Abstract
There are two mechanisms for the incorporation of B5 into the envelope of extracellular virions produced by orthopoxviruses, one that requires A33 and one that does not. We have hypothesized that the A33-dependent mechanism requires a direct interaction between A33 and B5. In this study, chimeric constructs of A33 and B5/B5-green fluorescent protein (GFP) were used to show that the two proteins interact through their lumenal domains and that the coiled-coil domain of B5 is sufficient for an interaction with A33. Furthermore, our experiments reveal that a transmembrane domain, not necessarily its own, is requisite for the lumenal domain of B5 to interact with A33. In contrast, the lumenal domain of A33 is sufficient for interaction with B5. Furthermore, the lumenal domain of A33 is sufficient to restore the proper localization of B5-GFP in infected cells. Taken together, our results demonstrate that the lumenal domains of A33 and B5 interact and that the interaction is required for the incorporation of B5-GFP into extracellular virions, whereas the incorporation of A33 is independent of B5. These results suggest that viral protein incorporation into extracellular virions is an active process requiring specific protein-protein interactions.
INTRODUCTION
Remarkably, orthopoxviruses produce two infectious forms that are morphologically and antigenically distinct (1, 35). Viral replication occurs entirely in the cytoplasm of infected cells in a specialized area known as the viral factory, where the first form of infectious virions, termed intracellular mature virions (IMV), is produced (7, 26). IMV represent the majority of progeny virions and are released only if the cell is lysed. A subset of IMV is transported along microtubules to the site of wrapping and obtains an additional double membrane envelope derived from the trans-Golgi network or early endosome (15, 40, 45). These wrapped virions are termed intracellular enveloped virions (IEV). IEV are transported along microtubules to the plasma membrane and released from the cytoplasm by fusion of their outermost membrane with the plasma membrane (13, 17, 33, 52). Virions retained on the cell surface are termed cell-associated enveloped virions (CEV). Some CEV are propelled away from infected cells by actin tails, a process that requires the viral A36 protein (16, 37, 44, 56). When CEV are released from the cell surface, they become extracellular enveloped virions (EEV). CEV are required for efficient cell-to-cell spread (43, 52), while EEV are required for long-range dissemination of virus (1, 30). CEV and EEV are collectively termed extracellular virions (EV) (27).
Eight proteins encoded by vaccinia virus, A33 (36), A34 (8), A36 (48), A56 (42), B5 (11, 55), F12 (47, 58), F13 (2), and K2 (46, 49), have been shown to be specific for IEV. Deletion of the gene that encodes any of these proteins, with the exception of A56 and K2, results in a small-plaque phenotype, implying that these proteins have a role in EV morphogenesis and/or infectivity. Several interactions among these proteins have been reported, and it appears that these interactions are required for the proper complement of viral proteins in the envelope of progeny EV (6, 10, 22, 29, 31, 32, 54, 57). Among these proteins, the integral membrane protein A33 has been reported to interact with both A36 (52, 57) and B5 (3, 31). A36 requires an interaction with A33 for incorporation into the outer IEV membrane (57), suggesting that A33 has the ability to target proteins to the EV envelope. B5 has been shown to target to virion membranes in the absence of A33 (5, 31). It has been reported that the incorporation of A33 into wrapped virions requires an interaction with B5 (31). Subsequently, we reported that chimeric B5-green fluorescent protein (GFP) requires an interaction with A33 for its incorporation into EV (5). This implies that there are two mechanisms for the incorporation of B5 into wrapping membranes; one is dependent on A33, and the other appears to be inhibited by the addition of GFP to B5. In an effort to clear up this controversy and determine the dependence of proper intracellular trafficking and virion incorporation on the B5-A33 interaction, we have characterized the A33-B5 interaction in detail. Our studies show that the trafficking and EV incorporation of A33 are independent of B5. In addition, we have mapped the region of interaction to the lumenal domains of both proteins and show that the lumenal domain of A33 is sufficient to properly target B5-GFP and mediate its incorporation into EV. These results, in conjunction with those of our companion paper (4), support the conclusion that incorporation of B5 into the EV membrane is an active process that is normally mediated through an interaction with A33.
MATERIALS AND METHODS
Cells and viruses.
HeLa and RK13 cells were maintained as previously described (50). The construction of vB5R-GFP (53), vB5R-GFP/ΔA33R (5), vTF7.3 (9), and vΔB5R (55) has been described previously.
Plasmid constructs.
The construction of pB5-GFP, pB5/cc/G/Δ-GFP (51), and pA33R-HA (3) has been described previously. To construct pΔ/cc/B5/Δ-GFP, overlapping primer pairs were designed to amplify the coding sequences for residues 1 to 28 and 242 to 303 of B5. These fragments were joined by a second PCR and subsequently cloned using engineered 5′ HindIII and 3′ NcoI sites in place of the full-length B5 coding sequence in pB5-GFP. A plasmid containing the coding sequence of influenza virus A/Chicken/Nanchang/3-120/01 neuraminidase (NA) was a gift from Toru Takimoto at the University of Rochester Medical Center. A chimeric form of A33 that has the cytoplasmic tail domain deleted and the transmembrane domain replaced with that of influenza virus Nanchang NA (Δ/NA/A33R-HA) was constructed by two-step overlapping PCR. For expression, the coding sequence of Δ/NA/A33R-HA was inserted under the control of the T7 promoter into pcDNA3 (Invitrogen) (pΔ/NA/A33R-HA). To construct B5 lacking the transmembrane and cytoplasmic tail domains (pB5RLD/cc), the sequence encoding amino acid residues 1 to 275 followed by a stop codon was amplified by PCR and inserted under the control of the T7 promoter into pcDNA3 (Invitrogen). To construct A33-hemagglutinin (A33-HA) lacking the transmembrane and cytoplasmic tail domains (A33R-HALD), the sequence encoding amino acid residues 61 to 185 followed by an HA epitope tag and a stop codon, which was preceded by the signal peptide sequence of vesicular stomatitis virus G (VSVG) (Indiana strain), was amplified by PCR and inserted under the control of the T7 promoter into pcDNA3 (Invitrogen) to yield pA33R-HALD. To place the coding sequence of A33R-HALD under the control of the normal A33R promoter, two separate PCRs were performed. The first reaction amplified the ∼500-bp upstream region of A33R, which contains the A33R promoter. The second reaction amplified the coding sequence of the soluble form of A33 with the HA epitope tag. The two resulting products were joined by an overlapping PCR, and this product was inserted into pCR2.1 (Invitrogen) to yield pLF A33R-HALD. The coding sequence of A33R-HA plus ∼500 bp of the upstream sequence, which would include the normal A33R promoter, was removed from pLF A33R-HA-118 (5) with ApaI and HindIII and inserted into pCR2.1 that had been digested in the same way. The resulting plasmid, pLF A33R-HA, was sequenced to verify its integrity. To construct pVSVG-GFP, the coding sequences of VSVG and enhanced GFP were amplified by PCR. PCR fragments were annealed using an overlapping PCR. The resulting PCR product was inserted into pCR2.1 (Invitrogen) and subcloned into pcDNA3 (Invitrogen) that had been digested with HindIII and XhoI. Oligonucleotides were designed to remove the coding sequence of GFP and add the coding sequence for a Strep-tag II (41) epitope followed by a stop codon for constructs B5-GFP, B5/cc/B5/Δ-GFP (51), and Δ/cc/B5/Δ-GFP using PCR. The resulting products, which contained the coding sequence for B5-Strep, B5-cc/B5/Δ-Strep, and Δ/cc/B5/Δ-Strep, respectively, were inserted into pCR2.1 (Invitrogen) and subcloned into pcDNA3 (Invitrogen) using standard cloning techniques. Similarly, an oligonucleotide was designed to add the coding sequence for Strep-tag II after residue 482 of the transmembrane domain of VSVG. This oligonucleotide was used with the overlapping oligonucleotides described above, which remove residues 29 to 241 of B5 in construct pΔ/cc/B5/Δ-GFP, in a two-step PCR to remove lumenal domain residues 29 to 241 and replace the coding sequence of GFP with the coding sequence for the Strep-tag II epitope, followed by a stop codon of construct B5/cc/G/Δ-GFP. The resulting product contained the coding sequence for Δ/cc/G/Δ-Strep. It was inserted into pCR2.1 (Invitrogen) and subcloned into pcDNA3 (Invitrogen) using standard cloning techniques. All constructs were verified by sequencing.
Immunofluorescence microscopy.
