Abstract
Cryoelectron microscopy reconstruction of Cryphonectria nitschkei virus 1, a double-stranded RNA (dsRNA) virus, shows that the capsid protein (60 copies/particle) is formed by a repeated helical core, indicative of gene duplication. This unusual organization is common to chrysoviruses. The arrangement of many of these putative α-helices is conserved in the totivirus L-A capsid protein, suggesting a shared motif. Our results indicate that a 120-subunit T=1 capsid is a conserved architecture that optimizes dsRNA replication and organization.
TEXT
Most double-stranded RNA (dsRNA) viruses have a specialized icosahedral capsid implicated in genome replication and transcription. This capsid has a triangulation number equal to 1 (T=1) and consists of 60 asymmetric dimers of a single, predominantly α-helical protein (10). With the exception of birnaviruses (16), these capsids are present in members of the families Reoviridae, Picobirnaviridae, Cystoviridae, Totiviridae, and Partitiviridae (14). Chrysoviruses are mycoviruses with a multipartite genome consisting of four monocistronic dsRNA segments (7), each separately encapsidated in a similar particle (5, 8). The capsids are authentic T=1 shells formed by only 60 copies of a single polypeptide.
We previously determined the three-dimensional (3D) structure of Penicillium chrysogenum virus (PcV) to an 8-Å resolution and found that the PcV capsid protein (CP) is a duplication of a mostly α-helical domain (10). The spatial arrangement of many of these putative α-helices resembles that in other fungal dsRNA virus CP, suggesting a conserved common ancestor (10). To test whether the apparently unusual fold signature of PcV is a feature shared by other chrysoviruses, we determined the 8-Å cryoelectron microscopy (cryo-EM) map of Cryphonectria nitschkei virus 1 (CnV1).
CnV1 virions were purified from C. nitschkei strain OB5-11 (ATTC 4105) by rate zonal centrifugation in a sucrose density gradient (5). Partial amino acid sequence of CnV1 CP from strain OB5-11 (accession no. DQ865187) showed that it is identical to the corresponding region of the fully sequenced CnV1 CP from strain BS-122 (accession no. GQ290645) (9). Whereas strain OB5-11 was available from the ATCC, strain BS-122 was not; we therefore used strain OB5-11 as a source of CnV1 virus particles. Cryo-EM data were collected in a Tecnai G2 electron microscope (200 kV) using a nominal magnification of ×62,000 (11). Micrographs were digitized with a Nikon Super CoolScan 9000 ED scanner at a 6.35-μm/pixel step size (1.024 Å/pixel at the sample level). Data were processed from 18,732 full capsid images with Xmipp software (17). Resolution was assessed using the 0.5 threshold criterion of the Fourier shell correlation from two independent half data sets. At the end of refinements, 90% initial particle images were included in the full CnV1 capsid 3D reconstruction (3DR). Graphics were prepared with Chimera (15). Amplitude correction of the CnV1 map was done using the X-ray L-A capsid map (PDB 1m1c) (12). CP subunits (and their halves) were segmented using Chimera and were iteratively refined to avoid subunit overlap or loose density. Capsid protein secondary structure elements (SSE) were identified using Gorgon (2). Various SSE prediction methods of the CnV1 and PcV CP sequences were used to test correlation with our structural subunit model (10). CnV1 CP halves were aligned manually as for PcV (10). X-ray structure of the L-A capsid Gag protein was manually aligned and refined to optimize the matched SSE models of CnV and PcV CP elements. The 3DR for CnV1 full capsids is deposited in the EMDB (www.ebi.ac.uk; accession no. emd-2062). Sedimentation velocity experiments were performed with the same CnV1 particle sample in an XL-I analytical ultracentrifuge (Beckman-Coulter), as described previously (5).
