Abstract
Smallpox vaccine is considered a gold standard of vaccines, as it is the only one that has led to the complete eradication of an infectious disease from the human population. B cell responses are critical for the protective immunity induced by the vaccine, yet their targeted epitopes recognized in humans remain poorly described. Here we describe the biochemical and structural characterization of one of the immunodominant vaccinia virus (VACV) antigens, D8, and its binding to the monoclonal antibody LA5, which is capable of neutralizing VACV in the presence of complement. The full-length D8 ectodomain was found to form a tetramer. We determined the crystal structure of the LA5 Fab-monomeric D8 complex at a resolution of 2.1 Å, as well as the unliganded structures of D8 and LA5-Fab at resolutions of 1.42 Å and 1.6 Å, respectively. D8 features a carbonic anhydrase (CAH) fold that has evolved to bind to the glycosaminoglycan (GAG) chondroitin sulfate (CS) on host cells. The central positively charged crevice of D8 was predicted to be the CS binding site by automated docking experiments. Furthermore, sequence alignment of various poxvirus D8 orthologs revealed that this crevice is structurally conserved. The D8 epitope is formed by 23 discontinuous residues that are spread across 80% of the D8 protein sequence. Interestingly, LA5 binds with a high-affinity lock-and-key mechanism above this crevice with an unusually large antibody-antigen interface, burying 2,434 Å2 of protein surface.
INTRODUCTION
Smallpox, which is caused by infection with the orthopoxvirus variola virus, was one of mankind's greatest plagues, and early vaccine development led to its complete eradication. Although variola virus is no longer a natural threat to human health, there is fear of its potential use as a biological weapon (2). In addition, the natural zoonotic ability of the related monkeypox virus to infect humans has led to concern that it could evolve into a global pathogen. Sporadic human outbreaks have been reported since 1970 (18, 45), and infected rodents exported to the United States caused a highly publicized human outbreak in 2003 (23, 28, 44).
For a better understanding of poxvirus immunity, we chose vaccinia virus (VACV) as a model, as it is the active component of the vaccine that led to the eradication of smallpox (1, 37). Vaccine-mediated protection against smallpox is mediated largely through the production of highly potent neutralizing antibodies (20). VACV contains approximately 25 integral or peripheral membrane proteins (12, 36), of which 17 have been implicated as functioning in virus entry and/or fusion (A27, A17, H3, D8, L1, A28, H2, A21, L5, G3, G9, A16, J5, F9, I2, A26, and O3) (36, 37). The entry-fusion complex (EFC), an essential component of VACV-induced cell-to-cell fusion and viral core penetration, is composed of the eight core proteins A16 (42), A21 (55), A28 (49), G3 (26), G9 (26), H2 (48), L5 (54), and O3 (47). Envelope proteins A27 (13, 25) and H3 (31) are involved in cell adhesion of the intracellular mature virion (MV) to the host cell glycosaminoglycan (GAG) heparan sulfate (HS). Similarly, VACV envelope protein D8, a 32-kDa type 1 membrane protein, binds to cell surface chondroitin sulfate (CS) but not to HS. While VACV infectivity in BALB/c mice is reduced in D8 deletion strains (46), D8-negative virus replicates efficiently in cultured cells (40). This suggests that VACV utilizes several alternate routes of host cell adhesion and infection, thus reducing the chances of antibody-mediated neutralization by targeting a single VACV envelope protein. Vaccination of humans with VACV can elicit antibodies against at least four major MV surface antigens (Ags) (A27, L1, D8, and H3) (6, 17). To date, of the immunodominant VACV envelope proteins (A27, A33, L1, D8, B5, and H3), only the structures of L1 and A33 have been determined (50, 52), and only one cocrystal structure exists with a directly neutralizing antibody (L1 plus monoclonal antibody [MAb] 7D11) (51). Therefore, information on the structural basis of neutralizing antibodies against the major VACV envelope proteins is very limited.
Sequence alignments revealed a significant homology of D8 to human carbonic anhydrases (CAHs) (36% identity over 85% of the D8 ectodomain sequence [40]) that convert CO2 into bicarbonate and back and thus support carbon dioxide removal within the lungs. The regulatory domain of neural tissue-specific phosphotyrosine-phosphatase receptors (PTPRs) (38) also adopts the CAH fold, suggesting that the CAH fold has evolved to carry out multiple functions. The CAH catalytic site coordinates a zinc cation that is required for enzymatic activity. The zinc binding residues are not conserved in D8 (41), and D8 is not catalytically active for CO2 conversion (27).
This study focuses on the biochemical and structural characterization of the D8 ectodomain and its binding to the mouse IgG2a antibody LA5. D8 is an immunodominant antigen in the smallpox vaccine, and recombinant D8 protein was used successfully, along with A27 and B5, in a multiprotein vaccination (7). Currently, 12 T cell epitopes for D8 have been deposited in the immune epitope database (www.immuneepitope.org), while no information about a D8 B cell epitope is available. We show that the LA5 monoclonal antibody (MAb) is neutralizing in the presence of complement. The structural characterization of the D8/LA5 interface allowed us to define the first discontinuous D8 B cell epitope and its targeting by the neutralizing antibody LA5. We hypothesize that the LA5-dependent complement recruitment against D8 competes with binding to CS and therefore blocks viral attachment to the D8-specific host-cell GAG CS.
MATERIALS AND METHODS
D8 expression and purification.
We expressed two forms of recombinant D8 ectodomains: D8 Δ265 (D8 residues 1 to 264), which includes the D8 unique cysteine at position 262, and D8 Δ262 (residues 1 to 261), which lacks this unpaired cysteine. D8 Δ265 was used for in-solution characterization and surface plasmon resonance (SPR) affinity measurements, while D8 Δ262 was engineered to obtain a homogeneous sample for crystallization. The D8 Δ262 DNA fragment was obtained from VACV (strain Acam2000) genomic DNA by amplifying the gene with 5′ and 3′ primers 5′-ATA TAT CAT ATG CGC CTA AAT CAA TGC ATG TCT AAC-3′ and 5′-ATA TAT GGA TCC TTA GTT CAT TGT TTT AAC ACA AAA ATA C-3′, subsequently restricted with NdeI and BamHI, and ligated into pET22b for cytosolic expression. For D8 Δ265, the 5′ and 3′ primers 5′-TTA CCA TGG CGC AAC AAC TAT CTC C-3′ and 5′-CTC TCG AGT TAT GAA AAA CAT GTC TCT CTC A-3′ were used for PCR amplification, and the corresponding DNA fragment was digested with NcoI and XhoI and cloned into pProEX-HTA (Invitrogen). The resulting expression plasmids were transformed into CodonPlus BL21 cells (Agilent). After culture growth to an optical density (OD) of 0.6, D8 expression was induced by induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C for 4 h. Cells from typically 1 liter of culture were spun down (10 min at 4,000 × g) and resuspended in 50 ml lysis buffer (50 mM Tris [pH 8.0], 5 mM EDTA). Cells were sheared with a Microfluidizer (3 rounds at 1400 bars; Microfluidics), and crude lysate was clarified by centrifugation (1 h at 50,000 × g). Expression resulted in soluble recombinant ectodomains with hexahistidine sequences at the N terminus for D8 Δ265 and at the C terminus for D8 Δ262, which were further purified by immobilized metal affinity chromatography (IMAC) (GE Healthcare). Supernatant was passed through an IMAC column, washed with three column volumes (one column volume = 5 ml) of low-imidazole buffer (LIB) (50 mM Tris [pH 8.0], 300 mM NaCl, 20 mM imidazole). After another wash at 50 mM imidazole, D8 was eluted with the same buffer containing ∼250 mM imidazole and subsequently dialyzed twice in 10 mM Tris (pH 8.0)–200 mM NaCl prior to size exclusion chromatography (SEC) for complex preparation and/or crystallization.
