to SsHADV-1 (74). Unknown recombinant geminivirus-related viruses may occur widely in the near-surface atmosphere, and their ecological role in local ecosystems needs further investigation.
The majority of the sequences related to geminiviruses were closely related to SsHADV-1, and these accounted for over 28% of the sequences related to geminiviruses, with up to 98% in RD. SsHADV-1 was originally isolated from a plant fungal pathogen, Sclerotinia sclerotiorum, and characterized as a fungal hypovirulent virus. Even though SsHADV-1 is phylogenetically related to geminiviruses based on Rep sequences, its virion architecture and genome organization are distinct from those of geminiviruses (74). Fungi have well-documented aerial dispersal mechanisms. For example, S. sclerotiorum is able to enhance spore dispersal to surrounding air by synchronizing the ejection of thousands of spores and has a wide host range including more than 450 species of plants (6, 46). Mycoviruses typically contain double-strand or single-strand RNA, whereas SsHADV-1 has a DNA genome, which is unusual compared with known mycoviruses (74). The high abundance of the sequences related to SsHADV-1 in the near-surface atmosphere at all sites suggests that little is known about the actual abundance and diversity of DNA mycoviruses. Phylogenetic analysis based on the Rep proteins of contigs related to geminiviruses indicated the presence of a high diversity and abundance of unknown geminiviruses, particularly SsHADV-1-related viruses. The presence of abundant novel ssDNA mycovirus-related viruses in the atmosphere might revolutionize our knowledge regarding viral ecology and evolution.
Circoviruses are generally small nonenveloped viruses that infect birds and pigs (63). Using viral metagenomic techniques, discoveries of novel circoviruses in many environments, including aquatic environments (32, 47), feces (31), and insects (37, 48), have increased. A number of ssDNA sequences from the four viromes were designated circovirus-like viruses and had distinct genotypes compared with previously known circoviruses with low amino acid identities (<50%) based on phylogenetic analysis of a Rep protein (Fig. 5B and 6), suggesting a great diversity of circovirus-related sequences in the air. As one of the constituents of airborne viruses, circovirus-related viruses may originate from companion or wild animals and from insects, which could be the next major source of airborne viruses after plants. Prokaryote-infecting viruses related to the viral family Microviridae were found in samples from the three sites. ssDNA microphages are known to be dominant in the Sargasso Sea, followed by microbialites (12) and the human intestine (44).
Harvested rainwater is an effective alternative water resource in regions experiencing increased population growth, where it has various uses, such as drinking water and crop irrigation. In spite of its potential benefits, there are doubts regarding harvested rainwater quality because it might contain unknown airborne microbes (2, 22, 75). Studies of microbial contamination in rainwater have typically focused on atmospheric deposition of airborne bacteria (14), but this is the first study to investigate the airborne viruses present in rainwater. This study found that the RD-rain virome had a similar composition to the RD virome, suggesting that rainfall provides direct deposition of airborne viruses from the atmosphere. This finding may have implications for the use of harvested rainwater, and further research on rainwater is required.
Vector-mediated viral transmission has long been regarded as the major route for plant viruses (35). Ng et al. found a diverse range of circulating plant viruses in insect vectors (36, 37), and it was not surprising that a similarly broad range of animal and plant viruses was found in the near-surface atmosphere. In addition, wind-borne microbes are typically transported more than a kilometer from their point of origin and greater than 5,000 km when associated with dust events (7, 43). Thus, aerial transmission could be accessible to a broad range of viruses. For the first time, this study performed quantitative and qualitative characterization of airborne viruses from three land use types. The results indicate that the atmospheric environment is a largely unexplored reservoir of a great diversity of novel plant-associated viruses, which enhances our understanding of plant viral ecology and aerobiology in temperate regions. Further studies of the broad range of airborne viral diversity will be required to address growing concerns regarding human health risks and crop productivity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Mid-Career Researcher Program (2011-0028854), the Public Welfare and Safety research program (2011-0020967), and National Junior Research Fellowship program (2012H1A8002930) through the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology. T.W.W. and M.-S.K. were supported by a Hi Seoul Science (Humanities) Fellowship funded by the Seoul Scholarship Foundation.
We thank the Jeju center, the Korea Basic Science Institute, for TEM analysis.