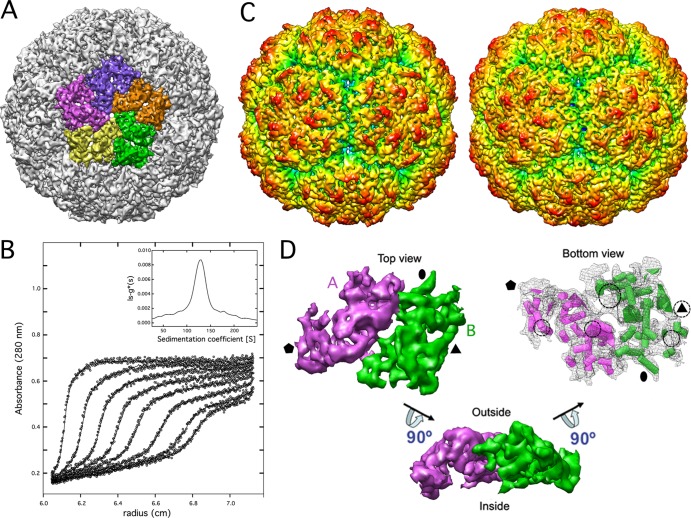
Three-dimensional reconstruction of full CnV1 was calculated to an 8.1-Å resolution (Fig. 1A). The most prominent features of the T=1 capsid (400-Å diameter) were 12 outwardly protruding pentons. CP boundaries were established based on its compactness and contacts with neighboring densities (Fig. 1A). Sedimentation velocity behavior of CnV1 particles was analyzed to determine their stoichiometry (Fig. 1B). The main species had a standard sedimentation coefficient of 135 S, corresponding to a true T=1 particle (60 copies of protein) with a frictional coefficient ratio of 1.3 ± 0.05, similar to that of PcV. Similarities between the uneven outer surfaces of CnV1 and PcV capsids were also clear (Fig. 1C). As with PcV, the asymmetric unit is formed by two similar ellipsoid-like structures (48 and 52% of total unit volume) (Fig. 1D, half-protein A, purple; half-protein B, green). These elements are arranged in two sets of five; five half-protein A structures surround the icosahedral 5-fold axis, and five half-protein B structures are intercalated between them, forming a pseudodecamer. This organization is reminiscent of the T=1 lattice in reo- and totivirus capsids, in which the asymmetric dimer is approximately parallel. In partiti- and picobirnavirus, the capsid protein dimer has a different quaternary organization with an almost-perfect local 2-fold symmetry (6, 13).
Fig 1.
Three-dimensional cryo-EM and stoichiometry of CnV1 virions. (A) CnV1 virion surface, viewed along an icosahedral 5-fold axis, showing the 5 structural subunits as a pentamer. (B) Analytical ultracentrifugation analysis of CnV1 virions. Symbols show sedimentation velocity profiles of CnV1 virions taken at different times (An50Ti rotor; 10,000 rpm, 20°C). Solid lines show best-fit distributions of the ls-g*(s) analysis. Apparent sedimentation coefficient distributions, ls-g*(s) (inset). (C) Radially color-coded outer surfaces of full capsids of CnV1 (left) and PcV (right), viewed along an icosahedral 2-fold axis. Maps are contoured 2 σ above mean density. (D) Segmented CnV1 CP monomer. Protein halves A (purple) and B (green) are indicated. The CnV1 asymmetric unit viewed from inside is shown as wire frames, with putative α-helices (cylinders) and β-sheets (planks). dsRNA interacts with five defined areas of the capsid inner surface (circles). Black symbols indicate icosahedral symmetry axes.
We studied the organization of the packaged RNA from our 3D cryoEM studies. Analysis of CnV1-full capsids showed that the inner surface of the protein shell is in close contact with the underlying density shell that is ascribed to encapsidated dsRNA. Numerous icosahedral positions are in contact with the genome, and specific RNA/protein interactions were evident in five areas per monomer (indicated in Fig. 1D, circles). This genome ordering suggests a dsRNA-mediated structural role for capsid stability, similar to PcV (10).
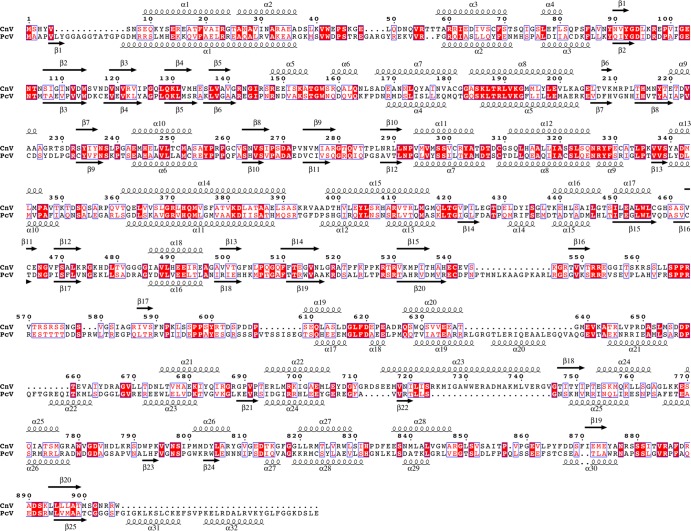
Sequence analysis of the PcV CP suggested that it has two roughly similar halves, with conserved segments in both halves of the protein (5). Plane sequence similarity analysis of the CnV1 CP showed no evidence of gene duplication, although the average identity between CP sequences of PcV (982 amino acids) versus CnV1 (906 residues) was 25% (average similarity, 40%) (Fig. 2). Multiple sequence alignment of CP amino acid sequences of several chrysoviruses indicated that the N-terminal halves align much better than the C-terminal halves (not shown). PcV and CnV1 SSE predictions of CP indicated a high α-helical content and aligned well, consistent with their cryo-EM maps. We found an unstructured central region that also divided the CnV CP into two parts, reflecting a structurally disordered region (amino acids ∼550 to ∼610). The unstructured region might represent a linker sequence between the two unequal halves of the CnV1 CP.