To analyze D8 oligomerization states, we compared the SEC profiles of D8 Δ265 in nonreducing (100 mM Tris [pH 8.0], 100 mM NaCl) and reducing environments; D8 Δ265 (1 mg/ml) was incubated on ice for 10 min in the presence of 10 mM dithiothreitol (DTT) before being subjected to SEC on Superdex S200 (GL10/300; GE Healthcare) under reducing conditions (100 mM Tris [pH 8.5], 100 mM NaCl, 10 mM DTT). For comparison, purified D8 Δ265 was also subjected to SEC under nonreducing conditions, in the absence of DTT. The disulfide-linked oligomeric nature of D8 was analyzed using nonreducing SDS-PAGE as well as native PAGE.
Antibody sequencing.
Total RNA from 5 × 106 hybridoma cells was isolated using the RNeasy Minikit according to the manufacturer's instructions (Qiagen). First-strand cDNA was amplified using the OneStep reverse transcription-PCR kit (Qiagen). The cycling profile used for the first-strand PCR was as follows: 1 cycle of 30 min at 50°C and 15 min at 95°C; 40 cycles of 30 s at 94°C, 30 s at 60°C, and 55 s at 72°C; and 1 cycle of 10 min at 72°C and a 4°C cool down. Second-strand PCR was performed using primers for heavy-chain IgG2a (5′MsVHE and 3′Cy2c outer) and light-chain kappa (5′mVkappa and 3′mCk) (53). PCR products were verified by gel electrophoresis, with ∼500-bp products for heavy chains and ∼450-bp products for light chains. PCR fragments were purified using the QIAquick PCR purification kit (Qiagen) and sequenced (5′ primer extension, using L and H chain-specific primers). Sequences include only V-D-J regions for heavy chains and V-J regions for light chains. Antibody germ line origin was determined using IMGT's V-Quest service (11). The LA5-Fab was sequenced as IGKV4-55*01 and IGHV1S127*01.
Viruses.
VACV Western Reserve strain (VACVWR) stocks were grown on HeLa cells in D-10 medium as described previously (6). The UV-inactivated vaccinia virus (UV-VACV) was prepared as described previously (16)).
Antibodies.
Mouse anti-D8 (clone LA5) and anti-H3 (clone 41) monoclonal antibodies (MAbs) were derived from a mouse infected with VACVWR or immunized with recombinant H3 protein as described previously (34, 35). For antibody purification, hybridomas were cultured and MAbs were purified from conditioned culture medium using a recombinant protein A-Sepharose Fast Flow gel (GE Healthcare) as described previously (4, 5, 34, 35). The purified antibodies were quantified by the Lowry method using bovine IgG (Pierce) as a standard and/or by OD at 280 nm (OD280) using the NanoDrop method and calculated using the following formula: concentration (mg/ml) = 1.35 × OD280. The MAbs were stored in aliquots at −80°C and diluted into phosphate-buffered saline (PBS) immediately prior to use as needed.
ELISA.
Flat-bottom 96-well microtiter plates (Nunc MaxiSorp) were coated overnight at 4°C with 100 μl of 1 μg/ml of recombinant D8 protein (D8) in PBS. Plates were washed with PBS plus 0.2% Tween 20 and blocked with PBS plus 0.5% bovine serum albumin and 0.2% Tween 20. Purified anti-D8 (clone LA5) and anti-H3 (clone 41) MAbs were screened using 3-fold-serial-dilution enzyme-linked immunosorbent assay (ELISA). The secondary antibody was streptavidin-horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG2a (Jackson ImmunoResearch Laboratories, West Grove, PA). The half-saturation binding concentration of the antibody corresponding to 50% maximum optical density (EC50) for anti-D8 monoclonal antibody was determined by sigmoidal dose-response nonlinear regression with variable slope (Prism 5.0). For whole vaccinia virus ELISA, plates coated overnight at 4°C with 100 μl/well of UV-inactivated MV VACVWR (5 × 107 PFU/ml and 51 μg/ml, prior to 1:25 dilution in PBS), as described previously (15, 57). Purified anti-D8 (LA5) and anti-H3 (clone 41) MAbs were used at 10 μg/ml. For linear peptide ELISA, biotinylated 20-mer peptides overlapping by 10 residues and covering the D8 protein sequence were synthesized by A&A Labs (San Diego, CA). Microtiter plates were coated with 100 μl of NeutrAvidin biotin binding protein (1 μg/ml) diluted in PBS overnight at 4°C (Thermo Scientific Pierce, Rockford, IL). The coated plates were then washed with washing buffer (PBS [pH 7.2] plus 0.5% Tween 20) and blocked with blocking buffer (PBS [pH 7.2] plus 0.1% bovine serum albumin plus 0.2% Tween 20) for 2 h at room temperature (RT). Plates were incubated with 100 μl of biotinylated peptides (200 ng/ml) in blocking buffer for 90 min at RT. Plates were washed and incubated with purified MAb (LA5) at 10 μg/ml for 90 min at RT. Plates were washed, and the bound antibody was detected by adding a streptavidin-horseradish peroxidase (HRP)-conjugated secondary antibody to mouse IgG (Invitrogen, CA) and incubation for 60 min at RT, followed by addition of o-phenylenediamine (OPD) substrate (Sigma-Aldrich).
VACV MV neutralization.
Vero E6 cells were seeded at 1.5 × 105 cells/well into 24-well Costar plates (Corning Inc., Corning, NY) and used on the following day (75 to 90% confluence). Two different VACV MV neutralization assays were used: (i) 1 h with no complement and (ii) 1 h with 1% baby rabbit complement (Cedarlane Laboratories, Burlington, Canada). Neutralization experiments were performed as described previously (34). The percentage of MV VACV neutralization was calculated and defined according to the formula [(PNV − PNS)/PNV] × 100, where PNV was the average number of plaques formed by MV VACVWR alone or MV VACVWR plus complement and PNS was the average number of plaques in the presence of each experimental sample.
SPR.
Surface plasmon resonance (SPR) studies were performed on a Biacore 3000 instrument (GE Healthcare) to determine IgG2a LA5/VAVC D8 equilibrium binding affinity. Rabbit anti-mouse IgGs (mouse antibody capture kit; GE Healthcare) were amine coupled to a CM5 chip using the amine coupling kit according to the manufacturer's suggestions (GE Healthcare). Mouse MAb LA5 was diluted in running buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20), and ∼300 response units (RU) was immobilized on the sensor chip. A mouse IgG1 MAb specific for a CD1d/glycolipid complex was used as a negative control for unspecific binding. Monomeric D8 Δ265 was isolated by SEC in SPR running buffer and passed over the LA5 immobilized sensor chip at 25°C with a flow rate of 30 μl/min using increasing concentrations of D8 (0.2 to 25.6 nM). Kinetic parameters were calculated using a simple Langmuir 1:1 model with the BIA evaluation software version 4.1.
Fab digestion and purification.
Purified LA5 MAb (mouse IgG2a) at 1 mg/ml was incubated with 1% (wt/wt) activated papain for 3 h at 37°C in digestion buffer (50 mM Tris, pH 7.0). Papain was activated by incubating 20.80 μl papain solution (Sigma) with 100 μl 10× papain buffer (1 M sodium acetate [NaOAc] [pH 5.5], 12 mM EDTA) and 100 μl cysteine (12.2 mg/ml) for 15 min at 37°C. The papain digestion was stopped by adding 20 mM iodoacetamide (IAA). Two volumes of 3 M NaCl–1.5 M glycine (pH 8.9) was added to the digestion mix for subsequent protein A purification to remove undigested IgG and Fc. The protein A flowthrough containing Fab was dialyzed against 50 mM NaOAc (pH 5.5) overnight and purified to homogeneity by cation-exchange chromatography using MonoS (GE Healthcare).