Footnotes
Published ahead of print 23 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adam RD. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed W, Gardner T, Toze S. 2011. Microbiological quality of roof-harvested rainwater and health risks: a review. J. Environ. Qual. 40:13–21 [DOI] [PubMed] [Google Scholar]
- 3. Angly FE, et al. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368 doi:10.1371/journal.pbio.0040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bai X, et al. 2006. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188:3682–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blomström AL. 2011. Viral metagenomics as an emerging and powerful tool in veterinary medicine. Vet. Q. 31:107–114 [DOI] [PubMed] [Google Scholar]
- 6. Boland GJ, Hall R. 1994. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16:93–108 [Google Scholar]
- 7. Bovallius A, Roffey R, Henningson E. 1980. Long-range transmission of bacteria. Ann. N. Y. Acad. Sci. 353:186–200 [DOI] [PubMed] [Google Scholar]
- 8. Bowers RM, McLetchie S, Knight R, Fierer N. 2011. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 5:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brodie EL, et al. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cecchi L, et al. 2010. Projections of the effects of climate change on allergic asthma: the contribution of aerobiology. Allergy 65:1073–1081 [DOI] [PubMed] [Google Scholar]
- 11. Delwart EL. 2007. Viral metagenomics. Rev. Med. Virol. 17:115–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Desnues C, et al. 2008. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452:340–343 [DOI] [PubMed] [Google Scholar]
- 13. Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504–510 [DOI] [PubMed] [Google Scholar]
- 14. Evans CA, Coombes PJ, Dunstan RH, Harrison T. 2009. Extensive bacterial diversity indicates the potential operation of a dynamic micro-ecology within domestic rainwater storage systems. Sci. Total Environ. 407:5206–5215 [DOI] [PubMed] [Google Scholar]
- 15. Fierer N, et al. 2008. Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol. 74:200–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finkbeiner SR, et al. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 4:e1000011 doi:10.1371/journal.ppat.1000011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Foxman EF, Iwasaki A. 2011. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat. Rev. Microbiol. 9:254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franzetti A, Gandolfi I, Gaspari E, Ambrosini R, Bestetti G. 2011. Seasonal variability of bacteria in fine and coarse urban air particulate matter. Appl. Microbiol. Biotechnol. 90:745–753 [DOI] [PubMed] [Google Scholar]
- 19. Gibbs MJ, Weiller GF. 1999. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc. Natl. Acad. Sci. U. S. A. 96:8022–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Griffin DW, Garrison VH, Herman JR, Shinn EA. 2001. African desert dust in the Caribbean atmosphere: microbiology and public health. Aerobiologia 17:203–213 [Google Scholar]
- 21. Handelsman J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heyworth JS, Glonek G, Maynard EJ, Baghurst PA, Finlay-Jones J. 2006. Consumption of untreated tank rainwater and gastroenteritis among young children in South Australia. Int. J. Epidemiol. 35:1051–1058 [DOI] [PubMed] [Google Scholar]
- 23. Hurst CJ, Murphy PA. 1996. The transmission and prevention of infectious disease, p 3–54, In Hurst CJ. (ed), Modeling disease transmission and its prevention by disinfection. Cambridge University Press, Cambridge, MA [Google Scholar]
- 24. Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Incardona NL, Maniloff J. 2000. Family Microviridae, p 277–284 In Van Regenmortel MHV, et al. (ed), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses Academic Press, San Diego, CA [Google Scholar]
- 26. Jaenicke R. 2005. Abundance of cellular material and proteins in the atmosphere. Science 308:73. [DOI] [PubMed] [Google Scholar]
- 27. Kim KH, Bae JW. 2011. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 77:7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim KH, et al. 2008. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 74:5975–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim MS, Park EJ, Roh SW, Bae JW. 2011. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77:8062–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuske CR. 2006. Current and emerging technologies for the study of bacteria in the outdoor air. Curr. Opin. Biotechnol. 17:291–296 [DOI] [PubMed] [Google Scholar]
- 31. Li L, et al. 2010. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84:1674–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lopez-Bueno A, et al. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858–861 [DOI] [PubMed] [Google Scholar]
- 33. Lowen AC, Mubareka S, Steel J, Palese P. 2007. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 3:1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lysholm F, et al. 2012. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS One 7:e30875 doi:10.1371/journal.pone.0030875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mansoor S, Briddon RW, Zafar Y, Stanley J. 2003. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8:128–134 [DOI] [PubMed] [Google Scholar]
- 36. Ng TF, et al. 2011. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS One 6:e19050 doi:10.1371/journal.pone.0019050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ng TF, et al. 2011. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS One 6:e20579 doi:10.1371/journal.pone.0020579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225 [DOI] [PubMed] [Google Scholar]