HeLa cells grown on glass coverslips were infected with either vB5R-GFP or vΔB5R at a multiplicity of infection (MOI) of 1.0. For in vivo trans complementation, HeLa cells infected with vB5R-GFP/ΔA33R at an MOI of 1.0 were transfected with either pLF A33R-HA or pLF A33R-HALD or mock transfected. The next day, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS). For intracellular staining, fixed cells were permeabilized with 0.1% Triton X-100 in PBS. Fixed or fixed and permeabilized cells were incubated with anti-A33 monoclonal antibody (MAb) 10F10, which was kindly provided by Jay Hooper (18), or with rabbit anti-HA antibody (Sigma), followed by Texas Red-conjugated donkey anti-mouse or anti-rabbit antibody, respectively (Jackson ImmunoResearch Laboratories). DNA in the nuclei and viral factories was stained with either 4′,6-diamidino-2-phenylindole (DAPI) or Hoechst as described previously (5). Cells were visualized and imaged as previously described (50). Images were minimally processed and pseudocolored using Adobe Photoshop software (Adobe Systems).
Immunoprecipitation and Western blotting.
HeLa cells infected with vTF7.3 at an MOI of 5.0 in the presence of 40 μg/ml of cytosine arabinoside (AraC; Sigma) were transfected with various plasmids containing the coding sequences of genes under the control of the vaccinia virus T7 promoter at 2 h p.i. The same amount of each construct was used in every transfection, and a total of 1 μg of total DNA was used for each transfection. In experiments where the total amount of constructs did not equal 1 μg, the difference was made up with pcDNA3. Transfection medium was removed at 4.5 h posttransfection and replaced with medium containing 25 μCi/ml of [35S]Met-Cys (Perkin-Elmer). For coimmunoprecipitation (CoIP) during infection, HeLa cells were infected with vB5R-GFP/ΔA33R at an MOI of 5.0 and transfected with either pLF A33R-HA or pLF A33R-HALD or mock transfected. The following day, cells were harvested by scraping, washed once in PBS, and lysed in radioimmunoprecipitation assay (RIPA) buffer (0.5× PBS, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 1% NP-40, 0.5% sodium deoxycholate) containing protease inhibitors as previously described (5). Immunoprecipitation was performed using an anti-HA MAb (Santa Cruz Biotechnology) as previously described (10). Proteins were resolved on 4 to 12% gradient or 12% acrylamide gels (Invitrogen) and detected by autoradiography or Western blotting. For Western blotting, proteins were transferred to nitrocellulose membranes. Membranes were incubated with a horseradish peroxidase (HRP)-conjugated anti-GFP antibody (Rockland), an HRP-conjugated anti-HA antibody (Roche), an anti-GFP MAb (Covance), an anti-HA MAb (Roche), or an anti-Strep-tag II MAb (Novagen). Unconjugated antibodies were followed with an appropriate HRP-conjugated anti-mouse or anti-rat antibody (Jackson ImmunoResearch Laboratories). Bound antibodies were detected by using chemiluminescent reagents (Pierce) and following the manufacturer's instructions.
Analysis of EEV.
RK13 cells were infected with vB5R-GFP, vΔB5R, or vB5R-GFP/ΔA33R at an MOI of 10.0. At 4 h p.i., the medium was replaced with medium containing [35S]Met-Cys (Perkin-Elmer). The next day, radiolabeled virions released into the medium were purified through a 36% sucrose cushion. The resulting viral pellets were lysed in RIPA buffer as described above. EEV lysates were equilibrated by scintillation counting, and equal counts were subjected to immunoprecipitation with an anti-A33 MAb. Antibody-protein complexes were pulled down as described previously (10). Immunoprecipitated proteins were analyzed by SDS-PAGE and detected by autoradiography.
Immunoelectron microscopy.
RK13 cells were infected with either vB5R-GFP or vΔB5R at an MOI of 10.0. The next day, virions released into the medium were purified as described above. Purified virions were adsorbed to Formvar-coated nickel grids and immunostained with either an anti-B5 MAb or an anti-A33 MAb, followed by an 18-nm colloidal gold-conjugated goat anti-rat or anti-mouse antibody, respectively (Jackson ImmunoResearch Laboratories). Virions were negatively stained and visualized using a Hitachi 7650 transmission electron microscope (TEM) with a Gatan 11-megapixel digital camera.
RESULTS
B5 is not required for proper subcellular localization of A33.
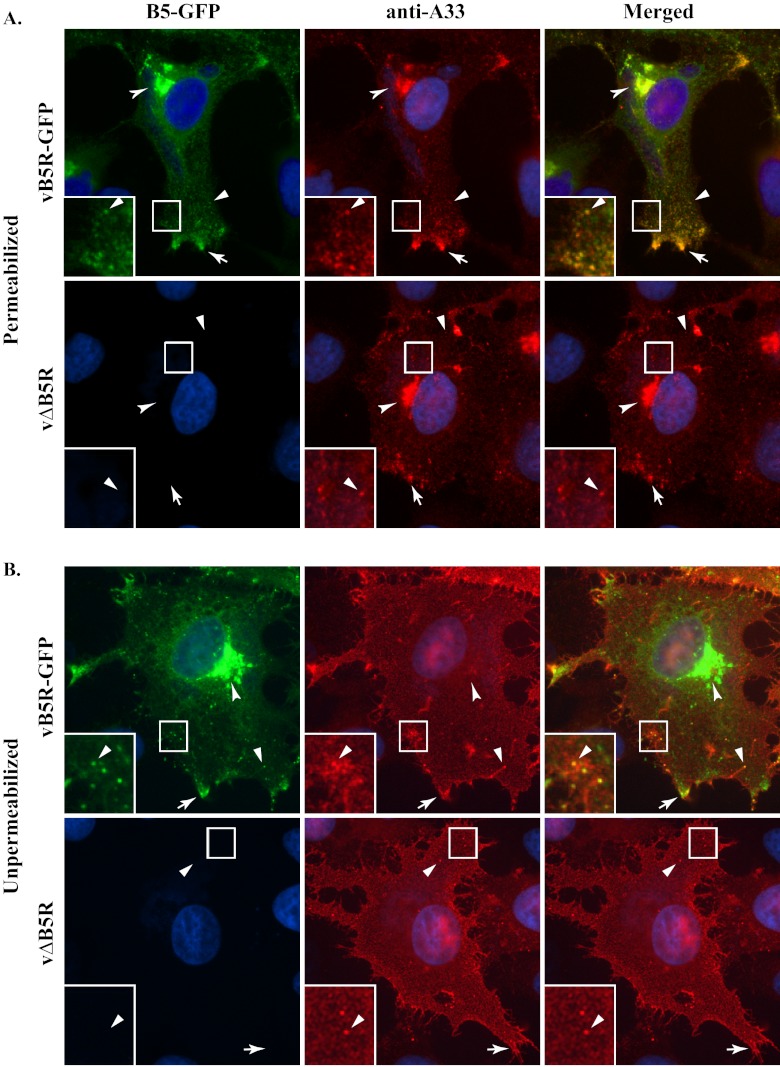
B5 is a type I integral membrane protein that has been shown to localize to EV in the absence of A33 (5, 21, 31). It has been reported that the interaction between A33 and B5 is required for the incorporation of A33 into EV (31). In contrast, a chimeric version of B5 (B5-GFP) was shown to require A33 for incorporation (5), indicating that there are two mechanisms for B5 incorporation, one that is dependent on A33 and one that is not. To investigate the dependence of these two proteins on each other for proper intracellular trafficking and virion incorporation, we first examined if A33 was properly localized during infection in the absence of B5. In cells infected with vB5R-GFP, A33 colocalized with B5-GFP at the site of wrapping, at the cell vertices, and on virion-sized particles (VSPs), which are three characteristic hallmarks of both B5 and B5-GFP in infected cells (Fig. 1A) that have been seen previously (53). Moreover, B5-GFP provides a convenient tool for studying the A33-dependent pathway because B5-GFP is dependent on A33 for incorporation whereas B5 is not. Therefore, B5-GFP was used in these studies. It should be noted that the localization pattern of B5-GFP observed in cells infected with vB5R-GFP is almost identical to the B5 localization pattern seen in cells infected with the parental virus, WR (5), indicating that fusion of GFP to B5 does not alter the proper localization of B5 when all of the viral proteins are present. Similarly, in cells infected with a virus that has B5R deleted (vΔB5R), A33 localized at the site of wrapping, at the cell vertices, and on VSPs in a pattern similar to that seen in cells infected with vB5R-GFP (Fig. 1A), demonstrating that proper subcellular localization of A33 is independent of B5. To look for A33 on CEV in the absence of B5, cells infected with vΔB5R were stained without permeabilization using an anti-A33 MAb. In cells infected with vB5R-GFP, A33 was found on the plasma membrane and concentrated with GFP-labeled VSPs (inset, Fig. 1B). Importantly, the site of wrapping was not stained, confirming that the cells were unpermeabilized. In the absence of B5, a similar staining pattern was seen with A33 on the plasma membrane and on VSPs (Fig. 1B), although the number of VSPs was greatly reduced. Thus, the proper subcellular targeting of A33 is independent of B5.