Fab-Ag complex preparation.
D8 and Fab were mixed together as homogeneous, monomeric species at an equimolar ratio and at a low concentration (<1 mg/ml). The D8-Fab complex was concentrated to a volume of 250 μl using centrifugal filtration devices (Amicon Ultra 15; 10-kDa-molecular-mass cutoff) and subjected to SEC purification (Superdex 200 GL10/300) to separate the D8-Fab complex from unbound D8 and/or Fab. Before crystallization, the nature of the complex was verified by nonreducing SDS-PAGE. Fractions corresponding to the complex were pooled and concentrated to ∼10 mg/ml using centrifugal filtration devices.
Crystallization and structure determination.
Initial crystallization experiments were carried out by sitting-drop vapor diffusion in a 96-well format, using the Phoenix liquid-handling robot (Art Robbins Instruments) and a panel of commercial sparse-matrix screens (PEG/Ion 1 and 2 from Hampton Research, Wizard 1 and 3 from Emerald Biosciences, JCSG+ Suite from Qiagen, and JBScreen 6 from Jena BioScience). Quality diffracting crystals of LA5-Fab were manually grown at room temperature (RT) by mixing 0.5 μl of protein solution at 5.0 mg/ml with 0.5 μl of precipitant (10% [wt/vol] polyethylene glycol [PEG] 3000, 100 mM phosphate-citrate [pH 4.2], and 200 mM NaCl). D8 and D8/LA5-Fab crystals were grown under similar conditions (D8, 0.2 M sodium iodide and 20% PEG 3350; D8/LA5-Fab, 0.2 M potassium thiocyanate and 20% PEG 3350). Protein concentrations were 9.1 and 5.0 mg/ml, respectively. Crystals were flash-cooled at 100 K in mother liquor containing 20% glycerol. Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamlines 7.1 and 11.1 and processed with the iMosflm software (3). Crystal structures were determined by molecular replacement (MR) with MOLREP as part of the CCP4 suite (14, 58), using a previously determined structure of the Fab of the anti-B5R (VACV) MAb B96 (unpublished) in combination with the homology model of D8 (based on Protein Data Bank [PDB] ID 3JXF) obtained with SWISS-MODEL (8) as search models.
Structures were refined by performing multiple runs of model building in Coot (21) followed by maximum-likelihood restrained refinement with Refmac (39). At later refinement stages, 10 cycles of translation/libration/screw-axis displacement (TLS) refinement were performed using the following five TLS domains assigned by the TLS motion determination server (TLSMD) (43): variable heavy (1 to 115), constant heavy (116 to 225), variable light (1 to 109), constant light 1 (110 to 186, and constant light 2 (187 to 225). One TLS group was assigned for the entire D8 molecule.
The D8/LA5-Fab complex structure was first determined, and the resulting coordinates for the Fab and D8 were subsequently used for structure determination of the separate D8 and LA5 Fab structures by MR. The quality of the models was examined with the program Molprobity (33). The D8/LA5-Fab complex, LA5 Fab, and D8 structures were refined to resolutions of 2.1 Å (Rcryst = 20.1%, Rfree = 25.2%), 1.6 Å (Rcryst = 19.8%, Rfree = 22.9%), and 1.42 Å (Rcryst = 17.4%, Rfree = 19.0%), respectively.
Data collection and refinement statistics for each of the three separate crystal structures are presented in Table S1 in the supplemental material. Figures were prepared using PyMOL (Schroedinger), and electrostatic surfaces were obtained by converting the pdb file to a pqr file using the PARSE force field in the pdbtopqr server (19) (http://kryptonite.nbcr.net/pdb2pqr/), which creates an adaptive Poisson-Boltzmann solver (apbs) input file. The apbs plug-in was used to generate the electrostatic surfaces in PyMOL.
D8/CS-4 docking.
Docking experiments were performed with Autodock Vina using a D8 pdbqt formatted structure file along with chondroitin 4-sulfate (C4S) di- and tetrasaccharides as potential ligands, extracted from the CS/cathepsin K complex structure (PDB code 3C9E) (30). A first docking experiment over the full D8 surface with the C4S disaccharide determined the positively charged crevice as the best-scored binding site. A second docking experiment using the tetrasaccharide included the area around the crevice as for the docking grid.
Accession numbers.
The coordinates and structure factors of D8, LA5-Fab, and the D8/LA5-Fab complex have been deposited in the Protein Data Bank (www.rcsb.org) with accession numbers 4E9O, 4EBQ, and 4ETQ, respectively. The nucleotide sequences of the LA5 antibody have been deposited in GenBank with accession numbers JQ815182 and JQ815183.
RESULTS
Biochemical characterization of D8.
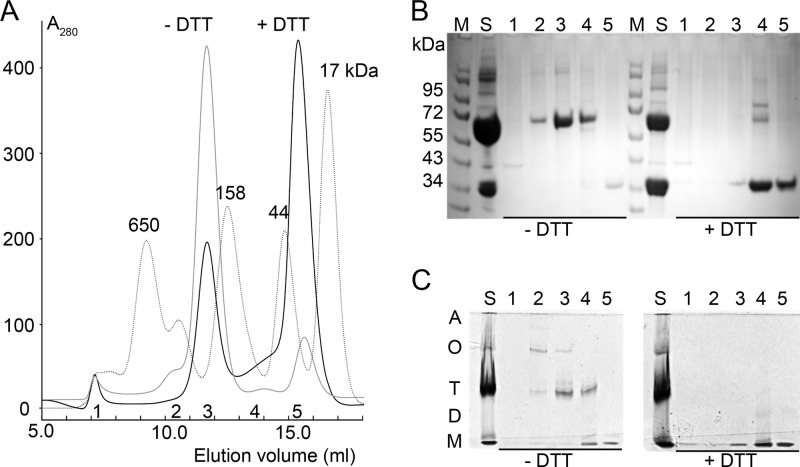
We prepared the D8 Δ265 ectodomain consisting of residues 1 to 264. During expression of D8 Δ265, we noticed a propensity for higher-molecular-weight (MW) oligomers detected by size exclusion chromatography (SEC) (Fig. 1A), indicating that D8 exists mostly as a tetramer. SEC carried out under nonreducing and reducing conditions indicated that the D8 tetramer forms by the noncovalent association of two disulfide-linked dimers. A minority of D8 also exists as octamers and dimers. An equilibrium exists between the tetramer and the monomer species, while the disulfide-linked dimer is hardly seen by SEC, supporting the notion of a rapid transition into the tetrameric state upon formation of the disulfide-linked dimer. The equilibrium can be shifted toward the D8 monomer species under reducing conditions, while the tetrameric form dominates under nonreducing conditions (Fig. 1A). Peak fractions for the tetramer and monomer were analyzed by nonreducing SDS-PAGE, as well as by nondenaturing gel electrophoresis (Fig. 1B and C). The SDS-PAGE illustrates the covalent nature of the dimer, since the protein migrates at approximately 70 kDa under nonreducing conditions and at 35 kDa when reduced. The intermolecular disulfide bond is likely formed through the only cysteine, C262, located near the C-terminal end of D8 Δ265, in the most membrane-proximal region of the ectodomain. As D8 Δ265 lacks only the C-terminal 10 residues of the full-length ectodomain (residues 265 to 274), the observed homotypic interaction of recombinant D8 Δ265 will likely also occur in the full-length D8 on the virus. We speculate that the oligomerization of D8 increases the viral binding avidity to CS, which would favor viral adhesion to cells expressing low levels of CS on the surface.
Fig 1.