Fig 2.
Sequence alignment and secondary structure consensus prediction of CnV1 and PcV CP amino acid sequences. Several SSE prediction methods (PsiPred, Jnet, Porter, Sable, Gor, Yaspin, and Profsec) were used to test correlation with our structural subunit model (10). A consensus SSE prediction was obtained by simple majority at each sequence position. Identical residues, white on red background; partially conserved residues, red. Arrows represent β-chains and spirals represent α-helices.
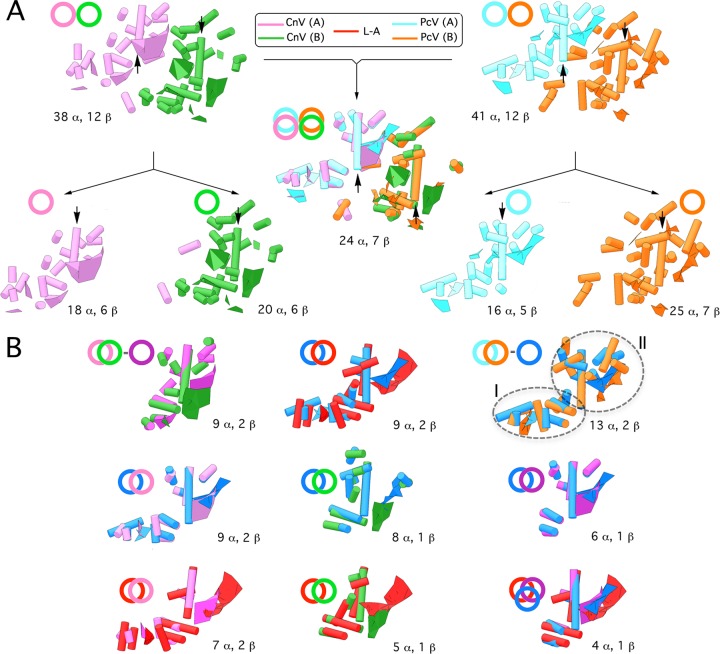
Structural comparison of secondary and tertiary structures of CP is currently used as a criterion to establish relatedness in the absence of sequence similarity (3, 4). The CnV1 CP has 38 rodlike densities, in addition to 12 planar regions (putative β-sheets); these SSE are also similar to PcV CP (Fig. 3A). Both CnV1 CP halves have a long putative α-helix tangential to the capsid surface, which we used as a reference for SSE comparisons (Fig. 3A, arrows). When CnV1 and PcV CP were superimposed, the relative spatial locations of 24 rodlike structures and 7 curved surfaces were close and required only minor local adjustments to overlap, indicating a similar structural signature of the asymmetric unit. Comparable analysis of CnV1 and PcV CP halves A and B (treated as a rigid body) showed substantial resemblance, as 9 (CnV1) and 13 (PcV) rods and two planks were superimposed. These SSE constitute the CnV1 and PcV conserved cores (Fig. 3B, top row). Overlaying L-A Gag on the PcV conserved core showed that 9 putative α-helices (of 13) and 2 sheetlike structures have similar spatial distributions (Fig. 3B, top row, center). Comparison of the PcV conserved core with each CnV1 half showed that the longest putative α-helix as well as other SSE (located mainly in domain II) are shared; furthermore, CnV1 and PcV conserved cores are nearly superimposed for 6 rods and one large curved surface (Fig. 3B, middle row). CnV1 and PcV CP elements have a similar structural signature and might thus derive from a gene duplication event. If that is the case, CnV1 CP half-protein B has less structural preservation and has diverged more rapidly than half-protein A.
Fig 3.