Fig 1.
B5 is not required for proper localization of A33. HeLa cells grown on glass coverslips were infected with the indicated viruses. The next day, fixed and permeabilized (A) or fixed (B) cells were immunostained with an anti-A33 MAb, followed by Texas Red-conjugated donkey anti-mouse antibody. The localization of B5-GFP or A33 at the site of wrapping (concave arrowheads), at the cell vertices (arrows), or at VSPs (arrowheads) is indicated. DNA in the nuclei and viral factories was stained with DAPI or Hoechst (blue). The overlap of B5-GFP (green) and A33 (red) is yellow. Boxed regions are enlarged in the lower left corners to highlight the A33-labeled VSPs.
A33 is efficiently incorporated into EEV in the absence of B5.
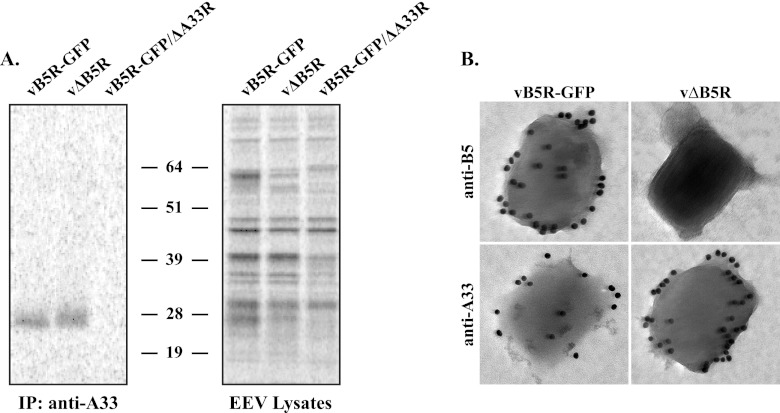
The previous results show that the cellular localization of A33 is unchanged in the absence of B5 and suggest that A33 may be incorporated into EV. To determine if the incorporation of A33 into EV is independent of B5, we looked for A33 in purified EEV released by cells infected with vΔB5R. We purified radiolabeled virions released by RK13 cells infected with vB5R-GFP, vΔB5R, or vB5R-GFP/ΔA33R. Purified virions were lysed, and equal amounts of radioactivity were subjected to immunoprecipitation with an anti-A33 MAb. A band of the size predicted for A33 was immunoprecipitated from EEV released by cells infected with either vB5R-GFP or vΔB5R but not from EEV released by cells infected with vB5R-GFP/ΔA33R (Fig. 2A), indicating that the incorporation of A33 into extracellular virions is independent of B5. The examination of equilibrated EEV lysates by SDS-PAGE showed that approximately equal amounts of protein were used in the assay (Fig. 2A).
Fig 2.
Incorporation of A33 into EEV does not require B5. (A) Immunoprecipitation (IP). Lysates of purified radiolabeled EEV released from cells infected with the indicated viruses were immunoprecipitated with an anti-A33 MAb. Protein-antibody complexes were resolved by SDS-PAGE, and proteins were detected by autoradiography. Equilibrated EEV lysates were analyzed to verify that equal amounts of EEV were used for immunoprecipitation. The molecular masses, in kilodaltons, and positions of marker proteins are shown. (B) Immunoelectron microscopy. EEV released from cells infected with the indicated viruses were stained with either an anti-B5 MAb or an anti-A33 MAb, followed by 18-nm colloidal gold-conjugated goat anti-rat or anti-mouse antibody, respectively. Immunogold-labeled EEV were negatively stained and visualized using a Hitachi 7650 TEM.
To verify our biochemical analysis, we performed immunoelectron microscopy of purified EEV to visualize the presence of A33 on extracellular virions. Purified EEV released from cells infected with either vB5R-GFP or vΔB5R were immunolabeled with either an anti-B5 or an anti-A33 MAb, followed by a colloidal-gold-conjugated goat anti-rat or anti-mouse antibody, respectively. B5-GFP and A33 were readily detected on EEV released from cells infected with vB5R-GFP (Fig. 2B). A33, but not B5, was detected on EEV released from cells infected with vΔB5R (Fig. 2B), supporting our biochemical analysis. Taken together, our data show that B5 is not required for proper subcellular localization and incorporation of A33 into wrapped virions.
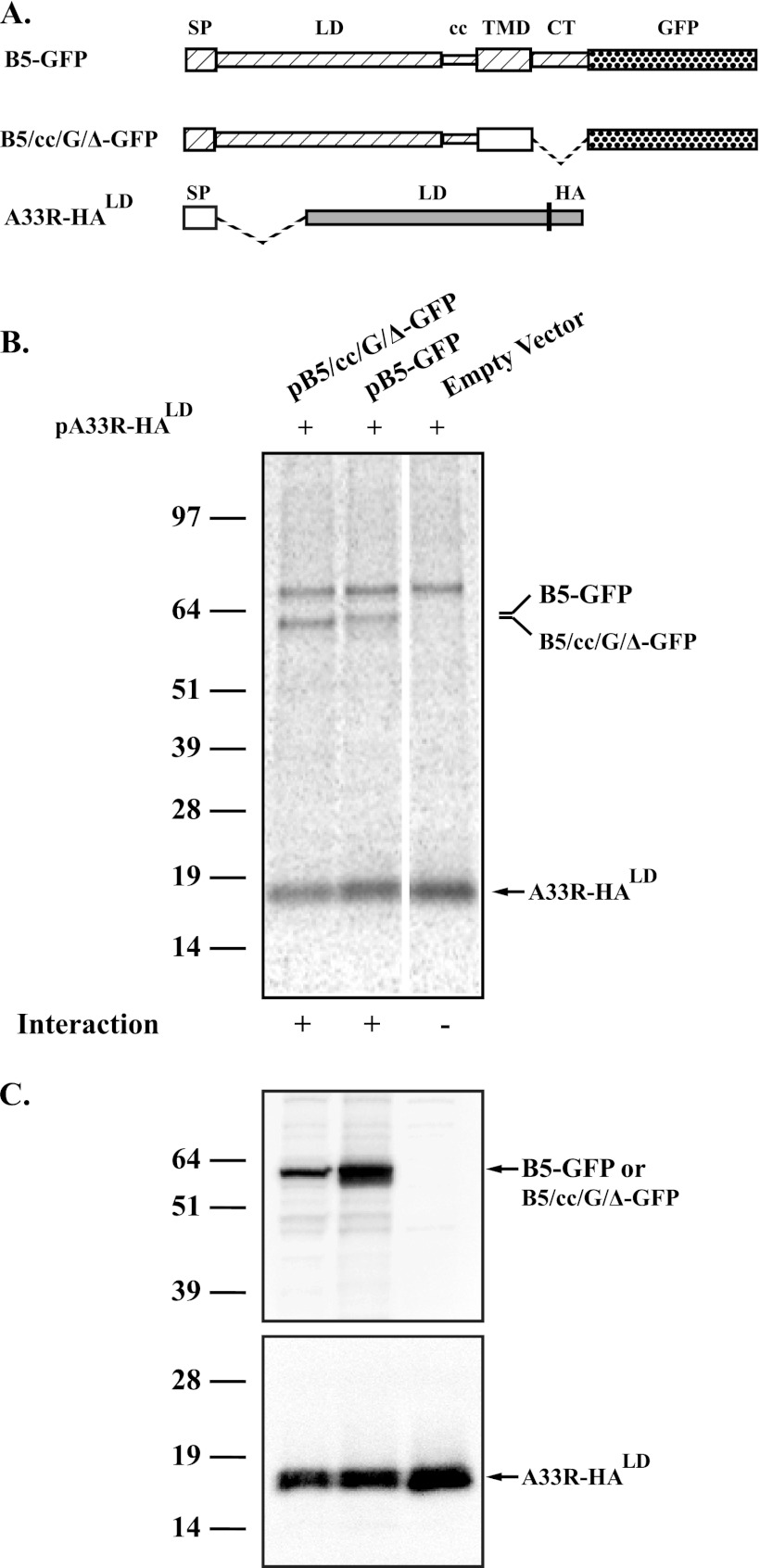
A33 and B5 interact through their lumenal domains.