Characterization of D8 Δ265. (A) Size exclusion chromatography (SEC) of D8 performed under nonreducing (−DTT) and reducing (+DTT) conditions. Molecular mass markers with sizes in kDa are shown as a reference (dotted lines). (B) Corresponding 8 to 16% SDS-PAGE to show migration of nonreduced and reduced D8 SEC samples (S) and peak fractions of the SEC (1, 2, 3, 4, and 5). (C) The corresponding nondenaturing PAGE (7.5%) confirms the nature of oligomeric states (A, aggregates; O, oligomers; T, tetramers; D, dimers; M, monomers).
VACV D8 crystal structure.
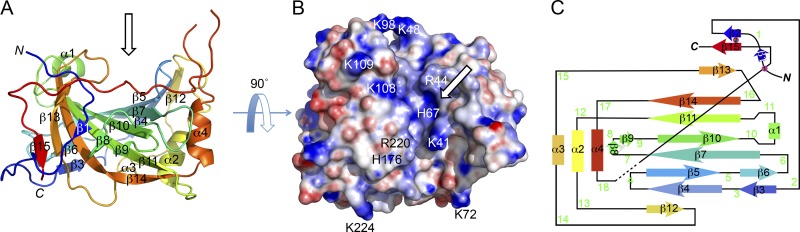
To increase the likelihood of crystallization, we prepared the truncated construct D8 Δ262 (residues 1 to 261), in which the C262 necessary for disulfide-mediated dimerization was eliminated. This construct exists predominantly as a D8 monomer (see Fig. S1 in the supplemental material). We then determined the structure of the monomeric VACV D8 ectodomain to a 1.42-Å resolution by X-ray crystallography (see Table S1 in the supplemental material). At the center of D8 is a twisted 10-stranded central β-sheet surrounded by four α-helices (Fig. 2A and C). The β-sheet is rather hydrophobic, and three-fourths of its surface is shielded from the solvent by the surrounding loops and helices. The β-sheet platform runs equatorially through D8, separating the bottom half from the top half. The top half is characterized by a highly positively charged crevice in the center of the protein, lined by loops 4/β5 (Lys41 and Arg44), 7 (His80), 16 (His176), and 18 (Arg220 and Lys224) on one side and loops 5 (Lys48) and 10/α1 (Lys98) at the other end (Fig. 2B and C). We have identified this positively charged crevice, by automated molecular docking routines (Autodock Vina [56]), as the potential binding site for CS, and it complements both the charge and shape of the elongated tetrasaccharide used for docking (see Fig. S2 in the supplemental material). The last 30 residues of the ectodomain are disordered in the structure and would be expected to protrude out of the bottom of full-length D8 and then connect to the viral membrane.
Fig 2.
D8 crystal structure and two-dimensional (2D) topology. (A) Cartoon representation of D8 in rainbow colors, from the N-terminal (blue) to the C-terminal (red) extremity. The core domain of D8 is a 10-stranded twisted β-sheet, surrounded by four α-helices and various loops. (B) Electrostatic surface representation of D8. The view is as in panel A but 90° rotated toward the reader to highlight the deep and positively charged cavity (indicated by arrows). (C) 2D topology of D8 using the same color code as in panel A. Loops are numbered in green.
LA5 is a complement-dependent neutralizing antibody that binds a conformational epitope.
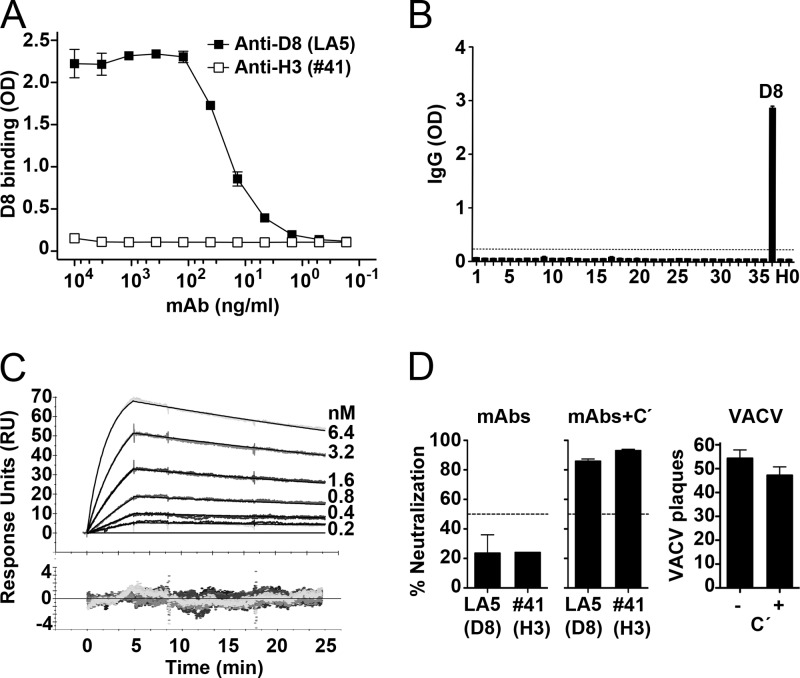
Recently, a number of MAbs targeting the D8 protein have been described (35). Importantly, these MAbs are derived from VACV-infected mice and represent the physiologic B cell response to the smallpox vaccine. One of these MAbs, LA5, is described here in more detail. LA5 is an IgG2a isotype (35) and binds recombinant D8 (Fig. 3A). LA5 failed to bind any of the 20-mer peptides spanning the entire D8 amino acid sequence (Fig. 3B), suggesting that MAb LA5 targets a discontinuous conformational epitope on D8. We used surface plasmon resonance (SPR) to determine the real-time binding kinetics of LA5 to recombinant D8. Recombinant, monomeric D8 binds to the immobilized LA5 MAb with an equilibrium dissociation constant (KD) of 0.18 nM, indicative of high-affinity binding (Fig. 3C). The binding is characterized by a fast association phase (kon = 10.8 × 105 M−1 s−1) and a very slow dissociation (koff = 1.92 × 10−4 s−1), resulting in a very stable noncovalent LA5-D8 complex.
Fig 3.
LA5 IgG specificity and neutralization activity. (A) MAb LA5 binds recombinant D8 by ELISA. An anti-H3 MAb (clone 41) was used as a negative control. (B) Peptide ELISA using overlapping 20-mer D8 peptides (peptides 1 to 35). Anti-D8 MAb LA5 showed reactivity to D8 but not to any of the overlapping D8 peptides, recombinant H3 (H), or a buffer-only control (0). (C) Surface plasmon resonance sensogram showing high-affinity binding (KD = 0.18 nM) of D8 to immobilized LA5 IgG. (D) In vitro VACV neutralization assays were done using anti-D8 MAb LA5 and anti-H3 MAb 41. Neutralization assays were done in the absence (left) or presence (middle) of complement (C′). VACV extracellular virions (EV) alone and plus complement were negative controls (right). Dashed lines indicate 50% neutralization. Data are representative of three or more independent experiments. Error bars indicate the standard error of the mean (SEM) in each group.
To determine if the binding of LA5 MAb to D8 is functional, we performed neutralization assays where LA5 was preincubated with VACVWR MV in the presence or absence of complement. Clone 41, an H3-specific antibody previously shown to neutralize VACV infection, was used as a positive control (16). As shown in Fig. 3D, neither LA5 nor clone 41 alone is able to prevent VACV infection after a 1-h preincubation of MAb plus VACV. However, upon addition of complement, 80 to 90% neutralization is observed for both MAbs. One percent complement alone has no effect on VACV (Fig. 3D). Our data suggest that complement is needed to increase the footprint of the anti-D8 Ig via C1q and C3, due to the presence of alternative VACV adhesion proteins. We conclude that LA5 binds native D8 protein on VACV virions and is able to neutralize VACV infection in the presence of complement.