Structural comparison of CnV, PcV, and L-A capsid proteins. (A) Superimposition (center) of CnV1 CP SSE (half-protein A, purple; half-protein B, green) (left) on PcV CP SSE (half-protein A, blue; half-protein B, orange) (right). Total numbers of SSE with close relative spatial locations are indicated (24 putative α-helices and 7 putative β-sheets). Black arrows indicate the ∼25 (purple)- and 35 (green)-Å rodlike densities of both CnV1 CP halves and the two ∼37-Å putative α-helices (blue and orange) of PcV CP elements. CnV1 and PcV CP are divided into their protein halves A and B (total number of SSE indicated, bottom). Circles and superimposed circles for each model (top) indicate structural superimpositions. (B) Superimposed views of conserved SSE in CnV1 protein halves A and B (conserved SSE, dark-purple circle) (top row, left). Superimposition of conserved SSE in PcV protein halves A and B (conserved SSE, dark-blue circle). The PcV conserved core is subdivided into domains I and II (top row, right). Superimposition of helical and planar regions of selected L-A Gag SSE (red) and conserved PcV SSE (dark blue) (top row, center). Superimposition of the PcV conserved core (dark blue) with CnV half-protein A (left), CnV1 half-protein B (center), or CnV1 conserved core (dark purple) (middle row). Superimposition of the L-A Gag conserved core (red), CnV1 half-protein A (left), and CnV1 half-protein B (center). Motif conserved in CnV1 and PcV halves with L-A Gag (right) (bottom row). These structural matchings preserve the spatial orientation of CnV1, PcV, and L-A capsid protein structural units within their capsids.
Overlaying L-A Gag on either CnV1 element showed many SSE with similar spatial distributions, including 7 or 5 putative α-helices (of 9) and 2 or 1 putative β-sheets (Fig. 3B, bottom row). Intersection among L-A Gag, CnV1, and PcV conserved cores indicates the shared motif for these three fungal viruses. Whereas the folds of the Reoviridae T=1 capsid proteins are similar, those of picobirna-, partiti-, and totivirus (as well as chrysovirus) have different structures. Tools such as the Structure Homology Program (SHP) (reviewed in reference 1) nevertheless indicate that dsRNA viruses (except birnavirus) are grouped in the same lineage, as their capsid proteins are similar in the positioning of numerous SSE. The systematic structural comparison between toti- and chrysovirus capsid proteins shows the gradual loss of nonshared SEE and might reveal the hallmark fold of dsRNA viruses.
ACKNOWLEDGMENTS
We thank C. Mark for editorial assistance.
This work was supported by grants from the Spanish Ministry of Science and Innovation (BFU2011-25902 to J.R.C. and BFU2011-29038 to J.L.C.), the NIH Intramural Research Program with support from the Center for Information Technology (to B.T.), and the Kentucky Science & Engineering Foundation (to S.A.G.).
Footnotes
Published ahead of print 16 May 2012
REFERENCES
- 1. Abrescia NG, Bamford DH, Grimes JM, Stuart DI. 2012. Structure unifies the viral universe. Annu. Rev. Biochem. doi:10.1146/annurev-biochem-060910-095130 [DOI] [PubMed] [Google Scholar]
- 2. Baker ML, et al. 2011. Modeling protein structure at near atomic resolutions with Gorgon. J. Struct. Biol. 174:360–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker ML, Jiang W, Rixon FJ, Chiu W. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967–14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655–663 [DOI] [PubMed] [Google Scholar]
- 5. Castón JR, et al. 2003. Three-dimensional structure of Penicillium chrysogenum virus: a double-stranded RNA virus with a genuine T=1 capsid. J. Mol. Biol. 331:417–431 [DOI] [PubMed] [Google Scholar]
- 6. Duquerroy S, et al. 2009. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. 28:1655–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghabrial SA, Jiang D, Castón JR. 2005. Chrysoviridae, p 591–595 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy, VIIIth report of the International Committee on Taxonomy of Viruses Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 8. Jiang D, Ghabrial SA. 2004. Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J. Gen. Virol. 85:2111–2121 [DOI] [PubMed] [Google Scholar]
- 9. Kim JM, et al. 2010. Nucleotide sequences of four segments of chrysovirus in Korean Cryphonectria nitschkei BS122 strain. Virus Genes 41:292–294 [DOI] [PubMed] [Google Scholar]
- 10. Luque D, et al. 2010. The T=1 capsid protein of Penicillium chrysogenum virus is formed by a repeated helix-rich core indicative of gene duplication. J. Virol. 84:7256–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luque D, et al. 2007. Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. J. Virol. 81:6869–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. 2002. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 9:725–728 [DOI] [PubMed] [Google Scholar]
- 13. Pan J, et al. 2009. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc. Natl. Acad. Sci. U. S. A. 106:4225–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patton JT. 2008. Segmented double-stranded RNA viruses. Structure and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 15. Pettersen EF, et al. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 16. Saugar I, et al. 2005. Structural polymorphism of the major capsid protein of a double-stranded RNA virus: an amphipathic alpha helix as a molecular switch. Structure 13:1007–1017 [DOI] [PubMed] [Google Scholar]
- 17. Scheres SH, Nunez-Ramírez R, Sorzano CO, Carazo JM, Marabini R. 2008. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3:977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]