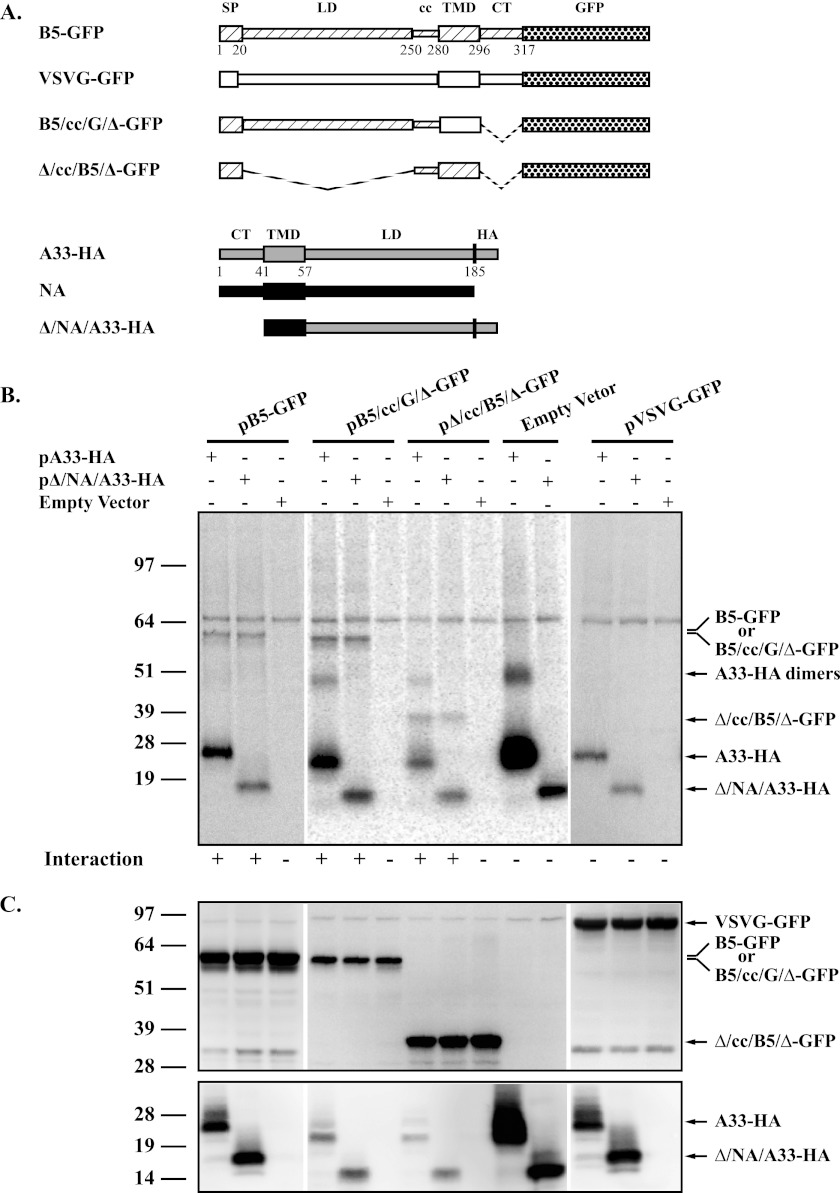
The results from the above-described experiments show that the localization and incorporation of A33 into EV are independent of B5. We have shown that A33 interacts with B5-GFP (3) and that the incorporation of B5-GFP into IEV/EV requires A33 (5). Therefore, we hypothesized that the interaction between A33 and B5-GFP was necessary for B5-GFP incorporation into EV. We next wanted to map the domains in A33 and B5 that are involved in interaction using CoIP. B5 and A33 are type I and type II integral membrane proteins, respectively (Fig. 3A) (21, 36). Chimeras were generated in which the various domains of B5 and A33 (lumenal, transmembrane, and cytoplasmic tail [LD, TMD, and CT, respectively]) were replaced with the corresponding domains from analogous non-poxvirus proteins that do not interact (the G protein from VSV and the NA protein from influenza A virus, respectively). Diagrammatic representations of the chimeras constructed are shown in Fig. 3A with each predicted domain (LD, TMD, and CT) depicted as a letter representing the protein it was derived from (B5, A33, G, and NA proteins) separated by a slash. For B5, the predicted coiled coil (CC) is also included, as VSVG is not predicted to encode such a structure. Interaction of the chimeras was tested for by coexpression using the vaccinia virus T7 expression system (9) in the presence of AraC, to inhibit viral late protein synthesis and reduce the production of endogenous untagged late proteins that may compete for interaction in our assay, followed by CoIP. As expected, a band corresponding to full-length B5-GFP was coimmunoprecipitated with A33-HA, confirming the interaction between B5 and A33 (Fig. 3B) (3). Similarly, a chimera that has the lumenal domain of A33 and the transmembrane domain of influenza NA (Δ/NA/A33-HA) also interacted with B5-GFP, suggesting that the two proteins interact through their lumenal domains (Fig. 3B).
Fig 3.
A33 and B5 interact through their lumenal domains. (A) Diagrammatic representation of constructs used in CoIP assays. Chimeras were generated from the signal peptide (SP), the coiled-coil (cc) domain, the lumenal domain (LD), and the transmembrane domain (TMD) of B5 (▨), VSVG (□), A33 (▨), and influenza A virus NA (■). Δ indicates deletion of the corresponding domain, which is represented in the diagram by a dashed line connecting included domains. B5 and A33 were tagged with either full-length GFP or an HA epitope, respectively. For B5 and A33, the number of the residue that starts the predicted domain is shown below the protein diagram. (B) CoIP. HeLa cells infected with vTF7.3 in the presence of AraC were transfected with either the indicated plasmids that expressed the constructs described in panel A or pcDNA3 (Empty Vector). At 4.5 h posttransfection, the medium was replaced with medium containing [35S]Met-Cys. Cells were harvested at 24 h posttransfection and lysed, and cell lysates were immunoprecipitated with an anti-HA MAb. Immune complexes were analyzed by SDS-PAGE, and proteins were detected by autoradiography. (C) Protein expression. Cell lysates used in panel B were subjected to SDS-PAGE and analyzed by Western blot assay using an anti-GFP (top) or anti-HA (bottom) MAb, followed by an HRP-conjugated donkey anti-mouse or anti-rat antibody, respectively. The molecular masses, in kilodaltons, and positions of marker proteins are shown on the left of the blots.
Next, we replaced the transmembrane domain of B5 with the transmembrane domain from VSVG to test the ability of the lumenal domain to interact with A33. This new construct (B5/cc/G/Δ-GFP) was able to interact with both full-length A33-HA and a construct that has only the lumenal domain of A33, Δ/NA/A33-HA, confirming that an interaction between their lumenal domains occurs (Fig. 3B). Amino acid residues 236 to 276 of B5 are predicted to form a coiled-coil structure. These structures are known to be involved in protein-protein interactions (19). A truncated version of B5R-GFP that encodes only the predicted coiled-coil and transmembrane domains of B5 fused to GFP, pΔ/cc/B5/Δ-GFP, was tested to determine its ability to interact with A33. Δ/cc/B5-GFP was coimmunoprecipitated with A33-HA (Fig. 3B), indicating that much of the lumenal domain and the cytoplasmic tail of B5 are not required for interaction with A33. Taken together, our results suggest that the coiled-coil domain of B5 interacts with the lumenal domain of A33. The expression of all constructs was verified by Western blotting using appropriate antibodies (Fig. 3C).
The lumenal domain of A33 is sufficient for interaction with B5.