Structure of the D8 ectodomain in complex with the Fab LA5.
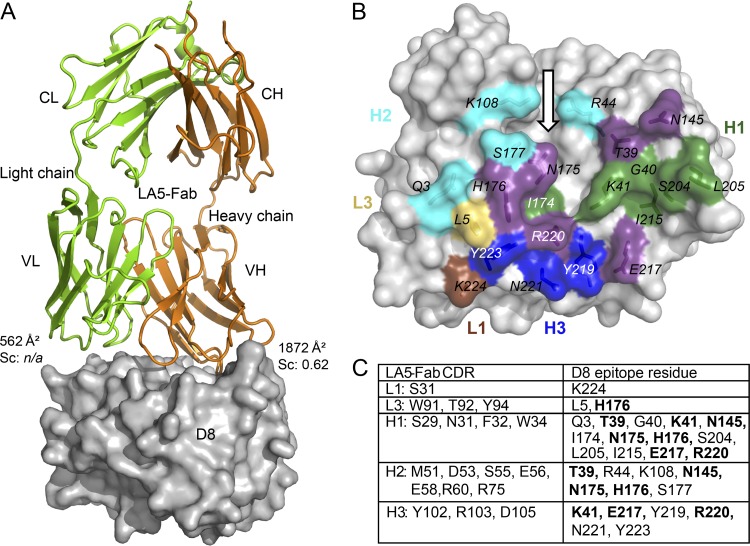
To understand the structural basis of the specificity of the MAb LA5 in recognizing D8, we determined the crystal structure of the complex between LA5-Fab and the D8 ectodomain to a resolution of 2.1 Å (Fig. 4; see Table S1 and Fig. S1 in the supplemental material). Two highly similar copies of the Fab/D8 complex (0.37-Å root mean square deviation [RMSD]) are present in the asymmetric unit (ASU) of the crystal, and therefore the structure is reported for only one of the two complexes. The LA5-Fab binds to a discontinuous, conformational epitope located around the positively charged crevice of D8 (Fig. 2B and 4B). A detailed list of all atom contacts between Fab and D8 is provided in Table S2 in the supplemental material.
Fig 4.
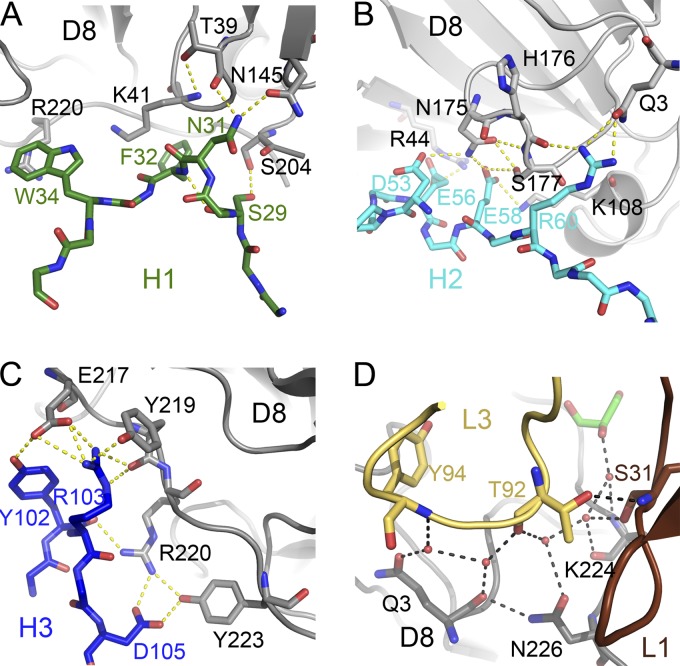
Structure of the D8/LA5-Fab complex. (A) Overall structure of the complex. The Fab is shown as a cartoon representation, with light chains (VL and CL) in green and heavy chains (VH and CH) in orange, while the molecular surface of D8 is shown in gray. The major contribution to the Ab-Ag interface comes from the heavy chain. The light chain buries only 562 Å2 of surface, while the heavy chain dominates binding, with 1,872 Å2. Shape complementarity (Sc) measures the geometric surface complementarity of protein-protein interfaces (29). As Sc becomes meaningless for surface areas that are too small, it has not been calculated for the light chain (n/a). (B) LA5-Fab footprint on D8 surface, colored by individual CDR loops. The CDRs were removed, but the D8 surface residues that are in direct contacts with individual CDRs of LA5 are colored as follows: L1, brown; L3, yellow; H1, green; H2, cyan; H3, blue; overlapping contacts, purple. The arrow indicates the location of the positively charged crevice mentioned in Fig. 2. (C) Summary of the residues involved in direct contacts. Bold residues may elicit contacts with multiple CDRs and correspond to the purple ones in panel B.
The binding interface between D8 and LA5 is very large, with a total of 2,434 Å2 of buried surface area (BSA). The heavy chain dominates the binding footprint, with a total of 1,872 Å2 of BSA, in contrast to the 562-Å2 BSA covered by the L chain (Fig. 4A). The LA5 D8 epitope is formed by 23 residues spanning a primary sequence distance of over 220 residues (Gln3, Leu5, Thr39, Gly40, Lys41, Arg44, Lys108, Asn145, Ile174, Asn175, His176, Ser177, Ser204, Leu205, Ile215, Glu217, Tyr219, Arg220, Asn221, Tyr223, and Lys224, plus N218 and N226 if including water-mediated contacts), with seven residues sharing contacts with at least two complementarity-determining region (CDR) loops of LA5-MAb (Thr39, Lys41, Asn145, Asn175, His176, Glu217, and Arg220) (Fig. 4B and C). Eighteen residues of LA5 contact the antigen D8, from 5 of the 6 CDRs of LA5 (L1, Ser31; L3, Trp91, Thr92, and Tyr94; H1, Ser29, Asn31, Phe32, and Trp34; H2, Met51, Asp53, Ser55, Glu56, Glu58, Arg60, and Arg75; H3, Tyr102, Arg103, and Asp105) (Fig. 4C). While the L chain forms only 1 polar interaction (H bond) and 15 van der Waals (VdW) contacts, the H chain binds through an intricate network of polar interactions (16 hydrogen bonds and 11 salt bridges), with an additional 115 VdW interactions contributing to the vast majority of the Fab-Ag interactions (Fig. 5; see Table S3 in the supplemental material). CDR H2 and H3 seem to contribute equally in terms of the number of interactions with D8, while eliciting some degree of difference in the nature of their interactions: CDR H3 electrostatic interactions with D8 are more frequent (7 versus 4) and are very localized and backed by the hydrophobicity of the surrounding interface, while the H2/D8 interface is spread out and less hydrophobic (Fig. 5B and C). CDR H1 contributes the least to the binding interface, with only three H bonds and 43 VdW interactions. Strikingly, a triad of negatively charged residues of CDR H2 (Asp53, Glu56, and Glu58) binds inside the positively charge crevice of D8. H2 Asp53 and Glu58 bind to Asn175 of D8, H2 E56 binds to Arg44, and H2 Glu58 binds to Lys108, indicating extensive electrostatic interaction between LA5H2 and D8 (Fig. 5B). Similarly, H3 residues Tyr102, Arg103, and Asp105 from several salt bridges and H bonds with D8 residues Glu217, Tyr219, Arg220, and Tyr223 (Fig. 5C). In conclusion, H2 and H3 together dominate and dictate the overall binding of LA5 to D8 and are responsible for the specificity of the antibody, as well as the overall stability of the complex. The light chain forms only one direct hydrogen bond with D8 (L3 Tyr94 with His176 of D8) (Fig. 5D). However, the most interesting feature about the light chain is the recruitment of highly ordered water molecules forming what we refer to as a water zipper (Fig. 5D; see Table S4 in the supplemental material). Four water molecules bridge D8 Gln3, Lys224, and Asn226 with L1 Ser31 and L3 Thr92 and Tyr94. This water zipper is also largely conserved in the other complex of the unit cell.