- 39. Park AW, Glass K. 2007. Dynamic patterns of avian and human influenza in East and Southeast Asia. Lancet Infect. Dis. 7:543–548 [DOI] [PubMed] [Google Scholar]
- 40. Park EJ, et al. 2011. Metagenomic analysis of the viral communities in fermented foods. Appl. Environ. Microbiol. 77:1284–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patel A, et al. 2007. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2:269–276 [DOI] [PubMed] [Google Scholar]
- 42. Phan TG, et al. 2011. The fecal viral flora of wild rodents. PLoS Pathog. 7:e1002218 doi:10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prospero JM, Blades E, Mathison G, Naidu R. 2005. Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 21:1–19 [Google Scholar]
- 44. Reyes A, et al. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rho M, Tang H, Ye Y. 2010. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 38:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roper M, et al. 2010. Dispersal of fungal spores on a cooperatively generated wind. Proc. Natl. Acad. Sci. U. S. A. 107:17474–17479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosario K, Duffy S, Breitbart M. 2009. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. virol. 90:2418–2424 [DOI] [PubMed] [Google Scholar]
- 48. Rosario K, et al. 2011. Dragonfly cyclovirus, a novel single-stranded DNA virus discovered in dragonflies (Odonata: Anisoptera). J. Gen. virol. 92:1302–1308 [DOI] [PubMed] [Google Scholar]
- 49. Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. 2009. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 11:2806–2820 [DOI] [PubMed] [Google Scholar]
- 50. Rothschild LJ, Mancinelli RL. 2001. Life in extreme environments. Nature 409:1092–1101 [DOI] [PubMed] [Google Scholar]
- 51. Roux S, et al. 2012. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One 7:e33641 doi:10.1371/journal.pone.0033641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 53. Sattar SA, Ijaz MK. 1987. Spread of viral infections by aerosols. Crit. Rev. Environ. Control 17:89–131 [Google Scholar]
- 54. Saunders K, Bedford ID, Stanley J. 2002. Adaptation from whitefly to leafhopper transmission of an autonomously replicating nanovirus-like DNA component associated with ageratum yellow vein disease. J. Gen. Virol. 83:907–913 [DOI] [PubMed] [Google Scholar]
- 55. Shaman J, Kohn M. 2009. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc. Natl. Acad. Sci. U. S. A. 106:3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shan T, et al. 2011. The fecal virome of pigs on a high-density farm. J. Virol. 85:11697–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stanley J. 2004. Subviral DNAs associated with geminivirus disease complexes. Vet. Microbiol. 98:121–129 [DOI] [PubMed] [Google Scholar]
- 58. Stanley J. 2005. Geminiviridae, p 301–326 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA [Google Scholar]
- 59. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tang JW. 2009. The effect of environmental parameters on the survival of airborne infectious agents. J. R. Soc. Interface 6(Suppl. 6):S737–S746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4:470–483 [DOI] [PubMed] [Google Scholar]
- 62. Todd D. 2005. Circoviridae, p 326–334 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA [Google Scholar]
- 63. Todd D. 2000. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol. 29:373–394 [DOI] [PubMed] [Google Scholar]
- 64. Tulman ER, et al. 2004. The genome of canarypox virus. J. Virol. 78:353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Varma A, Malathi VG. 2003. Emerging geminivirus problems: a serious threat to crop production. Ann. Appl. Biol. 142:145–164 [Google Scholar]
- 66. Verreault D, et al. 2011. Detection of airborne lactococcal bacteriophages in cheese manufacturing plants. Appl. Environ. Microbiol. 77:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vetten HJ. 2005. Nanoviridae, p 343–352 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA [Google Scholar]
- 68. Wang W, et al. 2010. Molecular monitoring of causative viruses in child acute respiratory infection in endemo-epidemic situations in Shanghai. J. Clin. Virol. 49:211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Weinbauer MG. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127–181 [DOI] [PubMed] [Google Scholar]
- 70. West JS, Atkins SD, Emberlin J, Fitt BD. 2008. PCR to predict risk of airborne disease. Trends Microbiol. 16:380–387 [DOI] [PubMed] [Google Scholar]
- 71. Wiegering V, et al. 2011. Gastroenteritis in childhood: a retrospective study of 650 hospitalized pediatric patients. Int. J. Infect. Dis. 15:e401–e407 [DOI] [PubMed] [Google Scholar]
- 72. Willner D, et al. 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370 doi:10.1371/journal.pone.0007370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Womack AM, Bohannan BJ, Green JL. 2010. Biodiversity and biogeography of the atmosphere. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:3645–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu X, et al. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U. S. A. 107:8387–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yuan T, Fengmin L, Puhai L. 2003. Economic analysis of rainwater harvesting and irrigation methods, with an example from China. Agr. Water Manage. 60:217–226 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.