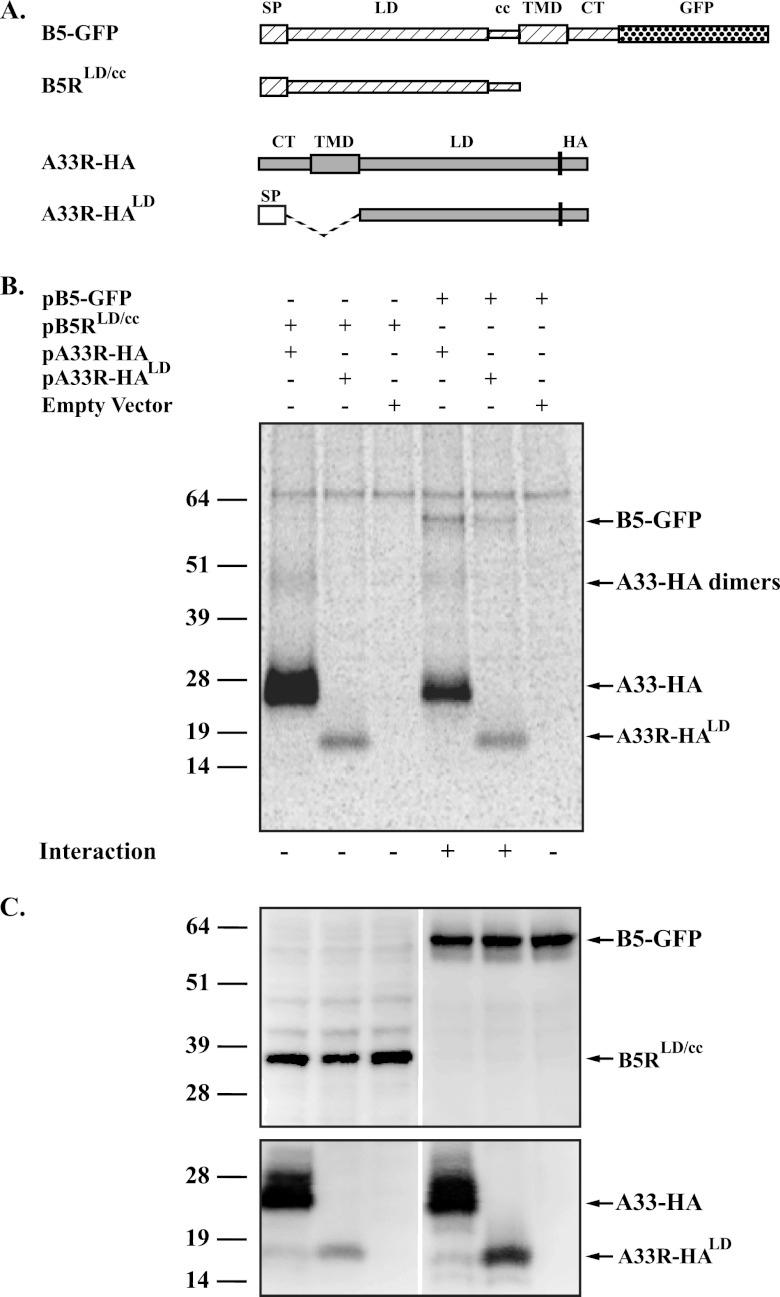
The above result indicates that A33 and B5 interact through their lumenal domains. To test if the lumenal domains are sufficient for interaction, constructs were made that express the predicted lumenal domains of B5 and A33-HA (pB5RLD/cc and pA33R-HALD, respectively) (Fig. 4A). Using the same T7 expression system as above, the new constructs were tested for the ability to interact using CoIP. A33-HALD brought down a band of the predicted size for full-length B5-GFP (Fig. 4B), demonstrating that the lumenal domain of A33 is sufficient for interaction. However, an interaction between B5LD/cc and either full-length A33-HA or A33-HALD was not detected (Fig. 4B), suggesting that anchorage of B5 via a transmembrane domain is required for the interaction to occur. Expression of B5LD/cc was verified by Western blotting using an anti-B5 MAb (Fig. 4C). Immunofluorescence microscopy indicated that the inability of the soluble proteins to interact was not due to dissimilar subcellular localization (data not shown).
Fig 4.
The lumenal domain of A33 is sufficient for interaction with B5-GFP. (A) Diagrammatic representation of constructs used for CoIP. Soluble B5 containing the signal peptide (SP), the lumenal domain (LD), and the coiled-coil (cc) domain of B5 (▨). Soluble A33-HA containing the cleavable signal peptide (SP) sequence of VSVG (□), followed by the predicted lumenal domain (LD) of A33 (▨) and the HA epitope tag. Connecting domains that were deleted are represented by a dashed line in the diagram (B). CoIP. HeLa cells infected with vTF7.3 in the presence of AraC were transfected with either the indicated plasmids that expressed the constructs described in panel A or pCDNA3 (Empty Vector). At 4.5 h posttransfection, the medium was replaced with medium containing [35S]Met-Cys. Cells were harvested at 24 h posttransfection and lysed. Cell lysates were immunoprecipitated with an anti-HA MAb, immune complexes were resolved by SDS-PAGE, and proteins were detected by autoradiography. (C) Protein expression. Cell lysates used in panel B were resolved by SDS-PAGE and analyzed by Western blot assay using an anti-B5 (top) or anti-HA (bottom) MAb, followed by an HRP-conjugated donkey anti-rat antibody. The molecular masses, in kilodaltons, and positions of marker proteins are shown on the left of the blots.
Membrane anchoring of B5 is required for interaction with the lumenal domain of A33.
We wanted to rule out the possibility that the transmembrane domain of B5 was required for interaction with A33. Therefore, we tested if the lumenal domain of B5, including the coiled coil, fused to the transmembrane domain of VSVG (B5/cc/G/Δ-GFP) could interact with A33-HALD by using CoIP (Fig. 5A). A33-HALD coimmunoprecipitated a band corresponding to B5/cc/G/Δ-GFP, indicating that the B5 transmembrane domain is not required for the interaction but membrane anchoring by an unrelated transmembrane domain is (Fig. 5B).
Fig 5.
Membrane anchoring of B5 is required for interaction with A33. (A) Diagrammatic representation of constructs used for CoIP. Chimeric B5-GFP was generated from the predicted lumenal domain (LD), the coiled-coil (cc) domain of B5 (▨), and the transmembrane domain (TMD) of VSVG (□). The A33 construct contains the lumenal domain (LD) of A33 (▨), preceded by the cleavable signal peptide (SP) sequence of VSVG (□). B5 and A33 were tagged with either full-length GFP or an HA epitope, respectively. Δ indicates deletion of the corresponding domain, which is represented by a dashed line in the diagram connecting included domains. (B) CoIP. HeLa cells infected with vTF7.3 in the presence of AraC were transfected with either the indicated plasmids that expressed the constructs described in panel A or pCDNA3 (Empty Vector). At 4.5 h posttransfection, the medium was replaced with medium containing [35S]Met-Cys. Cells were harvested at 24 h posttransfection and lysed. Cell lysates were immunoprecipitated with an anti-HA MAb, immune complexes were resolved by SDS-PAGE, and proteins were detected by autoradiography. (C) Protein expression. Cell lysates used in panel B were resolved by SDS-PAGE and analyzed by Western blot assay using an anti-GFP (top) or anti-HA (bottom) MAb, followed by an HRP-conjugated donkey anti-mouse or anti-rat antibody, respectively. The molecular masses, in kilodaltons, and positions of marker proteins are shown on the left of the blots.