Fig 5.
Details of D8/LA5 interactions. (A to C) Interactions between heavy-chain CDRs H1 (A), H2, (B), and H3 (C) and D8 residues (gray). (D) The light chain water zipper. Five water molecules (red spheres) mediate contacts between LA5 light chain and D8. One water molecule is in contact with a glycerol molecule and D8 K224.
Predicted CS and LA5 binding sites on D8 overlap.
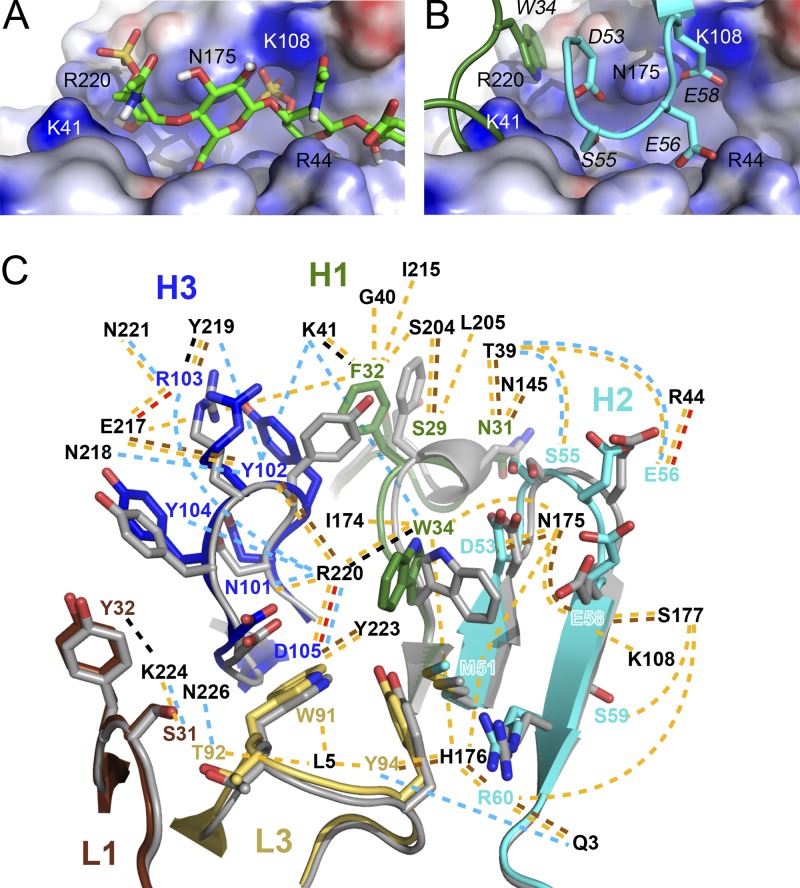
Most noticeable, the antibody binds with CDR H1 and H2 above the central crevice of D8, which was also predicted to bind CS (Fig. 6A and B). In the CS docking experiment, one of the two 4′ sulfates of the CS disaccharide binds in an electropositively charged pocket, lined by D8 residue Lys108, while the other 4′ sulfate sits above D8 residue Arg220 (Fig. 6B). While Arg220 is in contact with both CDR H1 and H2 of LA5, D8 residue Lys108 is directly engaged by H2 Glu58 (Fig. 6B). The LA5 footprint on D8 reveals that the stretch along the positive crevice of D8, between Arg220 and Lys108, is a major binding site for LA5, as both CDR H2 and H3 together bind to several D8 residues in that area (Asn175, His176, and Arg220) (Fig. 4). Therefore, the major energetic footprint of LA5 on D8 overlaps directly with the predicted binding site of CS and would likely outcompete CS binding to D8 through its high, subnanomolar binding affinity. As a result, LA5 likely prevents D8 binding to CS on host cells, thus leading to a reduced infectivity through the D8/CS axis. Loss of D8 activity, however, can be compensated for by VACV binding to HS through the adhesion molecules A27 and H3, as indicated by the failure of the antibody to neutralize in the absence of complement (Fig. 3D) and the normal infectivity of D8-deleted VACV in vitro.
Fig 6.
LA5 blocks D8 binding to CS and conformational changes upon antibody binding. (A) Details of chondroitin-4-sulfate binding to D8 as determined by Autodock Vina. (B) LA5 binds with H1 and H2 to the same area of D8. (C) Comparison of LA5 in the free form (gray) and when bound to LA5 (rainbow color). CDR coloring is as in Fig. 4. Dashed lines indicate VdW (yellow), salt bridge (red), H bond (brown), cation-pi (black), and water-mediated (light blue) interactions occurring upon binding of Ag to MAb, and contacting D8 residues are labeled.
Structural changes upon antibody binding.
The crystal structures of both the Fab LA5 and D8 in their unliganded form were also determined. These data sets give an opportunity to compare the structures of the individual unbound proteins to their conformation in the Ab-Ag complex. We first investigated whether the epitope is rigid in D8, which would favor a lock-and-key mechanism for the binding of LA5-Fab. Therefore, we compared the discontinuous epitopes in both complexes of the asymmetric unit of the crystal (see Fig. S3A in the supplemental material) (RMSD = 0.37 Å), as well as in D8 before and after LA5 Fab binding (see Fig. S3B in the supplemental material). When superimposing the unbound D8 epitope onto the one of the LA5-Fab bound complex, the RMSD is 0.56 Å over 186 atoms. Overall, the epitope-forming residues superimpose well, and no major structural changes occur in D8 upon antibody binding.
More noticeable structural changes are observed for the antibody upon binding to D8, notwithstanding that the backbone conformation of the CDR loops is preserved. A reorientation of side chains is observed only within the CDRs of the H chain, which correlates with the dominant role of the H chain in antigen binding. Notably, H1 Phe32 rotates about 45° to increase the hydrophobic and VdW interactions with D8, especially with residues Gly40, Lys41, Ser204, Ile215, and Glu217. H1 Trp34 is among the most dramatic changes as it rotates about 80° to form novel VdW interactions with D8 residues Ile174, Asn175, His176, as well as forming a highly energetic cation-pi interaction with Arg220 of D8. The third residue with a noticeable change in orientation is in CDR H3. Y102 tilts sideways to form two H bonds with E217 and Arg220 of D8 (Fig. 5C and 6C). Residues within CDR H2 are largely unchanged, except for Glu58 (Fig. 6C). Therefore, H2 has the optimal conformation to bind to the D8 crevice that is also predicted to bind CS, while residues of H3 have to reorient for a perfect fit, especially Tyr102 and Arg103, which form very localized H bonds and salt bridges with D8 (Glu217 and Tyr219). In summary, although the Fab LA5 largely appears to bind with a lock-and-key mechanism, slight changes have to occur to allow antigen binding. Those changes result in novel interactions between D8 and LA5 (VdW, cation-pi, and H bonds) and likely contribute to the affinity and stability of the antibody antigen complex.
Sequence alignment and structural comparison of D8 homologs.
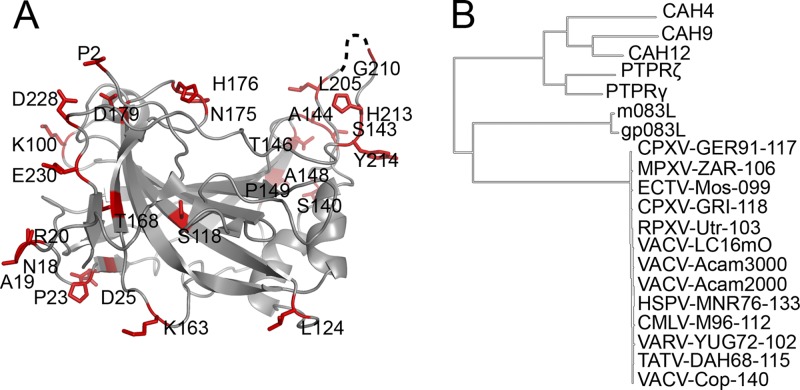
A phylogenetic alignment shows that D8 is widely conserved throughout the Poxviridae family (Fig. 7; see Fig. S4 in the supplemental material). It utilizes primarily the CAH fold originally defined in the carbonic anhydrase protein family. We analyzed sequence conservation of some of the most relevant orthologs of the Orthopoxvirus genus. Compared side by side, D8 sequences are nearly identical in vaccinia virus, variola virus, monkeypox virus, and other direct orthologs (Fig. 7; see Fig. S4 and Table S5 in the supplemental material). Initially, all 86 orthopoxvirus D8 representatives were extracted from the Poxviridae Bioinformatics Resource center (PBR) (http://www.poxvirus.org) and aligned against VACV Acam2000 D8, while 14 representative sequences are shown in the alignment (see Fig. S4 in the supplemental material). The variation spectrum is composed of 32 out of 304 residues in FL-D8, and 26 are located within the 234 residues of the D8 crystal structure. There are only substitutions and no deletions. Table S3 in the supplemental material lists all possible variations, along with their occurrence ratios, calculated on the 86 available orthopoxvirus D8 sequences. As a first observation, the Acam2000 D8 sequence is closest to the consensus sequence, as only five residues out of the 32 are underrepresented over the 86 orthopoxvirus sequences, and this property may contribute to vaccinia virus' broad-spectrum infectivity. We also observe that variations occur mostly for surface residues, but residues that form the proposed CS binding crevice are mostly conserved. (Fig. 7; see Fig. S4 in the supplemental material). This suggests that the central crevice is crucial for the function of the virus, which correlates with its predicted binding to CS. While MAb LA5 was raised against VACVWR D8, it likely exhibits cross-reactivity to most other orthopoxviruses.
Fig 7.
D8 orthopoxvirus sequence variations. (A) VACV D8 structure, with orthopoxvirus amino acid variations colored in red. Only three residues of the variation spectrum are also part of the D8 epitope targeted by LA5-MAb. (B) Phylogenetic tree/dendrogram of CAH domain-containing homologs, obtained using the SeaView PhyML algorithm (22). CAH-like homologs include orthopoxvirus and more distant myxoma virus and rabbit fibroma virus representatives (m083L and gp083L), human carbonic anhydrases (CAH) 4, 9, and 12, and extracytoplasmic anchoring domains of PTPRs. VACV, vaccinia virus; VARV, variola virus; HSPV, horsepox virus; CMLV, camelpox virus; MPXV, monkeypox virus; MYXV, myxoma virus; CPXV, cowpox virus; ECTV, ectromelia virus; RFV, rabbit fibroma virus; RPXV, rabbitpox virus; RFV rabbit fibroma virus; TATV, taterapox virus. For the orthopoxvirus family, all 85 sequences of orthopoxvirus D8 were extracted from the Poxvirus Bioinformatics Resource Center, and 14 representatives, which cover the full mutation spectrum, were selected for the final alignment in Fig. S4 in the supplemental material. Mutations are listed in Table S3 in the supplemental material along with their occurrence ratios.
DISCUSSION
Our study reports the structure of a major VACV surface protein, D8. We have identified a positively charged crevice running along the center of the CAH fold, which roughly corresponds to the so-called hydrophobic pocket that is adjacent to the catalytic sites of CAH proteins. This hydrophobic pocket confers specificity for CAHs to bind alternative substrates, such as phenyl esters (24) and phenylthiocarbamides (PTC), which are CAH inhibitors used for the treatment of glaucoma (9). The size, shape, and charge of the crevice make it a potential binding site for CS. An automated docking experiment using the CS tetrasaccharide from the cathepsin K structure (PDB code 3C9E) (30) positioned CS along the positively charged crevice of D8, with interactions similar to those observed for cathepsin K. Furthermore, we have shown that the full-length ectodomain of D8 exists predominantly as a tetramer. This tetramerization may be important for CS binding, as multivalent interactions generally increase affinity significantly. Disulfide-linked dimerization of D8 also occurs, in agreement with a recent observation from the Xiang lab. D8 antisera from VACV immunized mice contained antibodies that recognize a protein with an apparent molecular mass of 67 kDa, which disappeared upon the addition of a reducing agent and thus was presumed to be the disulfide-linked D8 dimer (see Fig. S5 in the supplemental material) (35).
Our understanding of antibody-antigen recognition is still limited, particularly in the context of physiological antibody responses to pathogen proteins that occur after infection or immunization with live pathogens. This study provides insights into the recognition of D8 by the neutralizing antibody LA5. The antibody binds across the whole width of D8 to a discontinuous epitope with a footprint (2,434 Å2 of BSA) that is at the far upper end of those of common antibody-antigen complexes (1,400 Å2 < BSA < 2,300 Å2 [32]). In comparison, a neutralizing antibody targeting the VACV protein L1 binds around the tip of L1, burying roughly 1,560 Å2. The LA5 binding footprint is dominated by the H chain, while the L chain binds to D8 through recruitment of a water zipper that complements the surface imperfection and distance between L chain and D8, and conserved water molecules have been observed in a variety of antibody-antigen complexes (10). A few conformational changes occur within the antibody upon binding to D8, while the D8 antigen is stabilized by its CAH fold and exhibits almost no conformational change. The absence of major conformational changes indicates a lock-and-key binding interaction between LA5 and D8, which is also supported by the fast association of the antibody as measured by SPR.
VACV neutralization with anti-D8 MAb is complement dependent, consistent with our previous finding for the antigens H3 and B5 (4, 5, 34) and suggesting that the virion is able to use two other attachment proteins (A27 and H3) to bind to host cell GAGs (13, 25, 31) even when D8 is targeted by antibodies. This is further supported by studies showing that D8 deletion strains replicate efficiently in tissue cultures (40, 46; Y. Xiang unpublished data), indicating that targeting D8 might not be sufficient for direct neutralization in vitro. In vivo, where GAG expression is more limited than on cell lines, the four attachment proteins (D8, H3, A27, and A26) may serve more specialized roles for facilitating VACV infection of cell types with differential GAG expression. Whether LA5 binding to D8 directly blocks the virion from interacting with CS or whether the crevice coincidentally forms the binding site for LA5 due to potentially increased immunogenicity is currently unknown. It is also conceivable that the high-affinity interaction combined with the large and robust footprint of LA5 on the D8 surface is a prerequisite for recruiting complement, resulting in effective neutralization. Conversely, tetramerization of D8 could potentially lead to an arrangement of D8 in which individual D8 molecules are covered by neutralizing anti-D8 MAbs, while other CS binding sites are not accessible to antibodies but can still bind CS. In that case, recruitment of complement to the D8-antibody complex would have an additional steric effect that further blocks access of D8 to CS. These possibilities can be examined in future studies.
A “holy grail” of infectious disease B cell biology research is the ability to predict antigen binding sites of antibodies. If such predictive power is possible, it will be so only if substantially more Ab-Ag structure complexes are known for baseline algorithm development, using real pathogen antigens and antibody responses generated under physiological conditions. The determination of a D8/LA5 structure is a significant step for those efforts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Howard Gray for helpful discussion and critical reading of the manuscript. We also thank Enrico Girardi for help with Autodock Vina. We thank the Stanford Synchrotron Radiation Lightsource (SSRL), especially beamlines 7-1 and 11-1, for remote data collection.