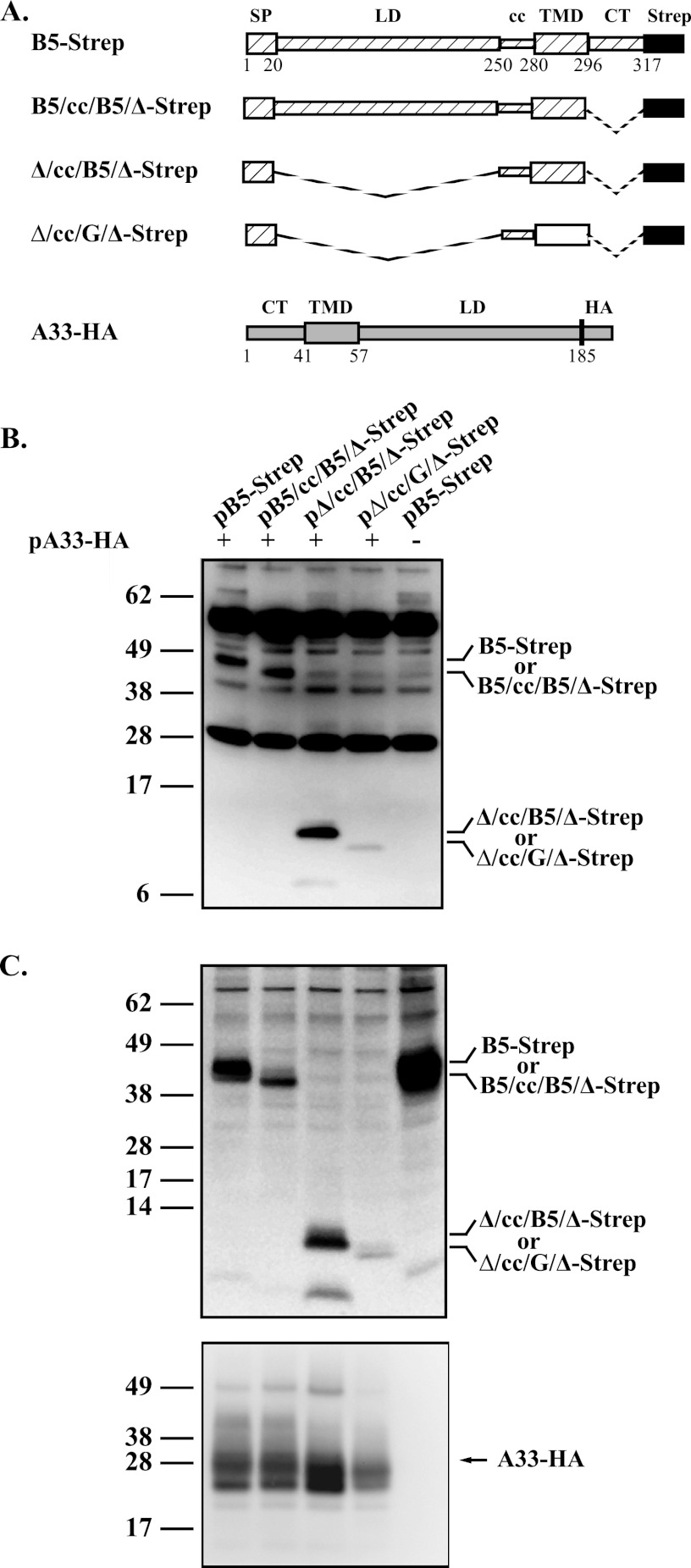
The coiled-coil domain of B5 is sufficient for interaction with A33.
Cumulatively, all of the previous results suggest that the coiled-coil region of B5 is sufficient for interaction with A33. However, these experiments were conducted using B5 constructs fused to GFP. Although it seems highly unlikely, we cannot rule out the possibility that GFP was influencing the interaction with A33. To directly test the coiled-coil region of B5 for interaction with A33 and to rule out any involvement of GFP, we made four new constructs. All of the new constructs have a COOH-terminal Strep-tag II epitope in place of GFP (Fig. 6A) and were tested for interaction with A33-HA. As had been seen with their GFP-containing counterparts, full-length B5, a truncated form missing the cytoplasmic tail, and a truncated form missing the cytoplasmic tail in addition to the lumenal domain (B5-Strep, B5/cc/B5/Δ-Strep, and Δ/cc/B5/Δ-Strep, respectively) were coimmunoprecipitated with A33-HA (Fig. 6B). Several nonspecific bands were detected in all of the samples; notably, the two at approximately 51 and 28 kDa likely represent the heavy and light chains, respectively, from the antibody used to perform the precipitation. A new construct that contains the predicted coiled-coil domain fused to the transmembrane domain of VSVG (Δ/cc/G/Δ-Strep) was also coimmunoprecipitated with A33-HA, although at much-reduced levels compared to B5-Strep. This reduction is most likely due to poor expression of the truncated form (Fig. 6C). An additional faster-migrating band was detected in the cell extract and pulldowns from cells that expressed Δ/cc/B5/Δ-Strep. We are unsure if this band was the result of cellular processing of the truncated form or of sample handling during analysis. Regardless, as VSVG is not known to interact with A33 (Fig. 3A), these results show that the coiled-coil domain of B5 is sufficient for interaction with A33.
Fig 6.
The coiled-coil domain of B5 is sufficient for interaction with A33. (A) Diagrammatic representation of constructs used in CoIP assays. Chimeras were generated from the signal peptide (SP), the coiled-coil (cc) domain, the lumenal domain (LD), or the transmembrane domain (TMD) of B5 (▨), VSVG (□), and A33 (▨). Δ indicates deletion of the corresponding domain, which is represented by a dashed line in the diagram. B5 and A33 were tagged with Strep-tag II and HA epitopes, respectively. For B5 and A33, the residue number that starts the predicted domain is shown below the protein diagram. (B) CoIP. HeLa cells infected with vTF7.3 in the presence of AraC were transfected with the indicated plasmids that expressed the constructs described in panel A. Cells were harvested at 24 h posttransfection and lysed, and cell lysates were immunoprecipitated with an anti-HA MAb. Immune complexes were analyzed by Western blot assay using an anti-Strep MAb (C) Protein expression. Cell lysates used in panel B were subjected to SDS-PAGE and analyzed by Western blot assay using an anti-Strep MAb, followed by HRP-conjugated donkey anti-mouse (top) or anti-HA (bottom) antiserum, followed by a Cy5-conjugated anti rabbit MAb. The molecular masses, in kilodaltons, and positions of marker proteins are shown on the left of the blots.
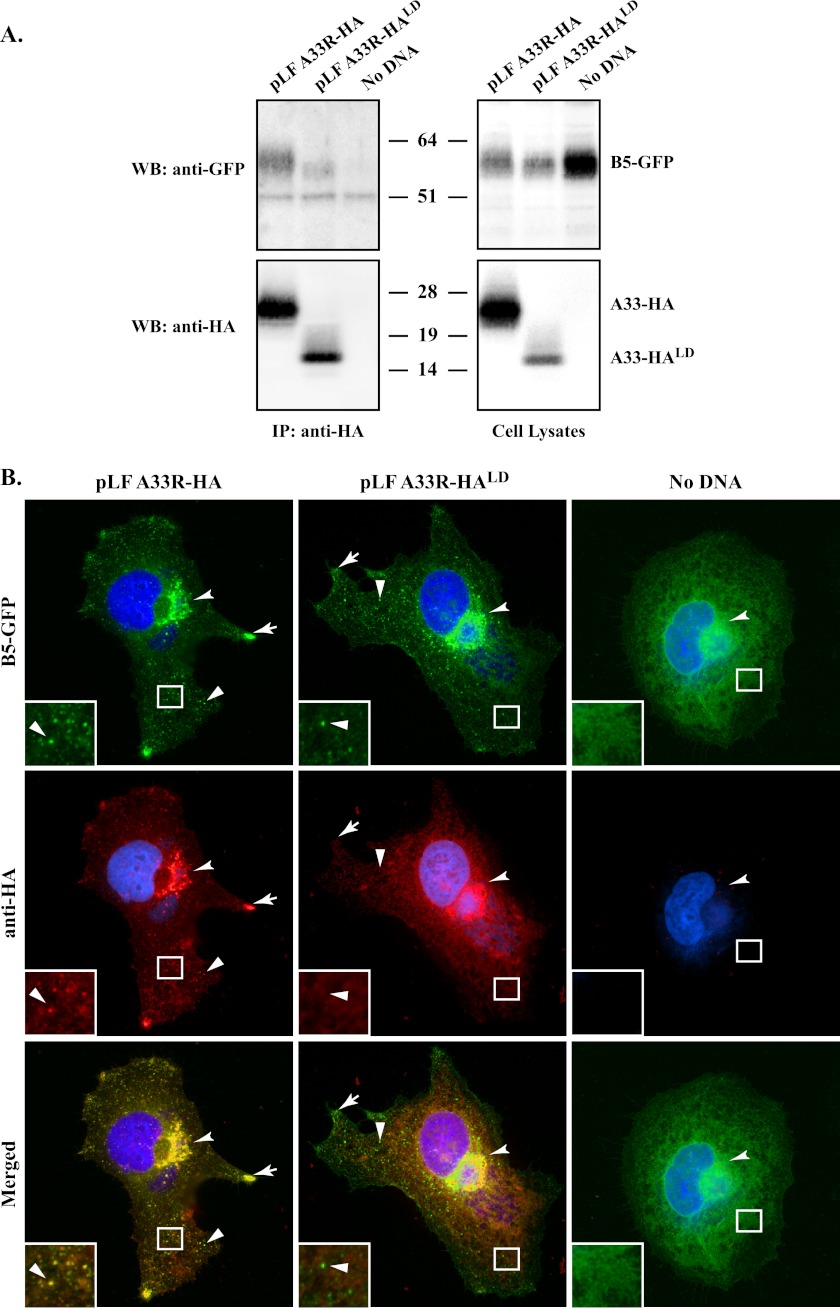
The lumenal domain of A33 is sufficient for interaction with B5-GFP and its incorporation into extracellular virions during infection.