This work is supported by NIH contract BAA-NIAID-DAIT-NIHAI2008031.
Footnotes
Published ahead of print 23 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Amanna IJ, Slifka MK, Crotty S. 2006. Immunity and immunological memory following smallpox vaccination. Immunol. Rev. 211:320–337 [DOI] [PubMed] [Google Scholar]
- 2. Atlas RM. 2002. Bioterriorism: from threat to reality. Annu. Rev. Microbiol. 56:167–185 [DOI] [PubMed] [Google Scholar]
- 3. Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67:271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benhnia MR, et al. 2009. Heavily isotype-dependent protective activities of human antibodies against vaccinia virus extracellular virion antigen B5. J. Virol. 83:12355–12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benhnia MR, et al. 2009. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J. Virol. 83:1201–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benhnia MR, et al. 2008. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J. Virol. 82:3751–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berhanu A, et al. 2008. Vaccination of BALB/c mice with Escherichia coli-expressed vaccinia virus proteins A27L, B5R, and D8L protects mice from lethal vaccinia virus challenge. J. Virol. 82:3517–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bordoli L, et al. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4:1–13 [DOI] [PubMed] [Google Scholar]
- 9. Boriack-Sjodin PA, et al. 1998. Structural analysis of inhibitor binding to human carbonic anhydrase II. Protein Sci. 7:2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Braden BC, Fields BA, Poljak RJ. 1995. Conservation of water molecules in an antibody-antigen interaction. J. Mol. Recognit. 8:317–325 [DOI] [PubMed] [Google Scholar]
- 11. Brochet X, Lefranc MP, Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J. and V-D-J. sequence analysis. Nucleic Acids Res. 36:W503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung CS, et al. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chung CS, Hsiao JC, Chang YS, Chang W. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 15. Crotty S, et al. 2003. Long-term B cell memory in humans after smallpox vaccination. J. Immunol. 171:4969–4973 [DOI] [PubMed] [Google Scholar]
- 16. Davies DH, et al. 2005. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 79:11724–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davies DH, et al. 2007. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 7:1678–1686 [DOI] [PubMed] [Google Scholar]
- 18. Di Giulio DB, Eckburg PB. 2004. Human monkeypox: an emerging zoonosis. Lancet Infect. Dis. 4:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32:W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edghill-Smith Y, et al. 2005. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 11:740–747 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 23. Hammarlund E, et al. 2005. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat. Med. 11:1005–1011 [DOI] [PubMed] [Google Scholar]
- 24. Host G, Martensson LG, Jonsson BH. 2006. Redesign of human carbonic anhydrase II for increased esterase activity and specificity towards esters with long acyl chains. Biochim. Biophys. Acta 1764:1601–1606 [DOI] [PubMed] [Google Scholar]
- 25. Hsiao JC, Chung CS, Chang W. 1998. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 72:8374–8379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Izmailyan RA, Huang CY, Mohammad S, Isaacs SN, Chang W. 2006. The envelope G3L protein is essential for entry of vaccinia virus into host cells. J. Virol. 80:8402–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janeczko RA, Rodriguez JF, Esteban M. 1987. Studies on the mechanism of entry of vaccinia virus in animal cells. Arch. Virol. 92:135–150 [DOI] [PubMed] [Google Scholar]
- 28. Karem KL, et al. 2007. Monkeypox-induced immunity and failure of childhood smallpox vaccination to provide complete protection. Clin. Vaccine Immunol. 14:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawrence MC, Colman PM. 1993. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234:946–950 [DOI] [PubMed] [Google Scholar]
- 30. Li Z, Kienetz M, Cherney MM, James MN, Bromme D. 2008. The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex. J. Mol. Biol. 383:78–91 [DOI] [PubMed] [Google Scholar]
- 31. Lin CL, Chung CS, Heine HG, Chang W. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lo Conte L, Chothia C, Janin J. 1999. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285:2177–2198 [DOI] [PubMed] [Google Scholar]
- 33. Lovell SC, et al. 2003. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50:437–450 [DOI] [PubMed] [Google Scholar]
- 34. McCausland MM, et al. 2010. Combination therapy of vaccinia virus infection with human anti-H3 and anti-B5 monoclonal antibodies in a small animal model. Antivir. Ther. 15:661–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meng X, et al. 2011. Generation and characterization of a large panel of murine monoclonal antibodies against vaccinia virus. Virology 409:271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moss B. 2007. Poxviridae: the viruses and their replication, p 2905–2946 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Hagerstown, MD [Google Scholar]
- 37. Moss B. 2011. Smallpox vaccines: targets of protective immunity. Immunological reviews. 239:8–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mourey RJ, Dixon JE. 1994. Protein tyrosine phosphatases: characterization of extracellular and intracellular domains. Curr. Opin. Genet. Dev. 4:31–39 [DOI] [PubMed] [Google Scholar]
- 39. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 40. Niles EG, Seto J. 1988. Vaccinia virus gene D8 encodes a virion transmembrane protein. J. Virol. 62:3772–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ohradanova A, et al. 2007. Reconstitution of carbonic anhydrase activity of the cell-surface-binding protein of vaccinia virus. Biochem. J. 407:61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ojeda S, Senkevich TG, Moss B. 2006. Entry of vaccinia virus and cell-cell fusion require a highly conserved cysteine-rich membrane protein encoded by the A16L gene. J. Virol. 80:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450 [DOI] [PubMed] [Google Scholar]
- 44. Reed KD, et al. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350:342–350 [DOI] [PubMed] [Google Scholar]
- 45. Regnery RL. 2007. Poxviruses and the passive quest for novel hosts. Curr. Top. Microbiol. Immunol. 315:345–361 [DOI] [PubMed] [Google Scholar]
- 46. Rodriguez JR, Rodriguez D, Esteban M. 1992. Insertional inactivation of the vaccinia virus 32-kilodalton gene is associated with attenuation in mice and reduction of viral gene expression in polarized epithelial cells. J. Virol. 66:183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Satheshkumar PS, Moss B. 2009. Characterization of a newly identified 35-amino-acid component of the vaccinia virus entry/fusion complex conserved in all chordopoxviruses. J. Virol. 83:12822–12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Senkevich TG, Moss B. 2005. Vaccinia virus H2 protein is an essential component of a complex involved in virus entry and cell-cell fusion. J. Virol. 79:4744–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Senkevich TG, Ward BM, Moss B. 2004. Vaccinia virus entry into cells is dependent on a virion surface protein encoded by the A28L gene. J. Virol. 78:2357–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Su HP, et al. 2005. The 1.51-angstrom structure of the poxvirus L1 protein, a target of potent neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 102:4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Su HP, Golden JW, Gittis AG, Hooper JW, Garboczi DN. 2007. Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein. Virology 368:331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Su HP, Singh K, Gittis AG, Garboczi DN. 2010. The structure of the poxvirus A33 protein reveals a dimer of unique C-type lectin-like domains. J. Virol. 84:2502–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tiller T, Busse CE, Wardemann H. 2009. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods 350:183–193 [DOI] [PubMed] [Google Scholar]
- 54. Townsley AC, Senkevich TG, Moss B. 2005. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell-cell fusion. J. Virol. 79:10988–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Townsley AC, Senkevich TG, Moss B. 2005. Vaccinia virus A21 virion membrane protein is required for cell entry and fusion. J. Virol. 79:9458–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsung K, Yim JH, Marti W, Buller RM, Norton JA. 1996. Gene expression and cytopathic effect of vaccinia virus inactivated by psoralen and long-wave UV light. J. Virol. 70:165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66:22–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.