The above CoIPs were carried out using the vaccinia virus T7 expression system in the absence of late viral protein synthesis and therefore viral morphogenesis. The use of fusion proteins, while convenient for the mapping of interaction domains, may not reflect interactions that happen during a typical infection. Therefore, we wanted to determine if an interaction between the lumenal domain of A33 and B5-GFP could be detected during infection. Cells were infected with a recombinant virus that has A33R deleted (vB5R-GFP/ΔA33R) and transfected with plasmids that contained either full-length A33R-HA or A33R-HALD (pLF A33R-HA or pLF A33R-HALD, respectively) under the control of the natural A33R promoter so that they would be expressed during infection. Once again, B5-GFP provides a convenient tool for studying the A33-dependent pathway because B5-GFP is dependent on A33 for proper localization and EV incorporation. Both A33-HA and A33-HALD coimmunoprecipitated B5-GFP (Fig. 7A), indicating that the lumenal domain of A33 interacted with B5-GFP during infection. The reduced amount of B5-GFP coimmunoprecipitated with A33-HALD compared to that of A33-HA is likely the result of the reduced expression of A33-HALD (Fig. 7A).
Fig 7.
The lumenal domain of A33 is sufficient for interaction with B5-GFP and its incorporation into IEV during infection. (A) CoIP. HeLa cells were infected with vB5R-GFP/ΔA33R at an MOI of 5.0 and transfected with the indicated plasmids. The next day, cells were harvested by scraping and lysed in RIPA buffer. Cell lysates were subjected to immunoprecipitation with an anti-HA MAb. Immune complexes (left blots) and cell lysates (right blots) were resolved by SDS-PAGE and analyzed by Western blot assays using HRP-conjugated anti-GFP and anti-HA antibodies. The molecular masses, in kilodaltons, and positions of marker proteins are shown. (B) In vivo trans-complementation analysis. HeLa cells grown on glass coverslips were infected with vB5R-GFP/ΔA33R at an MOI of 1.0 and transfected with the indicated plasmids. The next day, cells were fixed, permeabilized, and immunostained with a rabbit anti-HA antibody, followed by a Texas Red-conjugated donkey anti-rabbit antibody. Localization of B5-GFP, A33-HA, or A33-HALD at the site of wrapping (concave arrowheads), at the cell vertices (arrows), and at VSPs (arrowheads) is indicated. DNA in the nuclei and viral factories was stained with DAPI (blue). The overlap of B5-GFP (green) and A33-HA or A33-HALD (red) is yellow. Boxed regions are enlarged in the lower left corners to highlight the GFP-labeled VSPs.
In the absence of A33, B5-GFP is mistargeted during infection and EV that do not contain detectable amounts of B5-GFP are produced (5). Providing full-length A33 in trans restores the proper localization of B5-GFP to VSPs in the cytoplasm and cell vertices (5). We wanted to determine if the lumenal domain of A33 was sufficient to restore the localization of B5-GFP to VSPs. Therefore, we repeated the assay and looked at the localization of B5-GFP by using fluorescence microscopy. As shown in Fig. 7B, VSPs labeled with B5-GFP could be visualized in cells transfected with either full-length A33-HA or A33-HALD. In contrast, we were unable to detect B5-GFP-labeled VSPs in the cytoplasm of cells that were mock transfected (Fig. 7B). Interestingly, the B5-GFP-labeled VSPs produced in the presence of A33-HALD were not stained by the anti-HA antibody (Fig. 7B). Taken together, our results indicate that the lumenal domain of A33 is sufficient both for the interaction with B5-GFP and to drive its incorporation into EV.
DISCUSSION
Proper glycoprotein composition of the nascent viral envelope is important for efficient production and release of infectious wrapped virus. Of the eight virus-encoded IEV-specific proteins, only F13 has been predicted to have an enzymatic activity (20). Therefore, we theorized that the other IEV-specific proteins have structural roles during morphogenesis and thus the interactions among these proteins coordinate intracellular envelopment of IMV and ensure proper protein composition. Interactions between IEV-specific proteins have been described for the coordinated incorporation of proteins into IEV, i.e., A33 and A36 (54, 56), A33 and B5 (3, 31), A34 and B5 (10, 32), A36 and F12 (22), and E2 and F12 (19). The purposes of the present study were to examine the previously described interaction between A33 and B5 (3, 31) and determine which protein is dependent on the other for IEV incorporation. It was reported that in the absence of B5, colocalization of A33 with DNA staining particles in the cytoplasm was not observed, and it was concluded that the interaction between A33 and B5 is required for the incorporation of A33 into IEV (31). In contrast, when we looked directly at EV formed in the absence of B5, we were able to detect A33 (Fig. 1 and 2). Our results are supported by the findings of Röttger et al. (38). Using immunofluorescence microscopy, they found particles labeled with both A27 (an IMV-specific protein) and A33 in the cytoplasm of infected cells in the absence of B5 (38). In addition, they found that actin tails were formed in the absence of B5 (38), a process that requires A33 for the incorporation of A36 into IEV (56). Our results demonstrate that A33 does not require B5 for incorporation into EV but do not rule out the possibility that A33 is dependent on a different IEV-specific protein for its incorporation.
B5 has been reported to have a role in EV formation, as deletion of the gene causes a reduction in EV production (12, 21, 55). Previous studies have shown that the cytoplasmic tail (25) and a large portion of the extracellular domain of B5 (14) are not required for EV formation, indicating an important role for the transmembrane domain and the coiled-coil structure of B5. In addition, the coiled-coil structure, or stalk, has been reported to be critical for ligand-induced rupture of the outer EEV membrane (34). The fact that A33-HA interacts with Δ/cc/G/Δ-Strep, a construct that has only the coiled-coil domain of B5 and the transmembrane domain of VSVG, suggests that the coiled-coil domain of B5 is sufficient for interaction with A33 (Fig. 6). It seems likely that a transmembrane domain is required by B5 to coordinate its coiled-coil domain for interaction with A33. While the transmembrane domain may not be required for interaction with A33 and EV targeting, we cannot rule out a subsequent function in virion formation or infectivity. Similarly, while these domains are required for EV incorporation, it is still unclear exactly how they function during EV formation. Characterization of recombinant viruses expressing these truncated forms in place of the normal proteins should help in dissecting their functions.
In our companion paper (4) and elsewhere (31), A33 has been shown to interact with both B5 and B5-GFP during infection. It has also been shown that a recombinant virus expressing B5-GFP makes plaques comparable in size to the parental virus WR, which expresses a normal B5 (41). This suggests that interaction with A33 is the predominant mechanism of B5 incorporation into EV membranes. B5-GFP was shown to be targeted to the Golgi, the presumed site of intracellular envelopment, when expressed in the absence of other viral proteins (51). Therefore, it is unclear why B5 requires an interaction with both A34 (10, 32) and A33 for incorporation into IEV/EV (5). It is also unclear why only the lumenal domain of A33 is sufficient for proper targeting. One explanation is that interaction with A33 prevents aberrant interactions by B5 during its progression to the site of wrapping.
The exact role of the transmembrane domain of A33 remains to be determined. Previous work has shown that residues 5 to 40, which constitute most of the predicted cytoplasmic tail of A33, are not required for its incorporation into IEV (54). The lumenal domain of A33 is sufficient for B5 incorporation into IEV/EV, but A33-HALD was not detected on B5-GFP-labeled VSPs (Fig. 7B). This suggests that the interaction between B5 and A33 is either weak or transient. We can think of two possible explanations for the absence of A33-HALD on B5-GFP-labeled VSPs. The first is that the transmembrane domain of A33 is required for its incorporation into IEV/EVs. This implies that the A33-B5 interaction is disrupted before IEVs are formed and A33-HALD is excluded from IEV. Alternatively, the transmembrane domain of A33 is required for retention of A33 in EVs. A33-HALD should localize to the lumen of the endomembrane system and therefore the space between the two outer membranes on IEVs. When IEVs fuse with the plasma membrane, this space is exposed to the medium, and unless there is a mechanism to tether A33-HALD to the CEV, it would diffuse into the medium and be lost from the CEV surface. Recombinant viruses expressing A33-HALD should help determine which scenario is occurring.
A36, and therefore indirectly F12, requires an interaction with A33 for incorporation into IEV membranes (22, 54, 57). B5 is now the second viral protein shown to directly utilize an interaction with A33 for IEV/EV incorporation. The cytoplasmic tail of A33 has been shown to interact with A36, and the interaction is required for the incorporation of A36 into the wrapping membrane (54, 57). Our results demonstrate that the lumenal domain of A33 interacts with B5. This raises the possibility that these three proteins may exist as a complex. Indeed, mutations in A33 and B5 were found that have the same phenotype in the absence of A36, suggesting a commonality (23, 24). Subsequently, B5 was shown to be involved in an outside-in signaling cascade for actin tail formation via A36 (28). In addition, B5 has been reported to interact with F13 (31). It will be of interest to see if all of these proteins form a complex and, if so, how this complex functions in EV formation and determination of the final protein composition of IEV/EV membranes.
ACKNOWLEDGMENTS
We thank Jay Hooper for an anti-A33 MAb and Toru Takimoto for the influenza virus Nanchang NA plasmid. We also thank the Electron Microscope Research Core at the University of Rochester Medical Center.
This work was supported in part by NIH research grant AI067391. W.M.C. was supported by National Institute of Allergy and Infectious Diseases Molecular Pathogenesis of Bacteria and Viruses Training Grant T32 AI007362.
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Appleyard G, Hapel AJ, Boulter EA. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9–17 [DOI] [PubMed] [Google Scholar]
- 2. Blasco R, Moss B. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan WM, Kalkanoglu AE, Ward BM. 2010. The inability of vaccinia virus A33R protein to form intermolecular disulfide-bonded homodimers does not affect the production of infectious extracellular virus. Virology 408:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan WM, Ward BM. 2012. Increased interaction between vaccinia virus proteins A33 and B5 is detrimental to infectious extracellular enveloped virion production. J. Virol. 86:8232–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan WM, Ward BM. 2010. There is an A33-dependent mechanism for the incorporation of B5-GFP into vaccinia virus extracellular enveloped virions. Virology 402:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y, et al. 2009. Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis. Virol. J. 6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Condit RC, Moussatche N, Traktman P. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 66:31–124 [DOI] [PubMed] [Google Scholar]
- 8. Duncan SA, Smith GL. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 66:1610–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Earl PL, Moss B. 1991. Generation of recombinant vaccinia viruses, p 16.17.11–16.17.16 In Ausubel FM, et al. (ed), Current protocols in molecular biology, vol 2 Greene Publishing Associates & Wiley Interscience, New York, NY [Google Scholar]
- 10. Earley AK, Chan WM, Ward BM. 2008. The vaccinia virus B5 protein requires A34 for efficient intracellular trafficking from the endoplasmic reticulum to the site of wrapping and incorporation into progeny virions. J. Virol. 82:2161–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Engelstad M, Howard ST, Smith GL. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188:801–810 [DOI] [PubMed] [Google Scholar]
- 12. Engelstad M, Smith GL. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194:627–637 [DOI] [PubMed] [Google Scholar]
- 13. Geada MM, Galindo I, Lorenzo MM, Perdiguero B, Blasco R. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747–2760 [DOI] [PubMed] [Google Scholar]
- 14. Herrera E, del Mar Lorenzo M, Blasco R, Isaacs SN. 1998. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J. Virol. 72:294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hiller G, Weber K. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55:651–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. 1979. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology 98:142–153 [DOI] [PubMed] [Google Scholar]
- 17. Hollinshead M, et al. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hooper JW, Custer DM, Schmaljohn CS, Schmaljohn AL. 2000. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology 266:329–339 [DOI] [PubMed] [Google Scholar]
- 19. Hume AN, Tarafder AK, Ramalho JS, Sviderskaya EV, Seabra MC. 2006. A coiled-coil domain of melanophilin is essential for myosin Va recruitment and melanosome transport in melanocytes. Mol. Biol. Cell 17:4720–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Husain M, Moss B. 2002. Similarities in the induction of post-Golgi vesicles by the vaccinia virus F13L protein and phospholipase D. J. Virol. 76:7777–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Isaacs SN, Wolffe EJ, Payne LG, Moss B. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnston SC, Ward BM. 2009. Vaccinia virus protein F12 associates with intracellular enveloped virions through an interaction with A36. J. Virol. 83:1708–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katz E, Ward BM, Weisberg AS, Moss B. 2003. Mutations in the vaccinia virus A33R and B5R envelope proteins that enhance release of extracellular virions and eliminate formation of actin-containing microvilli without preventing tyrosine phosphorylation of the A36R protein. J. Virol. 77:12266–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katz E, Wolffe E, Moss B. 2002. Identification of second-site mutations that enhance release and spread of vaccinia virus. J. Virol. 76:11637–11644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lorenzo MD, Herrera E, Blasco R, Isaacs SN. 1998. Functional analysis of vaccinia virus B5R protein: role of the cytoplasmic tail. Virology 252:450–457 [DOI] [PubMed] [Google Scholar]
- 26. Moss B. 2001. Poxviridae: the viruses and their replication, p 2849–2883 In Fields BN, Knipe DM, Howley PM. (ed), Fields virology, fourth ed, vol 2 Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 27. Moss B. 2006. Poxvirus entry and membrane fusion. Virology 344:48–54 [DOI] [PubMed] [Google Scholar]
- 28. Newsome TP, Scaplehorn N, Way M. 2004. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science 306:124–129 [DOI] [PubMed] [Google Scholar]
- 29. Payne LG. 1992. Characterization of vaccinia virus glycoproteins by monoclonal antibody preparations. Virology 187:251–260 [DOI] [PubMed] [Google Scholar]
- 30. Payne LG. 1980. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 50:89–100 [DOI] [PubMed] [Google Scholar]
- 31. Perdiguero B, Blasco R. 2006. Interaction between vaccinia virus extracellular virus envelope A33 and B5 glycoproteins. J. Virol. 80:8763–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perdiguero B, Lorenzo MM, Blasco R. 2008. Vaccinia virus A34 glycoprotein determines the protein composition of the extracellular virus envelope. J. Virol. 82:2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rietdorf J, et al. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992–1000 [DOI] [PubMed] [Google Scholar]
- 34. Roberts KL, et al. 2009. Acidic residues in the membrane-proximal stalk region of vaccinia virus protein B5 are required for glycosaminoglycan-mediated disruption of the extracellular enveloped virus outer membrane. J. Gen. Virol. 90:1582–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roberts KL, Smith GL. 2008. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 16:472–479 [DOI] [PubMed] [Google Scholar]
- 36. Roper RL, Payne LG, Moss B. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roper RL, Wolffe EJ, Weisberg A, Moss B. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 72:4192–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Röttger S, Frischknecht F, Reckmann I, Smith GL, Way M. 1999. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 73:2863–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reference deleted. [Google Scholar]
- 40. Schmelz M, et al. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmidt TG, Skerra A. 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2:1528–1535 [DOI] [PubMed] [Google Scholar]
- 42. Shida H. 1986. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology 150:451–462 [DOI] [PubMed] [Google Scholar]
- 43. Smith GL, Vanderplasschen A, Law M. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915–2931 [DOI] [PubMed] [Google Scholar]
- 44. Stokes GV. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J. Virol. 18:636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163–178 [PubMed] [Google Scholar]
- 46. Turner PC, Moyer RW. 2006. The cowpox virus fusion regulator proteins SPI-3 and hemagglutinin interact in infected and uninfected cells. Virology 347:88–99 [DOI] [PubMed] [Google Scholar]
- 47. van Eijl H, Hollinshead M, Rodger G, Zhang WH, Smith GL. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83:195–207 [DOI] [PubMed] [Google Scholar]
- 48. van Eijl H, Hollinshead M, Smith GL. 2000. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology 271:26–36 [DOI] [PubMed] [Google Scholar]
- 49. Wagenaar TR, Moss B. 2007. Association of vaccinia virus fusion regulatory proteins with the multicomponent entry/fusion complex. J. Virol. 81:6286–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ward BM. 2005. Visualization and characterization of the intracellular movement of vaccinia virus intracellular mature virions. J. Virol. 79:4755–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ward BM, Moss B. 2000. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J. Virol. 74:3771–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ward BM, Moss B. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651–11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ward BM, Moss B. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ward BM, Weisberg AS, Moss B. 2003. Mapping and functional analysis of interaction sites within the cytoplasmic domains of the vaccinia virus A33R and A36R envelope proteins. J. Virol. 77:4113–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolffe EJ, Isaacs SN, Moss B. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 67:4732–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wolffe EJ, Weisberg AS, Moss B. 1998. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology 244:20–26 [DOI] [PubMed] [Google Scholar]
- 57. Wolffe EJ, Weisberg AS, Moss B. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang WH, Wilcock D, Smith GL. 2000. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J. Virol. 74:11654–11662 [DOI] [PMC free article] [PubMed] [Google Scholar]